Synthetic Biology Journal ›› 2020, Vol. 1 ›› Issue (1): 7-28.DOI: 10.12211/2096-8280.2020-057
• Invited Review • Previous Articles Next Articles
Significant research progress in synthetic biology
DING Mingzhu, LI Bingzhi, WANG Ying, XIE Zexiong, LIU Duo, YUAN Yingjin
- Frontiers Science Center for Synthetic Biology, Key Laboratory of Systems Bioengineering (Ministry of Education), School of Chemical Engineering and Technology, Tianjin University, Tianjin 300072, China
-
Received:2020-04-23Revised:2020-05-13Online:2020-09-24Published:2020-02-29 -
Contact:YUAN Yingjin
合成生物学重要研究方向进展
丁明珠, 李炳志, 王颖, 谢泽雄, 刘夺, 元英进
- 教育部合成生物学前沿科学中心,系统生物工程教育部重点实验室,天津大学化工学院,天津 300072
-
通讯作者:元英进 -
作者简介:丁明珠,女,博士,副研究员,研究方向为合成生物学,人工混菌体系设计合成。E-mail:mzding@tju.edu.cn
元英进(1963—),男,教授,博士生导师,研究方向为合成生物学及人工基因组化学合成。E-mail: yjyuan@tju.edu.cn -
基金资助:国家自然科学基金(21621004)
CLC Number:
Cite this article
DING Mingzhu, LI Bingzhi, WANG Ying, XIE Zexiong, LIU Duo, YUAN Yingjin. Significant research progress in synthetic biology[J]. Synthetic Biology Journal, 2020, 1(1): 7-28.
丁明珠, 李炳志, 王颖, 谢泽雄, 刘夺, 元英进. 合成生物学重要研究方向进展[J]. 合成生物学, 2020, 1(1): 7-28.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2020-057
| 1 | LEPROUST E M, PECK B J, SPIRIN K, et al. Synthesis of high-quality libraries of long (150mer) oligonucleotides by a novel depurination controlled process[J]. Nucleic Acids Research, 2010, 38 (8): 2522-2540. |
| 2 | PALLUK S, ARLOW D H, DE ROND T, et al. De novo DNA synthesis using polymerase-nucleotide conjugates[J]. Nature Biotechnology, 2018, 36 (7): 645-650. |
| 3 | ELOWITZ M B, LEIBLER S. A synthetic oscillatory network of transcriptional regulators[J]. Nature, 2000, 403 (6767): 335-338. |
| 4 | GARDNER T S, CANTOR C R, COLLINS J J. Construction of a genetic toggle switch in Escherichia coli [J]. Nature, 2000, 403 (6767): 339-342. |
| 5 | RANTASALO A, KUIVANEN J, PENTTILA M, et al. Synthetic toolkit for complex genetic circuit engineering in Saccharomyces cerevisiae [J]. ACS Synthtic Biology, 2018, 7 (6): 1573-1587. |
| 6 | ZONG Y, ZHANG H M, LYU C, et al. Insulated transcriptional elements enable precise design of genetic circuits[J]. Nature Communications, 2017, 8 (1): 52. |
| 7 | SHETTY R P, ENDY D, KNIGHT T F JR. Engineering BioBrick vectors from BioBrick parts[J]. J. Biol. Eng., 2008, 2: 5. |
| 8 | LEE T S, KRUPA R A, ZHANG F Z, et al. BglBrick vectors and datasheets: a synthetic biology platform for gene expression[J]. Journal of Biological Engineering, 2011, 5 (1): 12. |
| 9 | AGMON N, MITCHELL L A, CAI Y Z, et al. Yeast golden gate (yGG) for the efficient assembly of s-cerevisiae transcription Units[J]. ACS Synthetic Biology, 2015, 4 (7): 853-859. |
| 10 | WEBER E, ENGLER C, GRUETZNER R, et al. A modular cloning system for standardized assembly of multigene constructs[J]. PLoS One, 2011, 6 (2): e16765. |
| 11 | BERVOETS I, BREMPT M VAN, NEROM K VAN, et al. A sigma factor toolbox for orthogonal gene expression in Escherichia coli [J]. Nucleic Acids Research, 2018, 46 (4): 2133-2144. |
| 12 | REDDEN H, ALPER H S. The development and characterization of synthetic minimal yeast promoters[J]. Nature Communications, 2015, 6: e7810. |
| 13 | ROGERS J K, TAYLOR N D, CHURCH G M. Biosensor-based engineering of biosynthetic pathways[J]. Current Opinion in Biotechnology, 2016, 42: 84-91. |
| 14 | TEMME K, HILL R, SEGALL-SHAPIRO T H, et al. Modular control of multiple pathways using engineered orthogonal T7 polymerases[J]. Nucleic Acids Research, 2012, 40 (17): 8773-8781. |
| 15 | MORSE N J, WAGNER J M, REED K B, et al. T7 polymerase expression of guide RNAs in vivo allows exportable CRISPR-Cas9 editing in multiple yeast hosts[J]. ACS Synthetic Biology, 2018, 7 (4): 1075-1084. |
| 16 | LIAO W, LIU B, CHANG C C, et al. Functional characterization of insulation effect for synthetic gene circuits in mammalian cells[J]. ACS Synthetic Biology, 2018, 7 (2): 412-418. |
| 17 | BATEY R T, GILBERT S D, MONTANGE R K. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine[J]. Nature, 2004, 432 (7015): 411-415. |
| 18 | BADORREK C S, GHERGHE C M, WEEKS K M. Structure of an RNA switch that enforces stringent retroviral genomic RNA dimerization[J]. PNAS, 2006, 103 (37): 13640-13645. |
| 19 | MOON T S, LOU C B, TAMSIR A, et al. Genetic programs constructed from layered logic gates in single cells[J]. Nature, 2012, 491 (7423): 249-253. |
| 20 | SIUTI P, YAZBEK J, LU T K. Synthetic circuits integrating logic and memory in living cells[J]. Nature Biotechnology, 2013, 31 (5): 448-452. |
| 21 | STRICKER J, COOKSON S, BENNETT M R, et al. A fast, robust and tunable synthetic gene oscillator[J]. Nature, 2008, 456 (7221): 516-519. |
| 22 | ANDREWS L B, NIELSEN A A K, VOIGT C A. Cellular checkpoint control using programmable sequential logic[J]. Science, 2018, 361 (6408): eaap8987. |
| 23 | LIAO M J, DIN M O, TSINRING L, et al. Rock-paper-scissors: engineered population dynamics increase genetic stability[J]. Science, 2019, 365 (6457): 1045-1049. |
| 24 | POTVIN-TROTTIER L, LORD N D, VINNICOMBE G, et al. Synchronous long-term oscillations in a synthetic gene circuit[J]. Nature, 2016, 538 (7626): 514-517. |
| 25 | GAO X J, CHONG L S, KIM M S, et al. Programmable protein circuits in living cells[J]. Science, 2018, 361 (6408): 1252-1258. |
| 26 | ADAMALA K P, MARTIN-ALARCON D A, GUTHRIE-HONEA K R, et al. Engineering genetic circuit interactions within and between synthetic minimal cells[J]. Nature Chemistry, 2017, 9 (5): 431-439. |
| 27 | PURCELL O, WANG J, SIUTI P, et al. Encryption and steganography of synthetic gene circuits[J]. Nature Communications, 2018, 9 (1): 4942. |
| 28 | CHEN M T, WEISS R. Artificial cell-cell communication in yeast Saccharomyces cerevisiae using signaling elements from Arabidopsis thaliana [J]. Nature Biotechnology, 2005, 23 (12): 1551-1555. |
| 29 | YOU L C, COX R S, WEISS R, et al. Programmed population control by cell-cell communication and regulated killing[J]. Nature, 2004, 428 (6985): 868-871. |
| 30 | BALAGADDE F K, SONG H, OZAKI J, et al. A synthetic Escherichia coli predator-prey ecosystem[J]. Molecular Systems Biology, 2008, 4: 187. |
| 31 | LIU C L, FU X F, LIU L L, et al. Sequential establishment of stripe patterns in an expanding cell population[J]. Science, 2011, 334 (6053): 238-241. |
| 32 | TAMSIR A, TABOR J J, VOIGT C A. Robust multicellular computing using genetically encoded NOR gates and chemical 'wires'[J]. Nature, 2011, 469 (7329): 212-215. |
| 33 | REGOT S, MACIA J, CONDE N, et al. Distributed biological computation with multicellular engineered networks[J]. Nature, 2011, 469 (7329): 207-211. |
| 34 | TODA S, BLAUCH L R, TANG S K Y, et al. Programming self-organizing multicellular structures with synthetic cell-cell signaling[J]. Science, 2018, 361 (6398): 156-162. |
| 35 | ZHANG F Z, CAROTHERS J M, KEASLING J D. Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids[J]. Nature Biotechnology, 2012, 30 (4): 354-359. |
| 36 | CHOU H H, KEASLING J D. Programming adaptive control to evolve increased metabolite production[J]. Nature Communications, 2013, 4: 2595. |
| 37 | TAO R K, ZHAO Y Z, CHU H Y, et al. Genetically encoded fluorescent sensors reveal dynamic regulation of NADPH metabolism[J]. Nature Methods, 2017, 14 (9): 720-728. |
| 38 | TAY P K R, NGUYEN P Q, JOSHI N S. A Synthetic circuit for mercury bioremediation using self assembling functional amyloids[J]. ACS Synthetic Biology, 2017, 6 (10): 1841-1850. |
| 39 | WAN X Y, VOLPETTI F, PETROVA E, et al. Cascaded amplifying circuits enable ultrasensitive cellular sensors for toxic metals[J]. Nature Chemical Biology, 2019, 15 (5): 540-548. |
| 40 | GALLAGHER R R, PATEL J R, INTERIANO A L, et al. Multilayered genetic safeguards limit growth of microorganisms to defined environments[J]. Nucleic Acids Research, 2015, 43 (3): 1945-1954. |
| 41 | CHAN C T Y, LEE J W, CAMERON D E, et al. 'Deadman' and 'Passcode' microbial kill switches for bacterial containment[J]. Nature Chemical Biology, 2016, 12 (2): 82-86. |
| 42 | ROYBAL K T, RUPP L J, MORSUT L, et al. Precision tumor recognition by T cells with combinatorial antigen-sensing circuits[J]. Cell, 2016, 164 (4): 770-779. |
| 43 | ROYBAL K T, WILLIAMS J Z, MORSUT L, et al. Engineering T cells with customized therapeutic response programs using synthetic notch receptors[J]. Cell, 2016, 167 (2): 419-432. |
| 44 | MIMEE M, NADEAU P, HAYWARD A, et al. An ingestible bacterial-electronic system to monitor gastrointestinal health[J]. Science, 2018, 360 (6391): 915-918. |
| 45 | SHAO J W, XUE S, YU G L, et al. Smartphone-controlled optogenetically engineered cells enable semiautomatic glucose homeostasis in diabetic mice[J]. Science Translational Medicine, 2017, 9 (387): eaal2298. |
| 46 | TIGGES M, DENERVAUD N, GREBER D, et al. A synthetic low-frequency mammalian oscillator[J]. Nucleic Acids Research, 2010, 38 (8): 2702-2711. |
| 47 | TIGGES M, MARQUEZ-LAGO T T, STELLING J, et al. A tunable synthetic mammalian oscillator[J]. Nature, 2009, 457 (7227): 309-312. |
| 48 | GIBSON D G, GLASS J I, LARTIGUE C, et al. Creation of a bacterial cell controlled by a chemically synthesized genome[J]. Science, 2010, 329 (5987): 52-56. |
| 49 | HUTCHISON C A, CHUANG R Y, NOSKOV V N, et al. Design and synthesis of a minimal bacterial genome[J]. Science, 2016, 351 (6280): aad6253. |
| 50 | OSTROV N, LANDON M, GUELL M, et al. Design, synthesis, and testing toward a 57-codon genome[J]. Science, 2016, 353 (6301): 819-822. |
| 51 | FREDENS J, WANG K, DE LA TORRE D, et al. Total synthesis of Escherichia coli with a recoded genome[J]. Nature, 2019, 569 (7757): 514-518. |
| 52 | ANNALURU N, MULLER H, MITCHELL L A, et al. Total synthesis of a functional designer eukaryotic chromosome[J]. Science, 2014, 344 (6179): 55-58. |
| 53 | XIE Z X, LI B Z, MITCHELL L A, et al. Perfect designer chromosome Ⅴ and behavior of a ring derivative[J]. Science, 2017, 355 (6329): eaaf4704. |
| 54 | RICHARDSON S M, MITCHELL L A, STRACQUADANIO G, et al. Design of a synthetic yeast genome[J]. Science, 2017, 355 (6329): 1040-1044. |
| 55 | MITCHELL L A, WANG A, STRACQUADANIO G, et al. Synthesis, debugging, and effects of synthetic chromosome consolidation: syn Ⅵ and beyond[J]. Science, 2017, 355 (6329): eaaf4831. |
| 56 | SHEN Y, WANG Y, CHEN T, et al. Deep functional analysis of synII, a 770-kilobase synthetic yeast chromosome[J]. Science, 2017, 355 (6329): eaaf4791. |
| 57 | WU Y, LI B Z, ZHAO M, et al. Bug mapping and fitness testing of chemically synthesized chromosome X[J]. Science, 2017, 355 (6329): eaaf4706. |
| 58 | ZHANG W, ZHAO G, LUO Z, et al. Engineering the ribosomal DNA in a megabase synthetic chromosome[J]. Science, 2017, 355 (6329): eaaf3981. |
| 59 | XIE Z X, LIU D, LI B Z, et al. Design and chemical synthesis of eukaryotic chromosomes[J]. Chemical Society Reviews, 2017, 46 (23): 7191-7207. |
| 60 | ISAACS F J, CARR P A, WANG H H, et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement[J]. Science, 2011, 333 (6040): 348-353. |
| 61 | KOMOR A C, KIM Y B, PACKER M S, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage[J]. Nature, 2016, 533 (7603): 420-424. |
| 62 | GAUDELLI N M, KOMOR A C, REES H A, et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage[J]. Nature, 2017, 551 (7681): 464-471. |
| 63 | BAO Z, HAMEDIRAD M, XUE P, et al. Genome-scale engineering of Saccharomyces cerevisiae with single-nucleotide precision[J]. Nature Biotechnology, 2018, 36 (6): 505-508. |
| 64 | JIA B, WU Y, LI B Z, et al. Precise control of SCRaMbLE in synthetic haploid and diploid yeast[J]. Nature Communications, 2018, 9 (1): 1933. |
| 65 | WANG J, XIE Z X, MA Y, et al. Ring synthetic chromosome ⅤSCRaMbLE[J]. Nature Communications, 2018, 9 (1): 3783. |
| 66 | NAKAMURA C E, WHITED G M. Metabolic engineering for the microbial production of 1,3-propanediol[J]. Current Opinion in Biotechnology, 2003, 14 (5): 454-459. |
| 67 | RO D K, PARADISE E M, OUELLET M, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast[J]. Nature, 2006, 440 (7086): 940-943. |
| 68 | MARTIN V J, PITERA D J, WITHERS S T, et al. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids[J]. Nature Biotechnology, 2003, 21 (7): 796-802. |
| 69 | NAKAGAWA A, MATSUMURA E, KOYANAGI T, et al. Total biosynthesis of opiates by stepwise fermentation using engineered Escherichia coli [J]. Nature Communications, 2016, 7: 10390. |
| 70 | LUO X, REITER M A, D'ESPAUX L, et al. Complete biosynthesis of cannabinoids and their unnatural analogues in yeast[J]. Nature, 2019, 567 (7746): 123-126. |
| 71 | CHEN X L, GAO C, GUO L, et al. DCEO biotechnology: tools to design, construct, evaluate, and optimize the metabolic pathway for biosynthesis of chemicals[J]. Chemical Reviews, 2018, 118 (1): 4-72. |
| 72 | DU Y L, ALKHALAF L M, RYAN K S. In vitro reconstitution of indolmycin biosynthesis reveals the molecular basis of oxazolinone assembly[J]. PNAS, 2015, 112 (9): 2717-2722. |
| 73 | LUO Y, HUANG H, LIANG J, et al. Activation and characterization of a cryptic polycyclic tetramate macrolactam biosynthetic gene cluster[J]. Nature Communications, 2013, 4: 2894. |
| 74 | TU L, SU P, ZHANG Z, et al. GenomGenome of Tripterygium wilfordii and identification of cytochrome P450 involved in triptolide biosynthesis [J]. Nature Communications, 2020, 11 (1): 971. |
| 75 | LAU W, SATTELY E S. Six enzymes from mayapple that complete the biosynthetic pathway to the etoposide aglycone[J]. Science, 2015, 349 (6253): 1224-1228. |
| 76 | HAMMER S C, KNIGHT A M, ARNOLD F H. Design and evolution of enzymes for non-natural chemistry[J]. Current Opinion in Green and Sustainable Chemistry, 2017, 7: 23-30. |
| 77 | CHEN K, ARNOLD F H. Engineering new catalytic activities in enzymes[J]. Nature Catalysis, 2020, 3 (3): 203-213. |
| 78 | LEAVER-FAY A, TYKA M, LEWIS S M, et al. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules[J]. Methods Enzymol, 2011, 487: 545-574. |
| 79 | DOU J Y, VOROBIEVA A A, SHEFFLER W, et al. De novo design of a fluorescence-activating beta-barrel[J]. Nature, 2018, 561 (7724): 485-491. |
| 80 | BOYKEN S E, BENHAIM M A, BUSCH F, et al. De novo design of tunable, pH-driven conformational changes[J]. Science, 2019, 364 (6441): 658-664. |
| 81 | CHEN Z B, BOYKEN S E, JIA M X, et al. Programmable design of orthogonal protein heterodimers[J]. Nature, 2019, 565 (7737): 106-111. |
| 82 | HAMMER S C, KUBIK G, WATKINS E, et al. Anti-Markovnikov alkene oxidation by metal-oxo-mediated enzyme catalysis[J]. Science, 2017, 358 (6360): 215-218. |
| 83 | KAN S B J, HUANG X, GUMULYA Y, et al. Genetically programmed chiral organoborane synthesis[J]. Nature, 2017, 552 (7683): 132-136. |
| 84 | KAN S B, LEWIS R D, CHEN K, et al. Directed evolution of cytochrome c for carbon-silicon bond formation: bringing silicon to life[J]. Science, 2016, 354 (6315): 1048-1051. |
| 85 | IGNEA C, PONTINI M, MOTAWIA M S, et al. Synthesis of 11-carbon terpenoids in yeast using protein and metabolic engineering[J]. Nature Chemical Biology, 2018, 14 (12): 1090-1098. |
| 86 | FURUBAYASHI M, IKEZUMI M, TAKAICHI S, et al. A highly selective biosynthetic pathway to non-natural C50 carotenoids assembled from moderately selective enzymes[J]. Nature Communications, 2015, 6: 7534. |
| 87 | XIONG M, SCHNEIDERMAN D K, BATES F S, et al. Scalable production of mechanically tunable block polymers from sugar[J]. PNAS, 2014, 111 (23): 8357-8362. |
| 88 | TAI Y S, XIONG M, JAMBUNATHAN P, et al. Engineering nonphosphorylative metabolism to generate lignocellulose-derived products[J]. Nature Chemical Biology, 2016, 12 (4): 247-253. |
| 89 | YAO Y F, WANG C S, QIAO J, et al. Metabolic engineering of Escherichia coli for production of salvianic acid A via an artificial biosynthetic pathway[J]. Metabolic Engineering, 2013, 19: 79-87. |
| 90 | AJIKUMAR P K, XIAO W H, TYO K E, et al. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli [J]. Science, 2010, 330 (6000): 70-74. |
| 91 | XU P, GU Q, WANG W, et al. Modular optimization of multi-gene pathways for fatty acids production in E. coli [J]. Nature Communications, 2013, 4: 1409. |
| 92 | QIN J, ZHOU Y J, KRIVORUCHKO A, et al. Modular pathway rewiring of Saccharomyces cerevisiae enables high-level production of L-ornithine[J]. Nature Communications, 2015, 6: 8224. |
| 93 | XU P, QIAO K, AHN W S, et al. Engineering Yarrowia lipolytica as a platform for synthesis of drop-in transportation fuels and oleochemicals[J]. PNAS, 2016, 113 (39): 10848-10853. |
| 94 | FANG H, LI D, KANG J, et al. Metabolic engineering of Escherichia coli for de novo biosynthesis of vitamin B12 [J]. Nature Communications, 2018, 9 (1): 4917. |
| 95 | SRINIVASAN P, SMOLKE C D. Engineering a microbial biosynthesis platform for de novo production of tropane alkaloids[J]. Nature Communications, 2019, 10 (1): 3634. |
| 96 | SWEETLOVE L J, FERNIE A R. The role of dynamic enzyme assemblies and substrate channelling in metabolic regulation[J]. Nature Communications, 2018, 9 (1): 2136. |
| 97 | HAMMER S K, AVALOS J L. Harnessing yeast organelles for metabolic engineering[J]. Nature Chemical Biology, 2017, 13 (8): 823-832. |
| 98 | BERNER N, REUTTER K R, WOLF D H. Protein quality control of the endoplasmic reticulum and ubiquitin-proteasome-triggered degradation of aberrant proteins: yeast pioneers the path[J]. Annual Review of Biochemistry, 2018, 87: 751-782. |
| 99 | CHEN S J, WU X, WADAS B, et al. An N-end rule pathway that recognizes proline and destroys gluconeogenic enzymes[J]. Science, 2017, 355 (6323): aal3655. |
| 100 | PENG B, NIELSEN L K, KAMPRANIS S C, et al. Engineered protein degradation of farnesyl pyrophosphate synthase is an effective regulatory mechanism to increase monoterpene production in Saccharomyces cerevisiae [J]. Metabolic Engineering, 2018, 47: 83-93. |
| 101 | PENG B, PLAN M R, CHRYSANTHOPOULOS P, et al. A squalene synthase protein degradation method for improved sesquiterpene production in Saccharomyces cerevisiae [J]. Metabolic Engineering, 2017, 39: 209-219. |
| 102 | MORSUT L, ROYBAL K T, XIONG X, et al. Engineering customized cell sensing and response behaviors using synthetic notch receptors[J]. Cell, 2016, 164 (4): 780-791. |
| 103 | KENT R, DIXON N. Systematic evaluation of genetic and environmental factors affecting performance of translational riboswitches[J]. ACS Synthetic Biology, 2019, 8 (4): 884-901. |
| 104 | DAHLMAN J E, ABUDAYYEH O O, JOUNG J, et al. Orthogonal gene knockout and activation with a catalytically active Cas9 nuclease[J]. Nature Biotechnology, 2015, 33 (11): 1159-1161. |
| 105 | PANDIT A V, SRINIVASAN S, MAHADEVAN R. Redesigning metabolism based on orthogonality principles[J]. Nature Communications, 2017, 8: 15188. |
| 106 | DE LORENZO V. Beware of metaphors: chasses and orthogonality in synthetic biology[J]. Bioengineered Bugs, 2011, 2 (1): 3-7. |
| 107 | LIU H, LU T. Autonomous production of 1,4-butanediol via a de novo biosynthesis pathway in engineered Escherichia coli [J]. Metabolic Engineering, 2015, 29: 135-141. |
| 108 | IGNEA C, RAADAM M H, MOTAWIA M S, et al. Orthogonal monoterpenoid biosynthesis in yeast constructed on an isomeric substrate[J]. Nature Communications, 2019, 10 (1): 3799. |
| 109 | JI D, WANG L, HOU S, et al. Creation of bioorthogonal redox systems depending on nicotinamide flucytosine dinucleotide[J]. Journal of the American Chemical Society, 2011, 133 (51): 20857-20862. |
| 110 | LIU Y, FENG Y, WANG L, et al. Structural insights into phosphite dehydrogenase variants favoring a non-natural redox cofactor[J]. ACS Catalysis, 2019, 9 (3): 1883-1887. |
| 111 | WANG L, JI D, LIU Y, et al. Synthetic cofactor-linked metabolic circuits for selective energy transfer[J]. ACS Catalysis, 2017, 7 (3): 1977-1983. |
| 112 | GUO X, LIU Y, WANG Q, et al. Non-natural cofactor and formate-driven reductive carboxylation of pyruvate[J]. Angewandte Chemie International Edition, 2020, 59 (8): 3143-3146. |
| 113 | LIU G S, LI T, ZHOU W, et al. The yeast peroxisome: a dynamic storage depot and subcellular factory for squalene overproduction[J]. Metabolic Engineering, 2020, 57: 151-161. |
| 114 | BALDI N, DYKSTRA J C, LUTTIK M A H, et al. Functional expression of a bacterial alpha-ketoglutarate dehydrogenase in the cytosol of Saccharomyces cerevisiae [J]. Metabolic Engineering, 2019, 56: 190-197. |
| 115 | LV X, WANG F, ZHOU P, et al. Dual regulation of cytoplasmic and mitochondrial acetyl-CoA utilization for improved isoprene production in Saccharomyces cerevisiae [J]. Nature Communications, 2016, 7: 12851. |
| 116 | SADRE R, KUO P, CHEN J, et al. Cytosolic lipid droplets as engineered organelles for production and accumulation of terpenoid biomaterials in leaves[J]. Nature Communications, 2019, 10 (1): 853. |
| 117 | ZHAO C, KIM Y, ZENG Y, et al. Co-compartmentation of terpene biosynthesis and storage via synthetic droplet[J]. ACS Synthetic Biology, 2018, 7 (3): 774-781. |
| 118 | ZHOU Y J, BUIJS N A, ZHU Z, et al. Harnessing yeast peroxisomes for biosynthesis of fatty-acid-derived biofuels and chemicals with relieved side-pathway competition[J]. Journal of the American Chemical Society, 2016, 138 (47): 15368-15377. |
| 119 | KIM J E, JANG I S, SON S H, et al. Tailoring the Saccharomyces cerevisiae endoplasmic reticulum for functional assembly of terpene synthesis pathway[J]. Metabolic Engineering, 2019, 56: 50-59. |
| 120 | CHEN R, YANG S, ZHANG L, et al. Advanced strategies for production of natural products in yeast[J]. iScience, 2020, 23 (3): 100879. |
| 121 | KLEI I J VAN DER, VEENHUIS M. Yeast peroxisomes: function and biogenesis of a versatile cell organelle[J]. Trends Microbiol., 1997, 5 (12): 502-509. |
| 122 | CROSS L L, PAUDYAL R, KAMISUGI Y, et al. Towards designer organelles by subverting the peroxisomal import pathway[J]. Nature Communications, 2017, 8 (1): 454. |
| 123 | STEINKUHLER J, KNORR R L, ZHAO Z, et al. Controlled division of cell-sized vesicles by low densities of membrane-bound proteins[J]. Nature Communications, 2020, 11 (1): 905. |
| 124 | KERFELD C A, AUSSIGNARGUES C, ZARZYCKI J, et al. Bacterial microcompartments[J]. Nat. Rev. Microbiol., 2018, 16 (5): 277-290. |
| 125 | HAGEN A, SUTTER M, SLOAN N, et al. Programmed loading and rapid purification of engineered bacterial microcompartment shells[J]. Nature Communications, 2018, 9 (1): 2881. |
| 126 | LEE M J, MANTELL J, BROWN I R, et al. De novo targeting to the cytoplasmic and luminal side of bacterial microcompartments[J]. Nature Communications, 2018, 9 (1): 3413. |
| 127 | FERLEZ B, SUTTER M, KERFELD C A. A designed bacterial microcompartment shell with tunable composition and precision cargo loading[J]. Metabolic Engineering, 2019, 54: 286-291. |
| 128 | GIESSEN T W, SILVER P A. Widespread distribution of encapsulin nanocompartments reveals functional diversity[J]. Nature Microbiology, 2017, 2 (6): 17029. |
| 129 | BUGAJ L J, CHOKSI A T, MESUDA C K, et al. Optogenetic protein clustering and signaling activation in mammalian cells[J]. Nature Methods, 2013, 10 (3): 249-252. |
| 130 | GIL A A, LAPTENOK S P, IULIANO J N, et al. Photoactivation of the BLUF protein PixD Probed by the site-specific incorporation of fluorotyrosine residues[J]. Journal of the American Chemical Society, 2017, 139 (41): 14638-14648. |
| 131 | LAU Y H, GIESSEN T W, ALTENBURG W J, et al. Prokaryotic nanocompartments form synthetic organelles in a eukaryote[J]. Nature Communications, 2018, 9 (1): 1311. |
| 132 | ZHAO E M, SUEK N, WILSON M Z, et al. Light-based control of metabolic flux through assembly of synthetic organelles[J]. Nature Chemical Biology, 2019, 15 (6): 589-597. |
| 133 | RUGBJERG P, SOMMER M O A. Overcoming genetic heterogeneity in industrial fermentations[J]. Nature Biotechnology, 2019, 37 (8): 869-876. |
| 134 | RUGBJERG P, SARUP-LYTZEN K, NAGY M, et al. Synthetic addiction extends the productive life time of engineered Escherichia coli populations[J]. PNAS, 2018, 115 (10): 2347-2352. |
| 135 | XIAO Y, BOWEN C H, LIU D, et al. Exploiting nongenetic cell-to-cell variation for enhanced biosynthesis[J]. Nature Chemical Biology, 2016, 12 (5): 339-344. |
| 136 | RAMAN S, ROGERS J K, TAYLOR N D, et al. Evolution-guided optimization of biosynthetic pathways[J]. PNAS, 2014, 111 (50): 17803-17808. |
| 137 | GUPTA A, REIZMAN I M, REISCH C R, et al. Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit[J]. Nature Biotechnology, 2017, 35 (3): 273-279. |
| 138 | DAHL R H, ZHANG F, ALONSO-GUTIERREZ J, et al. Engineering dynamic pathway regulation using stress-response promoters[J]. Nature Biotechnology, 2013, 31 (11): 1039-1046. |
| 139 | LIANG C, ZHANG X, WU J, et al. Dynamic control of toxic natural product biosynthesis by an artificial regulatory circuit[J]. Metabolic Engineering, 2020, 57: 239-246. |
| 140 | CERONI F, BOO A, FURINI S, et al. Burden-driven feedback control of gene expression[J]. Nature Methods, 2018, 15 (5): 387-393. |
| 141 | JULLESSON D, DAVID F, PFLEGER B, et al. Impact of synthetic biology and metabolic engineering on industrial production of fine chemicals[J]. Biotechnology Advances, 2015, 33 (7): 1395-1402. |
| 142 | SANDBERG T E, SALAZAR M J, WENG L L, et al. The emergence of adaptive laboratory evolution as an efficient tool for biological discovery and industrial biotechnology[J]. Metabolic Engineering, 2019, 56: 1-16. |
| 143 | FLETCHER E, FEIZI A, BISSCHOPS M M M, et al. Evolutionary engineering reveals divergent paths when yeast is adapted to different acidic environments[J]. Metabolic Engineering, 2017, 39: 19-28. |
| 144 | DEATHERAGE D E, KEPNER J L, BENNETT A F, et al. Specificity of genome evolution in experimental populations of Escherichia coli evolved at different temperatures[J]. PNAS, 2017, 114 (10): E1904-E1912. |
| 145 | MUNDHADA H, SEOANE J M, SCHNEIDER K, et al. Increased production of L-serine in Escherichia coli through adaptive laboratory evolution[J]. Metabolic Engineering, 2017, 39: 141-150. |
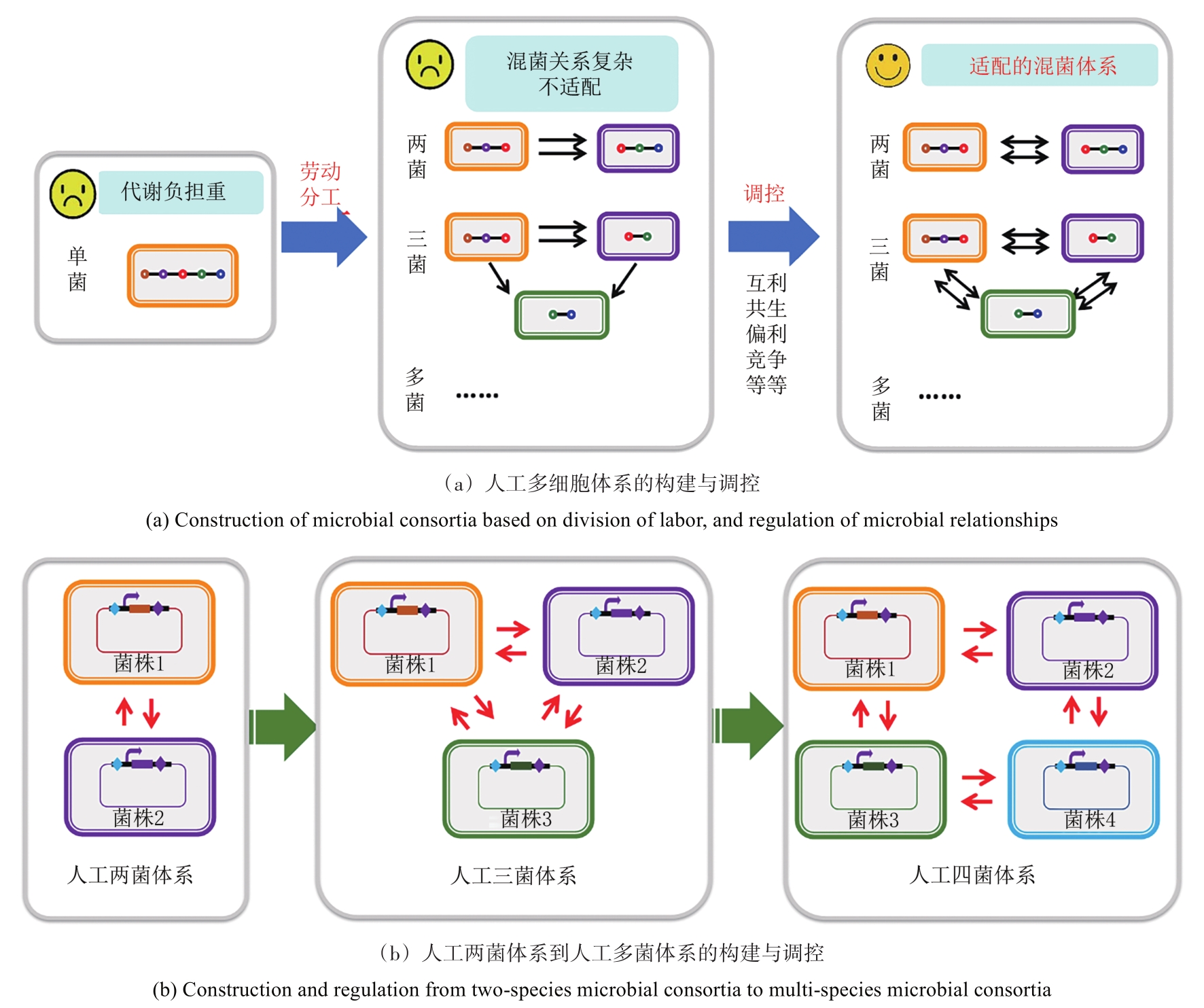
| 146 | RODRíGUEZ-VERDUGO A, CARRILLO-CISNEROS D, GONZáLEZ-GONZáLEZ A, et al. Different tradeoffs result from alternate genetic adaptations to a common environment[J]. PNAS, 2014, 111 (33): 12121. |
| 147 | CASPETA L, CHEN Y, GHIACI P, et al. Altered sterol composition renders yeast thermotolerant[J]. Science, 2014, 346 (6205): 75-78. |
| 148 | THORWALL S, SCHWARTZ C, CHARTRON J W, et al. Stress-tolerant non-conventional microbes enable next-generation chemical biosynthesis[J]. Nature Chemical Biology, 2020, 16 (2): 113-121. |
| 149 | CALERO P, NIKEL P I. Chasing bacterial chassis for metabolic engineering: a perspective review from classical to non-traditional microorganisms[J]. Microbial Biotechnology, 2019, 12 (1): 98-124. |
| 150 | KUMAR V, PARK S. Potential and limitations of Klebsiella pneumoniae as a microbial cell factory utilizing glycerol as the carbon source[J]. Biotechnology Advances, 2018, 36 (1): 150-167. |
| 151 | CHARUBIN K, BENNETT R K, FAST A G, et al. Engineering Clostridium organisms as microbial cell-factories: challenges & opportunities[J]. Metabolic Engineering, 2018, 50: 173-191. |
| 152 | TAN D, WU Q, CHEN J C, et al. Engineering Halomonas TD01 for the low-cost production of polyhydroxyalkanoates[J]. Metabolic Engineering, 2014, 26: 34-47. |
| 153 | BRENNER K, YOU L, ARNOLD F H. Engineering microbial consortia: a new frontier in synthetic biology[J]. Trends in Biotechnology, 2008, 26 (9): 483-489. |
| 154 | JONES J A, WANG X. Use of bacterial co-cultures for the efficient production of chemicals[J]. Current Opinion in Biotechnology, 2018, 53: 33-38. |
| 155 | SONG H, DING M Z, JIA X Q, et al. Synthetic microbial consortia: from systematic analysis to construction and applications[J]. Chemical Society Reviews, 2014, 43 (20): 6954-6981. |
| 156 | HAYS S G, PATRICK W G, ZIESACK M, et al. Better together: engineering and application of microbial symbioses[J]. Current Opinion in Biotechnology, 2015, 36: 40-49. |
| 157 | KONG W T, MELDGIN D R, COLLINS J J, et al. Designing microbial consortia with defined social interactions[J]. Nature Chemical Biology, 2018, 14 (8): 821-829. |
| 158 | JAROSZ D F, BROWN J C S, WALKER G A, et al. Cross-kingdom chemical communication drives a heritable, mutually beneficial prion-based transformation of metabolism[J]. Cell, 2014, 158 (5): 1083-1093. |
| 159 | ZELEZNIAK A, ANDREJEV S, PONOMAROVA O, et al. Metabolic dependencies drive species co-occurrence in diverse microbial communities[J]. PNAS, 2015, 112 (20): 6449-6454. |
| 160 | WINTERMUTE E H, SILVER P A. Dynamics in the mixed microbial concourse[J]. Genes & Development, 2010, 24 (23): 2603-2614. |
| 161 | BRENNER K, KARIG D K, WEISS R, et al. Engineered bidirectional communication mediates a consensus in a microbial biofilm consortium[J]. PNAS, 2007, 104 (44): 17300-17304. |
| 162 | FAUST K, RAES J. Microbial interactions: from networks to models[J]. Nature Reviews Microbiology, 2012, 10 (8): 538-550. |
| 163 | MCCARTY N S, LEDESMA-AMARO R. Synthetic biology tools to engineer microbial communities for biotechnology[J]. Trends in Biotechnology, 2019, 37 (2): 181-197. |
| 164 | SCOTT S R, DIN M O, BITTIHN P, et al. A stabilized microbial ecosystem of self-limiting bacteria using synthetic quorum-regulated[J]. Nature Microbiology, 2017, 2 (8): 17083. |
| 165 | SHOU W Y, RAM S, VILAR J M G. Synthetic cooperation in engineered yeast populations[J]. PNAS, 2007, 104 (6): 1877-1882. |
| 166 | HU B, DU J, ZOU R Y, et al. An environment-sensitive synthetic microbial ecosystem[J]. PLoS One, 2010, 5 (5): e10619. |
| 167 | WANG E X, DING M Z, MA Q, et al. Reorganization of a synthetic microbial consortium for one-step vitamin C fermentation[J]. Microbial Cell Factories, 2016, 15: 21. |
| 168 | ROELL G W, ZHA J, CARR R R, et al. Engineering microbial consortia by division of labor[J]. Microbial Cell Factories, 2019, 18 (1): 35. |
| 169 | LIU Y, DING M Z, LING W, et al. A three-species microbial consortium for power generation[J]. Energy & Environmental Science, 2017, 10 (7): 1600-1609. |
| 170 | ZHOU K, QIAO K J, EDGAR S, et al. Distributing a metabolic pathway among a microbial consortium enhances production of natural products[J]. Nature Biotechnology, 2015, 33 (4): 377-383. |
| 171 | JONES J A, VERNACCHIO V R, SINKOE A L, et al. Experimental and computational optimization of an Escherichia coli co-culture for the efficient production of flavonoids[J]. Metabolic Engineering, 2016, 35: 55-63. |
| 172 | PANDE S, SHITUT S, FREUND L, et al. Metabolic cross-feeding via intercellular nanotubes among bacteria[J]. Nature Communications, 2015, 6: 6238. |
| 173 | BENOMAR S, RANAVA D, CARDENAS M L, et al. Nutritional stress induces exchange of cell material and energetic coupling between bacterial species[J]. Nature Communications, 2015, 6: 6283. |
| 174 | WEBER W, BABA M, FUSSENEGGER M. Synthetic ecosystems based on airborne inter- and intrakingdom communication[J]. PNAS, 2007, 104 (25): 10435-10440. |
| 175 | CHEN Z Y, SUN X X, LI Y, et al. Metabolic engineering of Escherichia coli for microbial synthesis of monolignols[J]. Metabolic Engineering, 2017, 39: 102-109. |
| 176 | JONES J A, VERNACCHIO V R, COLLINS S M, et al. Complete biosynthesis of Anthocyanins using E. coli polycultures[J]. Mbio., 2017, 8 (3): e00621-00617. |
| 177 | QIAN X, CHEN L, SUI Y, et al. Biotechnological potential and applications of microbial consortia[J]. Biotechnology Advances, 2020, 40: 107500. |
| 178 | SGOBBA E, WENDISCH V F. Synthetic microbial consortia for small molecule production[J]. Current Opinion in Biotechnology, 2019, 62: 72-79. |
| 179 | ZHANG H, PEREIRA B, LI Z, et al. Engineering Escherichia coli coculture systems for the production of biochemical products[J]. PNAS, 2015, 112 (27): 8266-8271. |
| 180 | LIU X, LI X B, JIANG J L, et al. Convergent engineering of syntrophic Escherichia coli coculture for efficient production of glycosides[J]. Metabolic Engineering, 2018, 47: 243-253. |
| 181 | ZHAO C H, SINUMVAYO J P, ZHANG Y P, et al. Design and development of a "Y-shaped" microbial consortium capable of simultaneously utilizing biomass sugars for efficient production of butanol[J]. Metabolic Engineering, 2019, 55: 111-119. |
| 182 | AKDEMIR H, SILVA A, ZHA J, et al. Production of pyranoanthocyanins using Escherichia coli co-cultures[J]. Metabolic Engineering, 2019, 55: 290-298. |
| 183 | GUO X Y, LI Z H, WANG X N, et al. De novo phenol bioproduction from glucose using biosensor-assisted microbial coculture engineering[J]. Biotechnology and Bioengineering, 2019, 116 (12): 3349-3359. |
| 184 | MINTY J J, SINGER M E, SCHOLZ S A, et al. Design and characterization of synthetic fungal-bacterial consortia for direct production of isobutanol from cellulosic biomass[J]. PNAS, 2013, 110 (36): 14592-14597. |
| 185 | SCHOLZ S A, GRAVES I, MINTY J J, et al. Production of cellulosic organic acids via synthetic fungal consortia[J]. Biotechnology and Bioengineering, 2018, 115 (4): 1096-1100. |
| 186 | TSAI S L, GOYAL G, CHEN W. Surface display of a functional minicellulosome by intracellular complementation using a synthetic yeast consortium and its application to cellulose hydrolysis and ethanol production[J]. Applied and Environmental Microbiology, 2010, 76 (22): 7514-7520. |
| 187 | WEN Z Q, MINTON N P, ZHANG Y, et al. Enhanced solvent production by metabolic engineering of a twin-clostridial consortium[J]. Metabolic Engineering, 2017, 39: 38-48. |
| 188 | GILBERT E S, WALKER A W, KEASLING J D. A constructed microbial consortium for biodegradation of the organophosphorus insecticide parathion[J]. Applied Microbiology and Biotechnology, 2003, 61 (1): 77-81. |
| 189 | ZHANG H, YANG C, LI C K, et al. Functional assembly of a microbial consortium with autofluorescent and mineralizing activity for the biodegradation of organophosphates[J]. Journal of Agricultural and Food Chemistry, 2008, 56 (17): 7897-7902. |
| 190 | MARTINEZ I, MOHAMED M E S, ROZAS D, et al. Engineering synthetic bacterial consortia for enhanced desulfurization and revalorization of oil sulfur compounds[J]. Metabolic Engineering, 2016, 35: 46-54. |
| 191 | IBRAR M, ZHANG H. Construction of a hydrocarbon-degrading consortium and characterization of two new lipopeptides biosurfactants[J]. Science of the Total Environment, 2020, 714: 136400. |
| 192 | ERLICH Y, ZIELINSKI D. DNA Fountain enables a robust and efficient storage architecture[J]. Science, 2017, 355 (6328): 950-953. |
| 193 | ORGANICK L, ANG S D, CHEN Y J, et al. Random access in large-scale DNA data storage[J]. Nature Biotechnology, 2018, 36 (7): 660-660. |
| 194 | CEZE L, NIVALA J, STRAUSS K. Molecular digital data storage using DNA[J]. Nature Reviews Genetics, 2019, 20 (8): 456-466. |
| 195 | GOLDMAN N, BERTONE P, CHEN S Y, et al. Towards practical, high-capacity, low-maintenance information storage in synthesized DNA[J]. Nature, 2013, 494 (7435): 77-80. |
| 196 | CHURCH G M, GAO Y, KOSURI S. Next-generation digital information storage in DNA[J]. Science, 2012, 337 (6102): 1628. |
| 197 | KOCH J, GANTENBEIN S, MASANIA K, et al. A DNA-of-things storage architecture to create materials with embedded memory[J]. Nature Biotechnology, 2020, 38 (1): 39-43. |
| 198 | GRASS R N, HECKEL R, PUDDU M, et al. Robust chemical preservation of digital information on DNA in silica with error-correcting codes[J]. Angewandte Chemie-International Edition, 2015, 54 (8): 2552-2555. |
| 199 | TAKAHASHI C N, NGUYEN B H, STRAUSS K, et al. Demonstration of end-to-end automation of DNA data storage[J]. Scientific Reports, 2019, 9 (1): 4998. |
| 200 | CHOI Y, RYU T, LEE A C, et al. High information capacity DNA-based data storage with augmented encoding characters using degenerate bases[J]. Scientific Reports, 2019, 9 (1): 6582. |
| 201 | ANAVY L, VAKNIN I, ATAR O, et al. Data storage in DNA with fewer synthesis cycles using composite DNA letters[J]. Nature Biotechnology, 2019, 37 (10): 1237. |
| 202 | 陈为刚, 黄刚, 李炳志, 等. 音视频文件的DNA信息存储[J]. 中国科学:生命科学, 2020, 50 (1): 81-85. |
| CHEN W G, HUANG G, LI B Z, et al. DNA information storage for audio and video files (in Chinese)[J]. SCIENTIA SINICA Vitae, 2020, 50(1): 81-85. | |
| 203 | ROTHEMUND P W K. Folding DNA to create nanoscale shapes and patterns[J]. Nature, 2006, 440 (7082): 297-302. |
| 204 | 钱璐璐, 汪颖, 张钊, 等. DNA纳米结构仿中国地图[J]. 科学通报, 2006, 51 (24): 2860-2863. |
| QIAN L L, WANG Y, ZHANG Z, et al. Imating China map with DNA nanostructure[J]. Chinese Science Bulletin, 2006, 51 (24): 2860-2863. | |
| 205 | DOUGLAS S M, CHOU J J, SHIH W M. DNA-nanotube-induced alignment of membrane proteins for NMR structure determination[J]. PNAS, 2007, 104 (16): 6644-6648. |
| 206 | ANDERSEN E S, DONG M, NIELSEN M M, et al. Self-assembly of a nanoscale DNA box with a controllable lid[J]. Nature, 2009, 459 (7243): 73-76. |
| 207 | HAN D R, PAL S, NANGREAVE J, et al. DNA origami with complex curvatures in three-dimensional space[J]. Science, 2011, 332 (6027): 342-346. |
| 208 | DU Y, JIANG Q, BEZIERE N, et al. DNA-nanostructure-gold-nanorod hybrids for enhanced in vivo optoacoustic imaging and photothermal therapy[J]. Advanted Materials, 2016, 28 (45): 10000-10007. |
| 209 | ZHANG Y N, WANG F, CHAO J, et al. DNA origami cryptography for secure communication[J]. Nature Communications, 2019, 10 (1): 5469. |
| 210 | BAZRAFSHAN A, MEYER T A, SU H Q, et al. Tunable DNA origami motors translocate ballistically over μm distances at nm/s speeds[J]. Angewandte Chemie-International Edition, 2020, 132: 2-10. |
| 211 | YOUNG T S, SCHULTZ P G. Beyond the canonical 20 amino acids: expanding the genetic lexicon[J]. Journal of Biological Chemistry, 2010, 285 (15): 11039-11044. |
| 212 | DEEPANKUMAR K, SHON M, NADARAJAN S P, et al. Enhancing thermostability and organic solvent tolerance of w-transaminase through global incorporation of fluorotyrosine[J]. Advanced Synthesis & Catalysis, 2014, 356 (5): 993-998. |
| 213 | DRIENOVSKA I, MAYER C, DULSON C, et al. A designer enzyme for hydrazone and oxime formation featuring an unnatural catalytic aniline residue[J]. Nature Chemistry, 2018, 10 (9): 946-952. |
| 214 | ROVNER A J, HAIMOVICH A D, KATZ S R, et al. Recoded organisms engineered to depend on synthetic amino acids[J]. Nature, 2015, 518 (7537): 89-93. |
| 215 | MANDELL D J, LAJOIE M J, MEE M T, et al. Biocontainment of genetically modified organisms by synthetic protein design[J]. Nature, 2015, 518 (7537): 55-60. |
| 216 | YU Y, LV X, LI J, et al. Defining the role of tyrosine and rational tuning of oxidase activity by genetic incorporation of unnatural tyrosine analogs[J]. Journal of the American Chemical Society, 2015, 137 (14): 4594-4597. |
| 217 | YOUNG D D, SCHULTZ P G. Playing with the molecules of life[J]. ACS Chemical Biology, 2018, 13 (4): 854-870. |
| 218 | ZHANG Y, PTACIN J L, FISCHER E C, et al. A semi-synthetic organism that stores and retrieves increased genetic information[J]. Nature, 2017, 551 (7682): 644-647. |
| 219 | DUNN M R, JIMENEZ R M, CHAPUT J C. Analysis of aptamer discovery and technology[J]. Nature Reviews Chemistry, 2017, 1 (10): 0076. |
| 220 | CHEN T, HONGDILOKKUL N, LIU Z X, et al. The expanding world of DNA and RNA[J]. Current Opinion in Chemical Biology, 2016, 34: 80-87. |
| 221 | HOSHIKA S, LEAL N A, KIM M J, et al. Hachimoji DNA and RNA: a genetic system with eight building blocks[J]. Science, 2019, 363 (6429): 884-887. |
| 222 | EREMEEVA E, HERDEWIJN P. Non canonical genetic material[J]. Current Opinion in Biotechnology, 2019, 57: 25-33. |
| 223 | KUK S K, SINGH R K, NAM D H, et al. Photoelectrochemical reduction of carbon dioxide to methanol through a highly efficient enzyme cascade[J]. Angewandte Chemie-International Edition, 2017, 56 (14): 3827-3832. |
| 224 | SOKOL K P, ROBINSON W E, WARNAN J, et al. Bias-free photoelectrochemical water splitting with photosystem II on a dye-sensitized photoanode wired to hydrogenase[J]. Nature Energy, 2018, 3 (11): 944-951. |
| 225 | SAKIMOTO K K, WONG A B, YANG P D. Self-photosensitization of nonphotosynthetic bacteria for solar-to-chemical production[J]. Science, 2016, 351 (6268): 74-77. |
| 226 | GUO J L, SUASTEGUI M, SAKIMOTO K K, et al. Light-driven fine chemical production in yeast biohybrids[J]. Science, 2018, 362 (6416): 813-816. |
| 227 | WANG X Y, PU J H, AN B L, et al. Programming cells for dynamic assembly of Iiorganic nano-objects with spatiotemporal control[J]. Advanced Materials, 2018, 30 (16): e1705968. |
| 228 | WANG X Y, PU J H, LIU Y, et al. Immobilization of functional nano-objects in living engineered bacterial biofilms for catalytic applications[J]. National Science Review, 2019, 6 (5): 929-943. |
| 229 | JIN X, HONG S H. Cell-free protein synthesis for producing ‘difficult-to-express’ proteins[J]. Biochemical Engineering Journal, 2018, 138: 156-164. |
| 230 | SHINODA T, SHINYA N, ITO K, et al. Cell-free methods to produce structurally intact mammalian membrane proteins[J]. Scientific Reports, 2016, 6: 30442. |
| 231 | MARSHALL R, MAXWELL C S, COLLINS S P, et al. Rapid and scalable characterization of CRISPR technologies using an E. coli cell-free transcription-translation system[J]. Molecular Cell, 2018, 69 (1): 146-157.e3. |
| 232 | MARTIN R W, MAJEWSKA N I, CHEN C X, et al. Development of a CHO-based cell-free platform for synthesis of active monoclonal antibodies[J]. ACS Synthetic Biology, 2017, 6 (7): 1370-1379. |
| 233 | RAMOS-BENITEZ M J, LOPEZ-CRUZ L M, AGUAYO V, et al. Cell-free expression, purification and immunoreactivity assessment of recombinant Fasciola hepatica saposin-like protein-2[J]. Molecular Biology Reports, 2018, 45 (5): 1551-1556. |
| 234 | GAO W, CHO E, LIU Y, et al. Advances and challenges in cell-free incorporation of unnatural amino acids into proteins[J]. Front Pharmacol, 2019, 10: 611. |
| 235 | ZIMMERMAN E S, HEIBECK T H, GILL A, et al. Production of site-specific antibody-drug conjugates using optimized non-natural amino acids in a cell-free expression system[J]. Bioconjugate Chemistry, 2014, 25 (2): 351-361. |
| 236 | HOFFMANN B, LOHR F, LAGUERRE A, et al. Protein labeling strategies for liquid-state NMR spectroscopy using cell-free synthesis[J]. Progress in Nuclear Magnetic Resonance Spectroscopy, 2018, 105: 1-22. |
| 237 | PEREZ J G, STARK J C, JEWETT M C. Cell-free synthetic biology: engineering beyond the cell[J]. Cold Spring Harbor Perspectives in Biology, 2016, 8 (12): a023853. |
| 238 | ADACHI J, KATSURA K, SEKI E, et al. Cell-free protein synthesis using S30 extracts from Escherichia coli RFzero strains for efficient incorporation of non-natural amino acids into proteins[J]. International Journal of Molecular Sciences, 2019, 20 (3): 492. |
| [1] | YING Hanjie, LIU Dong, WANG Zhenyu, SHEN Tao, ZHUANG Wei, ZHU Chenjie. Exploring industrial biomanufacturing and the goal of “carbon neutrality” [J]. Synthetic Biology Journal, 2025, 6(1): 1-7. |
| [2] | GAO Ge, BIAN Qi, WANG Baojun. Synthetic genetic circuit engineering: principles, advances and prospects [J]. Synthetic Biology Journal, 2025, 6(1): 45-64. |
| [3] | LI Jiyuan, WU Guosheng. Two hypothesises for the origins of organisms from the synthetic biology perspective [J]. Synthetic Biology Journal, 2025, 6(1): 190-202. |
| [4] | JIAO Hongtao, QI Meng, SHAO Bin, JIANG Jinsong. Legal issues for the storage of DNA data [J]. Synthetic Biology Journal, 2025, 6(1): 177-189. |
| [5] | TANG Xinghua, LU Qianneng, HU Yilin. Philosophical reflections on synthetic biology in the Anthropocene [J]. Synthetic Biology Journal, 2025, 6(1): 203-212. |
| [6] | XU Huaisheng, SHI Xiaolong, LIU Xiaoguang, XU Miaomiao. Key technologies for DNA storage: encoding, error correction, random access, and security [J]. Synthetic Biology Journal, 2025, 6(1): 157-176. |
| [7] | SHI Ting, SONG Zhan, SONG Shiyi, ZHANG Yi-Heng P. Job. In vitro BioTransformation (ivBT): a new frontier of industrial biomanufacturing [J]. Synthetic Biology Journal, 2024, 5(6): 1437-1460. |
| [8] | CHAI Meng, WANG Fengqing, WEI Dongzhi. Synthesis of organic acids from lignocellulose by biotransformation [J]. Synthetic Biology Journal, 2024, 5(6): 1242-1263. |
| [9] | SHAO Mingwei, SUN Simian, YANG Shimao, CHEN Guoqiang. Bioproduction based on extremophiles [J]. Synthetic Biology Journal, 2024, 5(6): 1419-1436. |
| [10] | CHEN Yu, ZHANG Kang, QIU Yijing, CHENG Caiyun, YIN Jingjing, SONG Tianshun, XIE Jingjing. Progress of microbial electrosynthesis for conversion of CO2 [J]. Synthetic Biology Journal, 2024, 5(5): 1142-1168. |
| [11] | ZHENG Haotian, LI Chaofeng, LIU Liangxu, WANG Jiawei, LI Hengrun, NI Jun. Design, optimization and application of synthetic carbon-negative phototrophic community [J]. Synthetic Biology Journal, 2024, 5(5): 1189-1210. |
| [12] | CHEN Ziling, XIANG Yangfei. Integrated development of organoid technology and synthetic biology [J]. Synthetic Biology Journal, 2024, 5(4): 795-812. |
| [13] | CAI Bingyu, TAN Xiangtian, LI Wei. Advances in synthetic biology for engineering stem cell [J]. Synthetic Biology Journal, 2024, 5(4): 782-794. |
| [14] | XIE Huang, ZHENG Yilei, SU Yiting, RUAN Jingyi, LI Yongquan. An overview on reconstructing the biosynthetic system of actinomycetes for polyketides production [J]. Synthetic Biology Journal, 2024, 5(3): 612-630. |
| [15] | ZHA Wenlong, BU Lan, ZI Jiachen. Advances in synthetic biology for producing potent pharmaceutical ingredients of traditional Chinese medicine [J]. Synthetic Biology Journal, 2024, 5(3): 631-657. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||