Synthetic Biology Journal ›› 2023, Vol. 4 ›› Issue (3): 535-550.DOI: 10.12211/2096-8280.2022-066
• Invited Review • Previous Articles Next Articles
Data-driven prediction and design for enzymatic reactions
ZENG Tao, WU Ruibo
- School of Pharmaceutical Science,Sun Yat-Sen University,Guangzhou 510006,Guangdong,China
-
Received:2022-11-23Revised:2022-12-27Online:2023-07-05Published:2023-06-30 -
Contact:WU Ruibo
数据驱动的酶反应预测与设计
曾涛, 巫瑞波
- 中山大学药学院,广东 广州 510006
-
通讯作者:巫瑞波 -
作者简介:曾涛 (1995—),男,博士研究生。研究方向为计算驱动的生物合成路线设计与优化。 E-mail:zengt28@mail2.sysu.edu.cn巫瑞波 (1984—),男,教授,博士生导师。研究方向为基于多尺度模拟的萜类天然产物生物智造与药效挖掘。 E-mail:wurb3@mail.sysu.edu.cn -
基金资助:广东省重点研发计划(2022B1111080005)
CLC Number:
Cite this article
ZENG Tao, WU Ruibo. Data-driven prediction and design for enzymatic reactions[J]. Synthetic Biology Journal, 2023, 4(3): 535-550.
曾涛, 巫瑞波. 数据驱动的酶反应预测与设计[J]. 合成生物学, 2023, 4(3): 535-550.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2022-066
| 数据库 | 特点 | 网址 |
|---|---|---|
| KEGG[ | 具有物种、基因组、酶等多水平注释的合成(代谢)反应数据库 | https://www.kegg.jp/kegg |
| MetaCyc[ | 以全面的初级/次级代谢产物合成途径对反应进行注释 | https://metacyc.org |
| Rhea[ | 全面的生物酶反应数据库,与Uniprot高度关联 | https://www.rhea-db.org |
| BRENDA[ | 对酶的各项信息(如分类、反应、参数等)进行详细注释 | https://www.brenda-enzymes.org |
| SABIO-RK[ | 包含酶反应的动力学参数、反应条件等信息 | https://sabiork.h-its.org |
| Reactome[ | 综合的生物通路数据库,包括代谢、信号调控等通路数据 | https://reactome.org |
| PathBank[ | 以常见模式物种为基础的代谢、调控通路数据库 | http://www.pathbank.org |
| HMDB[ | 人体小分子代谢数据库,包含反应、MS、NMR谱图等信息 | https://hmdb.ca |
| MetaNetX[ | 整合了多个来源的生化反应数据库用于代谢网络模型构建 | https://www.metanetx.org |
| Reaxys[ | 从专利和文献搜集和整理的大量有机反应和酶反应路线(商业非开源) | https://www.reaxys.com |
Table 1 Databases of enzymatic reactions
| 数据库 | 特点 | 网址 |
|---|---|---|
| KEGG[ | 具有物种、基因组、酶等多水平注释的合成(代谢)反应数据库 | https://www.kegg.jp/kegg |
| MetaCyc[ | 以全面的初级/次级代谢产物合成途径对反应进行注释 | https://metacyc.org |
| Rhea[ | 全面的生物酶反应数据库,与Uniprot高度关联 | https://www.rhea-db.org |
| BRENDA[ | 对酶的各项信息(如分类、反应、参数等)进行详细注释 | https://www.brenda-enzymes.org |
| SABIO-RK[ | 包含酶反应的动力学参数、反应条件等信息 | https://sabiork.h-its.org |
| Reactome[ | 综合的生物通路数据库,包括代谢、信号调控等通路数据 | https://reactome.org |
| PathBank[ | 以常见模式物种为基础的代谢、调控通路数据库 | http://www.pathbank.org |
| HMDB[ | 人体小分子代谢数据库,包含反应、MS、NMR谱图等信息 | https://hmdb.ca |
| MetaNetX[ | 整合了多个来源的生化反应数据库用于代谢网络模型构建 | https://www.metanetx.org |
| Reaxys[ | 从专利和文献搜集和整理的大量有机反应和酶反应路线(商业非开源) | https://www.reaxys.com |

Fig. 2 Prediction of forward and backward enzymatic reactions[Prediction starts with an enzyme molecule (green node) to deduce its substrate or product (yellow nodes), the lines represent transformation reactions between two molecules, with arrow from substrate (enzyme) to product (a) and the reverse (b). A reaction network is developed after the iterative prediction in which both known (solid nodes) and unknown (hollow nodes) molecules are included. The forward prediction is generally random while a target (blue node, such as a building block) is specified in the backward prediction, and the exploration will lead to the target with the help of iterative algorithms.]
| 反应预测与酶设计工具 | ||||
|---|---|---|---|---|
| 基于相似性 | 基于反应规则 | 基于机器学习 | ||
| 正向反应预测 | BioSynther[ (http://www.rxnfinder.org/) | ATLASx[ (https://lcsb-databases.epfl.ch/Atlas2) BCSExplorer[ (http://www.rxnfinder.org/) | Reymond等[ (https://github.com/reymondgroup/OpenNMT-py) Kavraki等[ (https:// github.com/KavrakiLab/MetaTrans) | |
| 逆合成预测 | PrecursorFinder[ (http://www.rxnfinder.org/) | RetroPath[ (https://github.com/brsynth/RetroPathRL) RetroBioCat[ (https://retrobiocat.com) | BioNavi-NP[ (http://biopathnavi.qmclab.com/) Probst等[ (https://github.com/rxn4chemistry/biocatalysis-model) | |
| 酶搜索和设计 | EC-BLAST[ (https://www.ebi.ac.uk/) | Selenzyme[ (http://selenzyme.synbiochem.co.uk/) BridgIT[ (https://lcsb-databases.epfl.ch/Atlas2) E-zyme2[ (https://www.genome.jp/tools/e-zyme2/) | Faulon等[ (tool not available) Ranganathan等[ (https://github.com/ranganathanlab/bmDCA) | |
| 酶功能与性质预测工具 | ||||
| 酶功能预测 | 功能 分类 | DeepEC[ Araki等[ MTDNN[ | ||
| 功能 优化 | ECNet[ Gitter等[ | |||
| 酶反应性质预测 | Lercher等[ Palsson等[ DLKcat[ | |||
Table 2 Tools for the prediction and design of enzymatic reactions
| 反应预测与酶设计工具 | ||||
|---|---|---|---|---|
| 基于相似性 | 基于反应规则 | 基于机器学习 | ||
| 正向反应预测 | BioSynther[ (http://www.rxnfinder.org/) | ATLASx[ (https://lcsb-databases.epfl.ch/Atlas2) BCSExplorer[ (http://www.rxnfinder.org/) | Reymond等[ (https://github.com/reymondgroup/OpenNMT-py) Kavraki等[ (https:// github.com/KavrakiLab/MetaTrans) | |
| 逆合成预测 | PrecursorFinder[ (http://www.rxnfinder.org/) | RetroPath[ (https://github.com/brsynth/RetroPathRL) RetroBioCat[ (https://retrobiocat.com) | BioNavi-NP[ (http://biopathnavi.qmclab.com/) Probst等[ (https://github.com/rxn4chemistry/biocatalysis-model) | |
| 酶搜索和设计 | EC-BLAST[ (https://www.ebi.ac.uk/) | Selenzyme[ (http://selenzyme.synbiochem.co.uk/) BridgIT[ (https://lcsb-databases.epfl.ch/Atlas2) E-zyme2[ (https://www.genome.jp/tools/e-zyme2/) | Faulon等[ (tool not available) Ranganathan等[ (https://github.com/ranganathanlab/bmDCA) | |
| 酶功能与性质预测工具 | ||||
| 酶功能预测 | 功能 分类 | DeepEC[ Araki等[ MTDNN[ | ||
| 功能 优化 | ECNet[ Gitter等[ | |||
| 酶反应性质预测 | Lercher等[ Palsson等[ DLKcat[ | |||
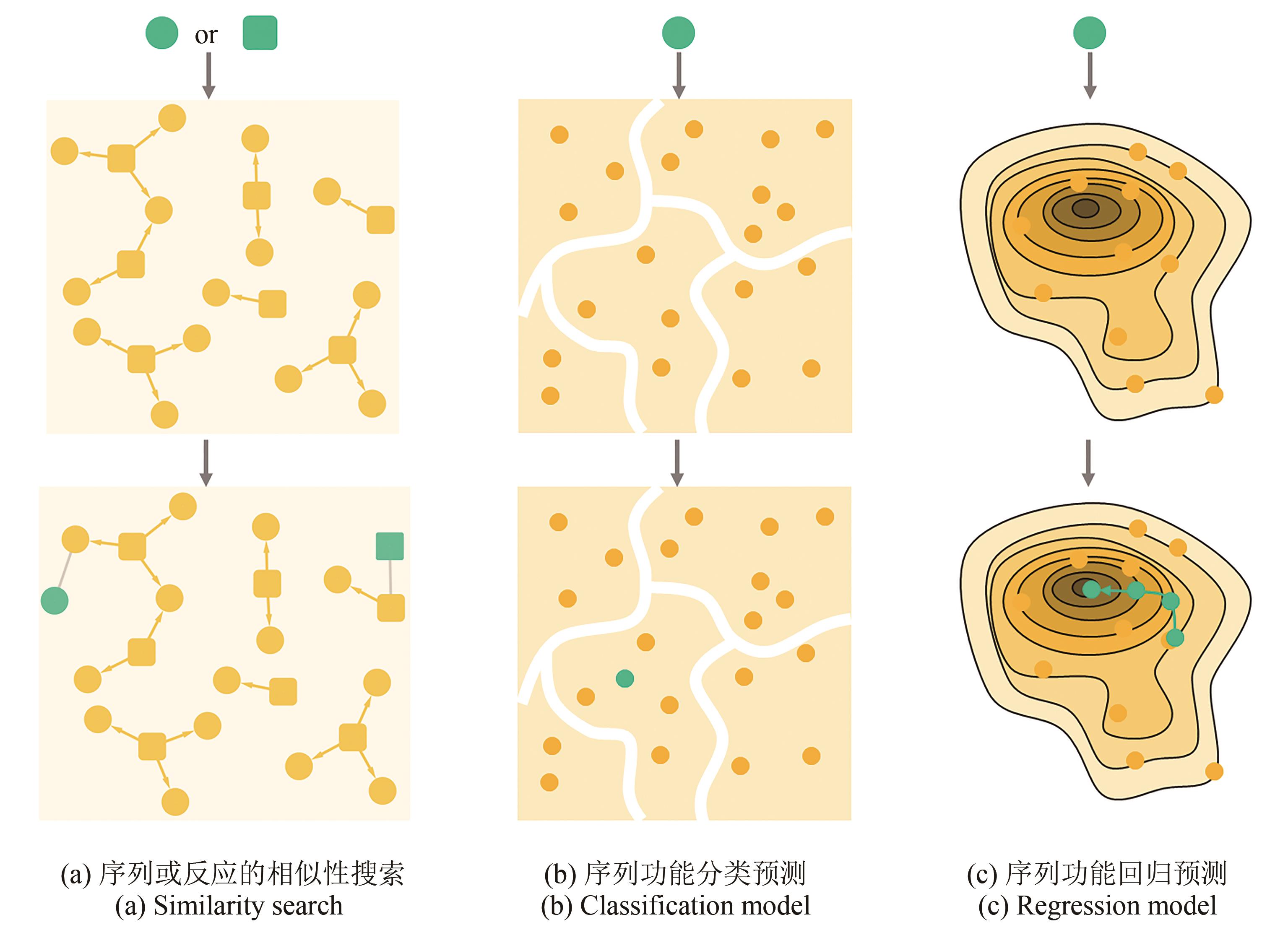
Fig. 3 Models for searching and predicting enzymes[Circular and square nodes represent sequences and reactions, respectively, and yellow filling indicates known data while green filling mean objects to be predicted. Similarity search (a) is to find a similar object in known enzyme-reaction pairs (connected nodes) to predict reactions (or enzymes) for target object. Classification model (b) is trained by enzymes with known function (usually discrete), in which the classification rule (white boundary) is clarified, and then the model can be used to classify an enzyme with unknown function. Regression model (c) is adapted to draw fitness landscape to predict continues variables such as the activity or stability of enzymes, which can then be used for enzyme design.]
| 1 | BENKOVIC S J, HAMMES-SCHIFFER S. A perspective on enzyme catalysis[J]. Science, 2003, 301(5637): 1196-1202. |
| 2 | BORNSCHEUER U T, BUCHHOLZ K. Highlights in biocatalysis-historical landmarks and current trends[J]. Engineering in Life Sciences, 2005, 5(4): 309-323. |
| 3 | SHELDON R A, WOODLEY J M. Role of biocatalysis in sustainable chemistry[J]. Chemical Reviews, 2018, 118(2): 801-838. |
| 4 | ZIMMERMAN J B, ANASTAS P T, ERYTHROPEL H C, et al. Designing for a green chemistry future[J]. Science, 2020, 367(6476): 397-400. |
| 5 | WINKLER C K, SCHRITTWIESER J H, KROUTIL W. Power of biocatalysis for organic synthesis[J]. ACS Central Science, 2021, 7(1): 55-71. |
| 6 | LIN G M, WARDEN-ROTHMAN R, VOIGT C A. Retrosynthetic design of metabolic pathways to chemicals not found in nature[J]. Current Opinion in Systems Biology, 2019, 14: 82-107. |
| 7 | SENN H M, THIEL W. QM/MM methods for biomolecular systems[J]. Angewandte Chemie International Edition, 2009, 48(7): 1198-1229. |
| 8 | ZHA W L, ZHANG F, SHAO J Q, et al. Rationally engineering santalene synthase to readjust the component ratio of sandalwood oil[J]. Nature Communications, 2022, 13: 2508. |
| 9 | WANG Y, MALACO MOROTTI A L, XIAO Y R, et al. Decoding the cytochrome P450 catalytic activity in divergence of benzophenone and xanthone biosynthetic pathways[J]. ACS Catalysis, 2022, 12(21): 13630-13637. |
| 10 | LIANG M M, ZHANG F, XU J X, et al. A conserved mechanism affecting hydride shifting and deprotonation in the synthesis of hopane triterpenes as compositions of wax in oat[J]. Proceedings of the National Academy of Sciences of the United States of America, 2022, 119(12): e2118709119. |
| 11 | VAN DIJK E L, AUGER H, JASZCZYSZYN Y, et al. Ten years of next-generation sequencing technology[J]. Trends in Genetics, 2014, 30(9): 418-426. |
| 12 | WANG H Y, GUO H, WANG N, et al. Toward the heterologous biosynthesis of plant natural products: gene discovery and characterization[J]. ACS Synthetic Biology, 2021, 10(11): 2784-2795. |
| 13 | JUMPER J, EVANS R, PRITZEL A, et al. Highly accurate protein structure prediction with AlphaFold[J]. Nature, 2021, 596(7873): 583-589. |
| 14 | PEARCE R, ZHANG Y. Deep learning techniques have significantly impacted protein structure prediction and protein design[J]. Current Opinion in Structural Biology, 2021, 68: 194-207. |
| 15 | CUI Y L, SUN J Y, WU B. Computational enzyme redesign: large jumps in function[J]. Trends in Chemistry, 2022, 4(5): 409-419. |
| 16 | SHELDON R A, PEREIRA P C. Biocatalysis engineering: the big picture[J]. Chemical Society Reviews, 2017, 46(10): 2678-2691. |
| 17 | JANG W D, KIM G B, KIM Y J, et al. Applications of artificial intelligence to enzyme and pathway design for metabolic engineering[J]. Current Opinion in Biotechnology, 2022, 73: 101-107. |
| 18 | HADADI N, HATZIMANIKATIS V Design of computational retrobiosynthesis tools for the design of de novo synthetic pathways[J]. Current Opinion in Chemical Biology, 2015, 28: 99-104. |
| 19 | 周静茹, 刘鹏, 夏建业, 等. 基于约束的基因组规模代谢网络模型构建方法研究进展[J]. 生物工程学报, 2021, 37(5): 1526-1540. |
| ZHOU J R, LIU P, XIA J Y, et al. Advances in the development of constraint-based genome-scale metabolic network models[J]. Chinese Journal of Biotechnology, 2021, 37(5): 1526-1540. | |
| 20 | MACKLIN D N, RUGGERO N A, COVERT M W. The future of whole-cell modeling[J]. Current Opinion in Biotechnology, 2014, 28: 111-115. |
| 21 | MAZURENKO S, PROKOP Z, DAMBORSKY J. Machine learning in enzyme engineering[J]. ACS Catalysis, 2020, 10(2): 1210-1223. |
| 22 | LIAO X P, MA H W, TANG Y J. Artificial intelligence: a solution to involution of design-build-test-learn cycle[J]. Current Opinion in Biotechnology, 2022, 75: 102712. |
| 23 | KANEHISA M, GOTO S. KEGG: Kyoto encyclopedia of genes and genomes[J]. Nucleic Acids Research, 2000, 28(1): 27-30. |
| 24 | CASPI R, BILLINGTON R, KESELER I M, et al. The MetaCyc database of metabolic pathways and enzymes-a 2019 update[J]. Nucleic Acids Research, 2020, 48(D1): D445-D453. |
| 25 | BANSAL P, MORGAT A, AXELSEN K B, et al. Rhea, the reaction knowledgebase in 2022[J]. Nucleic Acids Research, 2022, 50(D1): D693-D700. |
| 26 | CHANG A, JESKE L, ULBRICH S, et al. BRENDA, the ELIXIR core data resource in 2021: new developments and updates[J]. Nucleic Acids Research, 2021, 49(D1): D498-D508. |
| 27 | WITTIG U, REY M, WEIDEMANN A, et al. SABIO-RK: an updated resource for manually curated biochemical reaction kinetics[J]. Nucleic Acids Research, 2018, 46(D1): D656-D660. |
| 28 | GILLESPIE M, JASSAL B, STEPHAN R, et al. The reactome pathway knowledgebase 2022 [J]. Nucleic Acids Research, 2022, 50(D1): D687-D692. |
| 29 | WISHART D S, LI C, MARCU A, et al. PathBank: a comprehensive pathway database for model organisms[J]. Nucleic Acids Research, 2020, 48(D1): D470-D478. |
| 30 | WISHART D S, GUO A C, OLER E, et al. HMDB 5.0: the human metabolome database for 2022[J]. Nucleic Acids Research, 2022, 50(D1): D622-D631. |
| 31 | MORETTI S, TRAN V D T, MEHL F, et al. MetaNetX/MNXref: unified namespace for metabolites and biochemical reactions in the context of metabolic models[J]. Nucleic Acids Research, 2021, 49(D1): D570-D574. |
| 32 | LAWSON A J, SWIENTY-BUSCH J, GÉOUI T, et al. The making of reaxys—towards unobstructed access to relevant chemistry information[M]//ACS Symposium Series: The Future of the History of Chemical Information. Washington, DC: American Chemical Society, 2014: 127-148. |
| 33 | CONSORTIUM T U. UniProt: a worldwide hub of protein knowledge[J]. Nucleic Acids Research, 2019, 47(D1): D506-D515. |
| 34 | XU Y J, LIN K J, WANG S W, et al. Deep learning for molecular generation[J]. Future Medicinal Chemistry, 2019, 11(6): 567-597. |
| 35 | ELTON D C, BOUKOUVALAS Z, FUGE M D, et al. Deep learning for molecular design—a review of the state of the art[J]. Molecular Systems Design & Engineering, 2019, 4(4): 828-849. |
| 36 | HAGHIGHATLARI M, LI J, HEIDAR-ZADEH F, et al. Learning to make chemical predictions: the interplay of feature representation, data, and machine learning methods[J]. Chem, 2020, 6(7): 1527-1542. |
| 37 | SENIOR A W, EVANS R, JUMPER J, et al. Improved protein structure prediction using potentials from deep learning[J]. Nature, 2020, 577(7792): 706-710. |
| 38 | LANDRUM G. RDKit: Open-source cheminformatics software[EB/OL][2022-12-01]. . |
| 39 | The Gene Ontology Consortium. The gene ontology resource: enriching a GOld mine[J]. Nucleic Acids Research, 2021, 49(D1): D325-D334. |
| 40 | MOHAMMADIPEYHANI H, HAFNER J, SVESHNIKOVA A, et al. Expanding biochemical knowledge and illuminating metabolic dark matter with ATLASx[J]. Nature Communications, 2022, 13: 1560. |
| 41 | HATZIMANIKATIS V, LI C H, IONITA J A, et al. Exploring the diversity of complex metabolic networks[J]. Bioinformatics, 2005, 21(8): 1603-1609. |
| 42 | HAFNER J, PAYNE J, MOHAMMADIPEYHANI H, et al. A computational workflow for the expansion of heterologous biosynthetic pathways to natural product derivatives[J]. Nature Communications, 2021, 12: 1760. |
| 43 | TIAN Y, WU L, YUAN L, et al. BCSExplorer: a customized biosynthetic chemical space explorer with multifunctional objective function analysis[J]. Bioinformatics, 2020, 36(5): 1642-1643. |
| 44 | TU W Z, ZHANG H R, LIU J, et al. BioSynther: a customized biosynthetic potential explorer[J]. Bioinformatics, 2016, 32(3): 472-473. |
| 45 | KREUTTER D, SCHWALLER P, REYMOND J L. Predicting enzymatic reactions with a molecular transformer[J]. Chemical Science, 2021, 12(25): 8648-8659. |
| 46 | LITSA E E, DAS P, KAVRAKI L E. Prediction of drug metabolites using neural machine translation[J]. Chemical Science, 2020, 11(47): 12777-12788. |
| 47 | YUAN L, TIAN Y, DING S Z, et al. PrecursorFinder: a customized biosynthetic precursor explorer[J]. Bioinformatics, 2019, 35(9): 1603-1604. |
| 48 | KOCH M, DUIGOU T, FAULON J L. Reinforcement learning for bioretrosynthesis[J]. ACS Synthetic Biology, 2020, 9(1): 157-168. |
| 49 | FINNIGAN W, HEPWORTH L J, FLITSCH S L, et al. RetroBioCat as a computer-aided synthesis planning tool for biocatalytic reactions and cascades[J]. Nature Catalysis, 2021, 4(2): 98-104. |
| 50 | ZHENG S J, ZENG T, LI C T, et al. Deep learning driven biosynthetic pathways navigation for natural products with BioNavi-NP[J]. Nature Communications, 2022, 13: 3342. |
| 51 | PROBST D, MANICA M, NANA TEUKAM Y G, et al. Biocatalysed synthesis planning using data-driven learning[J]. Nature Communications, 2022, 13: 964. |
| 52 | RAHMAN S A, CUESTA S M, FURNHAM N, et al. EC-BLAST: a tool to automatically search and compare enzyme reactions[J]. Nature Methods, 2014, 11(2): 171-174. |
| 53 | CARBONELL P, WONG J, SWAINSTON N, et al. Selenzyme: enzyme selection tool for pathway design[J]. Bioinformatics, 2018, 34(12): 2153-2154. |
| 54 | HADADI N, MOHAMMADIPEYHANI H, MISKOVIC L, et al. Enzyme annotation for orphan and novel reactions using knowledge of substrate reactive sites[J]. Proceedings of the National Academy of Sciences of the United States of America, 2019, 116(15): 7298-7307. |
| 55 | MORIYA Y, YAMADA T, OKUDA S, et al. Identification of enzyme genes using chemical structure alignments of substrate-product pairs[J]. Journal of Chemical Information and Modeling, 2016, 56(3): 510-516. |
| 56 | MELLOR J, GRIGORAS I, CARBONELL P, et al. Semisupervised Gaussian process for automated enzyme search[J]. ACS Synthetic Biology, 2016, 5(6): 518-528. |
| 57 | RUSS W P, FIGLIUZZI M, STOCKER C, et al. An evolution-based model for designing chorismate mutase enzymes[J]. Science, 2020, 369(6502): 440-445. |
| 58 | RYU J Y, KIM H U, LEE S Y. Deep learning enables high-quality and high-throughput prediction of enzyme commission numbers[J]. Proceedings of the National Academy of Sciences of the United States of America, 2019, 116(28): 13996-14001. |
| 59 | WATANABE N, MURATA M, OGAWA T, et al. Exploration and evaluation of machine learning-based models for predicting enzymatic reactions[J]. Journal of Chemical Information and Modeling, 2020, 60(3): 1833-1843. |
| 60 | FA R, COZZETTO D, WAN C, et al. Predicting human protein function with multi-task deep neural networks[J]. PLoS One, 2018, 13(6): e0198216. |
| 61 | LUO Y N, JIANG G D, YU T H, et al. ECNet is an evolutionary context-integrated deep learning framework for protein engineering[J]. Nature Communications, 2021, 12: 5743. |
| 62 | GELMAN S, FAHLBERG S A, HEINZELMAN P, et al. Neural networks to learn protein sequence-function relationships from deep mutational scanning data[J]. Proceedings of the National Academy of Sciences of the United States of America, 2021, 118(48): e2104878118. |
| 63 | KROLL A, ENGQVIST M K M, HECKMANN D, et al. Deep learning allows genome-scale prediction of Michaelis constants from structural features[J]. PLoS Biology, 2021, 19(10): e3001402. |
| 64 | HECKMANN D, LLOYD C J, MIH N, et al. Machine learning applied to enzyme turnover numbers reveals protein structural correlates and improves metabolic models[J]. Nature Communications, 2018, 9: 5252. |
| 65 | LI F R, YUAN L, LU H Z, et al. Deep learning-based kcat prediction enables improved enzyme-constrained model reconstruction[J]. Nature Catalysis, 2022, 5(8): 662-672. |
| 66 | VASWANI A, SHAZEER N, PARMAR N, et al. Attention is all you need[C]//Proceedings of the 31st International Conference on Neural Information Processing Systems. December 4-9, 2017, Long Beach, California, USA. New York: ACM, 2017: 6000-6010. |
| 67 | COREY E J. General methods for the construction of complex molecules[J]. Pure and Applied Chemistry, 1967, 14(1): 19-38. |
| 68 | COREY E J, WIPKE W T. Computer-assisted design of complex organic syntheses[J]. Science, 1969, 166(3902): 178-192. |
| 69 | DELÉPINE B, DUIGOU T, CARBONELL P, et al. RetroPath2.0: a retrosynthesis workflow for metabolic engineers[J]. Metabolic Engineering, 2018, 45: 158-170. |
| 70 | SEGLER M H S, PREUSS M, WALLER M P. Planning chemical syntheses with deep neural networks and symbolic AI[J]. Nature, 2018, 555(7698): 604-610. |
| 71 | LIU B W, RAMSUNDAR B, KAWTHEKAR P, et al. Retrosynthetic reaction prediction using neural sequence-to-sequence models[J]. ACS Central Science, 2017, 3(10): 1103-1113. |
| 72 | CHEN B H, LI C T, DAI H J, et al. Retro*: learning retrosynthetic planning with neural guided a* search[C]//Proceedings of the 37th International Conference on Machine Learning. New York: ACM, 2020: 1608-1616. |
| 73 | 张建志, 付立豪, 唐婷, 等. 基于合成生物学策略的酶蛋白元件规模化挖掘[J]. 合成生物学, 2020, 1(3): 319-336. |
| ZHANG J Z, FU L H, TANG T, et al. Scalable mining of proteins for biocatalysis via synthetic biology[J]. Synthetic Biology Journal, 2020, 1(3): 319-336. | |
| 74 | FIGLIUZZI M, BARRAT-CHARLAIX P, WEIGT M. How pairwise coevolutionary models capture the collective residue variability in proteins?[J]. Molecular Biology and Evolution, 2018, 35(4): 1018-1027. |
| 75 | HUANG P S, BOYKEN S E, BAKER D. The coming of age of de novo protein design[J]. Nature, 2016, 537(7620): 320-327. |
| 76 | DAUPARAS J, ANISHCHENKO I, BENNETT N, et al. Robust deep learning-based protein sequence design using ProteinMPNN[J]. Science, 2022, 378(6615): 49-56. |
| 77 | LIU Y F, ZHANG L, WANG W L, et al. Rotamer-free protein sequence design based on deep learning and self-consistency[J]. Nature Computational Science, 2022, 2(7): 451-462. |
| 78 | JIANG L, ALTHOFF E A, CLEMENTE F R, et al. De novo computational design of Retro-Oldol enzymes[J]. Science, 2008, 319(5868): 1387-1391. |
| 79 | ALTSCHUL S F, GISH W, MILLER W, et al. Basic local alignment search tool[J]. Journal of Molecular Biology, 1990, 215(3): 403-410. |
| 80 | LI Z R, LIN H H, HAN L Y, et al. PROFEAT: a web server for computing structural and physicochemical features of proteins and peptides from amino acid sequence[J]. Nucleic Acids Research, 2006, 34(): W32-W37. |
| 81 | VAVRICKA C J, TAKAHASHI S, WATANABE N, et al. Machine learning discovery of missing links that mediate alternative branches to plant alkaloids[J]. Nature Communications, 2022, 13(1): 1405. |
| 82 | MISTRY J, CHUGURANSKY S, WILLIAMS L, et al. Pfam: the protein families database in 2021[J]. Nucleic Acids Research, 2021, 49(D1): D412-D419. |
| 83 | YANG K K, WU Z, ARNOLD F H. Machine-learning-guided directed evolution for protein engineering[J]. Nature Methods, 2019, 16(8): 687-694. |
| 84 | WITTMANN B J, JOHNSTON K E, WU Z, et al. Advances in machine learning for directed evolution[J]. Current Opinion in Structural Biology, 2021, 69: 11-18. |
| 85 | KLUMPP S, SCOTT M, PEDERSEN S, et al. Molecular crowding limits translation and cell growth[J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(42): 16754-16759. |
| 86 | CHEN Y, NIELSEN J. Energy metabolism controls phenotypes by protein efficiency and allocation[J]. Proceedings of the National Academy of Sciences of the United States of America, 2019, 116(35): 17592-17597. |
| 87 | ALLEY E C, KHIMULYA G, BISWAS S, et al. Unified rational protein engineering with sequence-based deep representation learning[J]. Nature Methods, 2019, 16(12): 1315-1322. |
| 88 | BORGER S, LIEBERMEISTER W, KLIPP E. Prediction of enzyme kinetic parameters based on statistical learning[J]. Genome Informatics International Conference on Genome Informatics, 2006, 17(1): 80-87. |
| 89 | YAN S M, SHI D Q, NONG H, et al. Predicting Km values of beta-glucosidases using cellobiose as substrate[J]. Interdisciplinary Sciences: Computational Life Sciences, 2012, 4(1): 46-53. |
| 90 | LI F R, CHEN Y, ANTON M, et al. GotEnzymes: an extensive database of enzyme parameter predictions[J]. Nucleic Acids Research, 2023, 51(D1): D583-D586. |
| 91 | MACKLIN D N, AHN-HORST T A, CHOI H, et al. Simultaneous cross-evaluation of heterogeneous E. coli datasets via mechanistic simulation[J]. Science, 2020, 369(6502): eaav3751. |
| 92 | THORNBURG Z R, BIANCHI D M, BRIER T A, et al. Fundamental behaviors emerge from simulations of a living minimal cell[J]. Cell, 2022, 185(2): 345-360.e28. |
| 93 | ENGQVIST M K M. Correlating enzyme annotations with a large set of microbial growth temperatures reveals metabolic adaptations to growth at diverse temperatures[J]. BMC Microbiology, 2018, 18(1): 177. |
| 94 | LI G, HU Y T, ZRIMEC J, et al. Bayesian genome scale modelling identifies thermal determinants of yeast metabolism[J]. Nature Communications, 2021, 12: 190. |
| 95 | REMBEZA E, ENGQVIST M K M. Experimental and computational investigation of enzyme functional annotations uncovers misannotation in the EC 1.1.3.15 enzyme class[J]. PLoS Computational Biology, 2021, 17(9): e1009446. |
| 96 | RAO R, BHATTACHARYA N, THOMAS N, et al. Evaluating protein transfer learning with TAPE[J]. Advances in Neural Information Processing Systems, 2019, 32: 9689-9701. |
| 97 | RIVES A, MEIER J, SERCU T, et al. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences[J]. Proceedings of the National Academy of Sciences of the United States of America, 2021, 118(15): e2016239118. |
| 98 | JAEGER S, FULLE S, TURK S. Mol2vec: unsupervised machine learning approach with chemical intuition[J]. Journal of Chemical Information and Modeling, 2018, 58(1): 27-35. |
| 99 | UNSAL S, ATAS H, ALBAYRAK M, et al. Learning functional properties of proteins with language models[J]. Nature Machine Intelligence, 2022, 4(3): 227-245. |
| 100 | COLEY C W, ROGERS L, GREEN W H, et al. Computer-assisted retrosynthesis based on molecular similarity[J]. ACS Central Science, 2017, 3(12): 1237-1245. |
| 101 | FERRUZ N, SCHMIDT S, HÖCKER B. ProtGPT2 is a deep unsupervised language model for protein design[J]. Nature Communications, 2022, 13(1): 4348. |
| 102 | SHIN J E, RIESSELMAN A J, KOLLASCH A W, et al. Protein design and variant prediction using autoregressive generative models[J]. Nature Communications, 2021, 12: 2403. |
| 103 | REPECKA D, JAUNISKIS V, KARPUS L, et al. Expanding functional protein sequence spaces using generative adversarial networks[J]. Nature Machine Intelligence, 2021, 3(4): 324-333. |
| 104 | BISWAS S, KHIMULYA G, ALLEY E C, et al. Low-N protein engineering with data-efficient deep learning[J]. Nature Methods, 2021, 18(4): 389-396. |
| 105 | LUO S T, SU Y F, PENG X G, et al. Antigen-specific antibody design and optimization with diffusion-based generative models for protein structures[EB/OL]. bioRxiv, 2022[2022-12-01]. . |
| 106 | WANG L, TITOV A, MCGIBBON R, et al. Discovering chemistry with an ab initio nanoreactor[J]. Nature Chemistry, 2014, 6(12): 1044-1048. |
| 107 | SIMM G N, VAUCHER A C, REIHER M. Exploration of reaction pathways and chemical transformation networks[J]. The Journal of Physical Chemistry A, 2019, 123(2): 385-399. |
| 108 | WANG Y H, XU H C, ZOU J, et al. Catalytic role of carbonyl oxygens and water in selinadiene synthase[J]. Nature Catalysis, 2022, 5(2): 128-135. |
| 109 | ZANGHELLINI A, JIANG L, WOLLACOTT A M, et al. New algorithms and an in silico benchmark for computational enzyme design[J]. Protein Science, 2006, 15(12): 2785-2794. |
| 110 | LEAVER-FAY A, TYKA M, LEWIS S M, et al. Chapter nineteen-Rosetta3: an object-oriented software suite for the simulation and design of macromolecules[M]// Methods in enzymology. Pittsburgh, PA, USA: Academic Press, 2011, 487: 545-574. |
| 111 | SIEGEL J B, SMITH A L, POUST S, et al. Computational protein design enables a novel one-carbon assimilation pathway[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(12): 3704-3709. |
| 112 | VERGES A, CAMBON E, BARBE S, et al. Computer-aided engineering of a transglycosylase for the glucosylation of an unnatural disaccharide of relevance for bacterial antigen synthesis[J]. ACS Catalysis, 2015, 5(2): 1186-1198. |
| 113 | CUI Y L, WANG Y H, TIAN W Y, et al. Development of a versatile and efficient C-N lyase platform for asymmetric hydroamination via computational enzyme redesign[J]. Nature Catalysis, 2021, 4(5): 364-373. |
| 114 | ZENG T, HESS B A, ZHANG F, et al. Bio-inspired chemical space exploration of terpenoids[J]. Briefings in Bioinformatics, 2022, 23(5): bbac197. |
| 115 | ZHANG L F, HAN J Q, WANG H, et al. Deep potential molecular dynamics: a scalable model with the accuracy of quantum mechanics[J]. Physical Review Letters, 2018, 120(14): 143001. |
| 116 | HU Q N, DENG Z, HU H N, et al. RxnFinder: biochemical reaction search engines using molecular structures, molecular fragments and reaction similarity[J]. Bioinformatics, 2011, 27(17): 2465-2467. |
| [1] | ZHONG Quanzhou, SHAN Yiyi, PEI Qingyun, JIN Yanyun, WANG Yihan, MENG Luyuan, WANG Xinyun, ZHANG Yuxin, LIU Kunyuan, WANG Huizhong, FENG Shangguo. Research progress in the production of α-arbutin through biosynthesis [J]. Synthetic Biology Journal, 2025, 6(1): 118-135. |
| [2] | ZHU Fanghuan, CEN Xuecong, CHEN Zhen. Research progress of diols production by microbes [J]. Synthetic Biology Journal, 2024, 5(6): 1367-1385. |
| [3] | LIU Yining, PU Wei, YANG Jinxing, WANG Yu. Recent advances in the biosynthesis of ω-amino acids and lactams [J]. Synthetic Biology Journal, 2024, 5(6): 1350-1366. |
| [4] | ZHENG Haotian, LI Chaofeng, LIU Liangxu, WANG Jiawei, LI Hengrun, NI Jun. Design, optimization and application of synthetic carbon-negative phototrophic community [J]. Synthetic Biology Journal, 2024, 5(5): 1189-1210. |
| [5] | CHENG Xiaolei, LIU Tiangang, TAO Hui. Recent research progress in non-canonical biosynthesis of terpenoids [J]. Synthetic Biology Journal, 2024, 5(5): 1050-1071. |
| [6] | XIA Kongchen, XU Weihua, WU Qi. Recent advances in photo-induced promiscuous enzymatic reactions [J]. Synthetic Biology Journal, 2024, 5(5): 997-1020. |
| [7] | LIU Zijian, MU Baiyang, DUAN Zhiqiang, WANG Xuan, LU Xiaojie. Advances in the development of DNA-compatible chemistries [J]. Synthetic Biology Journal, 2024, 5(5): 1102-1124. |
| [8] | ZHANG Shouqi, WANG Tao, KONG Yao, ZOU Jiasheng, LIU Yuanning, XU Zhengren. Chemoenzymatic synthesis of natural products: evolution of synthetic methodology and strategy [J]. Synthetic Biology Journal, 2024, 5(5): 913-940. |
| [9] | XIE Xiangqian, GUO Wen, WANG Huan, LI Jin. Biosynthesis and chemical synthesis of ribosomally synthesized and post-translationally modified peptides containing aminovinyl cysteine [J]. Synthetic Biology Journal, 2024, 5(5): 981-996. |
| [10] | TANG Zhijun, HU Youcai, LIU Wen. Enzymatic (4+2)- and (2+2)-cycloaddition reactions: fundamentals and applications of regio- and stereoselectivity [J]. Synthetic Biology Journal, 2024, 5(3): 401-407. |
| [11] | ZHANG Jun, JIN Shixue, YUN Qian, QU Xudong. Biosynthesis of the unnatural extender units with polyketides and their structural modifications for applications in medicines [J]. Synthetic Biology Journal, 2024, 5(3): 561-570. |
| [12] | CHEN Xiwei, ZHANG Huaran, ZOU Yi. Biosynthesis and metabolic engineering of fungal non-ribosomal peptides [J]. Synthetic Biology Journal, 2024, 5(3): 571-592. |
| [13] | FENG Jin, PAN Haixue, TANG Gongli. Research advances in biosynthesis of natural product drugs within the past decade [J]. Synthetic Biology Journal, 2024, 5(3): 408-446. |
| [14] | XI Mengyu, HU Yiling, GU Yucheng, GE Huiming. Genome mining-directed discovery for natural medicinal products [J]. Synthetic Biology Journal, 2024, 5(3): 447-473. |
| [15] | SHI Xinjie, DU Yiling. Research advances in the biosynthesis of nonribosomal peptides within the bisintercalator family as anticancer drugs [J]. Synthetic Biology Journal, 2024, 5(3): 593-611. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||

