Synthetic Biology Journal ›› 2023, Vol. 4 ›› Issue (2): 373-393.DOI: 10.12211/2096-8280.2022-063
• Invited Review • Previous Articles Next Articles
Synthetic biology and engineered T cell therapy
XIE Junhong1, HE Jingjing2, ZHOU Penghui3
- 1.National Center for International Research of Bio-targeting Theranostics,Guangxi Key Laboratory of Bio-targeting Theranostics,Guangxi Medical University,Nanning 530021,Guangxi,China
2.Medical Research Department,Guangdong Provincial People’s Hospital,Guangdong Academy of Medical Sciences,Guangzhou 510055,Guangdong,China
3.State Key Laboratory of Oncology in Southern China,Sun Yat-sen University Cancer Center,Guangzhou 510060,Guangdong,China
-
Received:2022-11-17Revised:2023-02-01Online:2023-04-27Published:2023-04-30 -
Contact:HE Jingjing, ZHOU Penghui
合成生物学与工程化T细胞治疗
谢君鸿1, 何晶晶2, 周鹏辉3
- 1.广西医科大学,广西生物靶向诊治重点实验室,国家生物靶向诊治国际联合研究中心,广西 南宁 530021
2.广东医学科学院,广东省人民医院医学研究部,广东 广州 510055
3.中山大学肿瘤防治中心,华南肿瘤学国家重点实验室,广东 广州 510060
-
通讯作者:何晶晶,周鹏辉 -
作者简介:谢君鸿 (1996—),男,硕士研究生,研究方向为肿瘤免疫治疗。E-mail:jhxietw@163.com何晶晶 (1987—),博士,特聘副研究员。主要研究方向为肿瘤免疫学和肿瘤免疫治疗。E-mail:hejingjing@gdph.org.cn周鹏辉 (1974—),男,博士,教授,博士生导师。研究方向为肿瘤免疫学和肿瘤免疫治疗。E-mail:zhouph@sysucc.org.cn -
基金资助:国家自然科学基金(82130086);广东省自然科学基金(2017A030308007);广东省特支计划项目(2016LJ06S464);广东省引进创新创业团队(2016ZT06S638)
CLC Number:
Cite this article
XIE Junhong, HE Jingjing, ZHOU Penghui. Synthetic biology and engineered T cell therapy[J]. Synthetic Biology Journal, 2023, 4(2): 373-393.
谢君鸿, 何晶晶, 周鹏辉. 合成生物学与工程化T细胞治疗[J]. 合成生物学, 2023, 4(2): 373-393.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2022-063
| 133 | SU S, HU B, SHAO J, et al. CRISPR-Cas9 mediated efficient PD-1 disruption on human primary T cells from cancer patients[J]. Scientific Reports, 2016, 6: 20070. |
| 134 | MAROTTE L, SIMON S, VIGNARD V, et al. Increased antitumor efficacy of PD-1-deficient melanoma-specific human lymphocytes[J]. Journal for Immunotherapy of Cancer, 2020, 8(1): e000311. |
| 135 | EYQUEM J, MANSILLA-SOTO J, GIAVRIDIS T, et al. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection[J]. Nature, 2017, 543(7643): 113-117. |
| 136 | ROTH T L, PUIG-SAUS C, YU R, et al. Reprogramming human T cell function and specificity with non-viral genome targeting[J]. Nature, 2018, 559(7714): 405-409. |
| 137 | MATTHEW B, DONG, DONG M B, WANG G C, CHOW R D, et al. Systematic immunotherapy target discovery using genome-scale in vivo CRISPR screens in CD8 T cells[J]. Cell, 2019, 178(5): 1189-1204.e23. |
| 138 | MASTAGLIO S, GENOVESE P, MAGNANI Z, et al. NY-ESO-1 TCR single edited stem and central memory T cells to treat multiple myeloma without graft-versus-host disease[J]. Blood, 2017, 130(5): 606-618. |
| 139 | BERDIEN B, MOCK U, ATANACKOVIC D, et al. TALEN-mediated editing of endogenous T-cell receptors facilitates efficient reprogramming of T lymphocytes by lentiviral gene transfer[J]. Gene Therapy, 2014, 21(6): 539-548. |
| 140 | POIROT L, PHILIP B, SCHIFFER-MANNIOUI C, et al. Multiplex genome-edited T-cell manufacturing platform for "off-the-shelf" adoptive T-cell immunotherapies[J]. Cancer Research, 2015, 75(18): 3853-3864. |
| 141 | OSBORN M J, WEBBER B R, KNIPPING F, et al. Evaluation of TCR gene editing achieved by TALENs, CRISPR/Cas9, and megaTAL nucleases[J]. Molecular Therapy, 2016, 24(3): 570-581. |
| 142 | LEGUT M, DOLTON G, MAIN A A, et al. CRISPR-mediated TCR replacement generates superior anticancer transgenic T cells[J]. Blood, 2018, 131(3): 311-322. |
| 143 | REN J T, LIU X J, FANG C Y, et al. Multiplex genome editing to generate universal CAR T cells resistant to PD1 inhibition[J]. Clinical Cancer Research, 2017, 23(9): 2255-2266. |
| 144 | SHI L, MENG T Y, ZHANG Z L, et al. CRISPR knock out CTLA-4 enhances the anti-tumor activity of cytotoxic T lymphocytes[J]. Gene, 2017, 636: 36-41. |
| 145 | STADTMAUER E A, FRAIETTA J A, DAVIS M M, et al. CRISPR-engineered T cells in patients with refractory cancer[J]. Science, 2020, 367(6481): eaba7365. |
| 146 | WANG D, ZHANG F, GAO, G P. CRISPR-based therapeutic genome editing: strategies and in vivo delivery by AAV vectors[J]. Cell, 2020, 181(1): 136-150. |
| 147 | SATHER B D, ROMANO IBARRA G S, SOMMER K, et al. Efficient modification of CCR5 in primary human hematopoietic cells using a megaTAL nuclease and AAV donor template[J]. Science Translational Medicine, 2015, 7(307): 307ra156. |
| 148 | HALE M, LEE B, HONAKER Y, et al. Homology-directed recombination for enhanced engineering of chimeric antigen receptor T cells[J]. Molecular Therapy Methods & Clinical Development, 2017, 4: 192-203. |
| 149 | SACHDEVA M, BUSSER B W, TEMBURNI S, et al. Repurposing endogenous immune pathways to tailor and control chimeric antigen receptor T cell functionality[J]. Nature Communications, 2019, 10: 5100. |
| 150 | YANG L F, YIN J L, WU J L, et al. Engineering genetic devices for in vivo control of therapeutic T cell activity triggered by the dietary molecule resveratrol[J]. Proceedings of the National Academy of Sciences of the United States of America, 2021, 118(34): e2106612118. |
| 151 | KOBAYASHI A, NOBILI A, NEIER S C, et al. Light-controllable binary switch activation of CAR T cells[J]. ChemMedChem, 2022, 17(12): e202100722. |
| 152 | NOBLES C L, SHERRILL-MIX S, EVERETT J K, et al. CD19-targeting CAR T cell immunotherapy outcomes correlate with genomic modification by vector integration[J]. The Journal of Clinical Investigation, 2020, 130(2): 673-685. |
| 153 | FRAIETTA J A, NOBLES C L, SAMMONS M A, et al. Disruption of TET2 promotes the therapeutic efficacy of CD19-targeted T cells[J]. Nature, 2018, 558(7709): 307-312. |
| 154 | SHIFRUT E, CARNEVALE J, TOBIN V, et al. Genome-wide CRISPR screens in primary human T cells reveal key regulators of immune function[J]. Cell, 2018, 175(7): 1958-1971.e15. |
| 155 | GRUPP S A, PRAK E L, BOYER J, et al. Adoptive transfer of autologous T cells improves T-cell repertoire diversity and long-term B-cell function in pediatric patients with neuroblastoma[J]. Clinical Cancer Research, 2012, 18(24): 6732-6741. |
| 156 | KALOS M, LEVINE B L, PORTER D L, et al. T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia[J]. Science Translational Medicine, 2011, 3(95): 95ra73. |
| 1 | ROSENBERG S A, PACKARD B S, AEBERSOLD P M, et al. Use of tumor-infiltrating lymphocytes and interleukin-2 in the immunotherapy of patients with metastatic melanoma. A preliminary report[J]. The New England Journal of Medicine, 1988, 319(25): 1676-1680. |
| 2 | WATANABE K, NISHIKAWA H. Engineering strategies for broad application of TCR-T- and CAR-T-cell therapies[J]. International Immunology, 2021, 33(11): 551-562. |
| 3 | WEBER E W, MAUS M V, MACKALL C L. The emerging landscape of immune cell therapies[J]. Cell, 2020, 181(1): 46-62. |
| 4 | GRUPP S A, KALOS M, BARRETT D, et al. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia[J]. The New England Journal of Medicine, 2013, 368(16): 1509-1518. |
| 5 | KERSTEN M J, SPANJAART A M, THIEBLEMONT C. CD19-directed CAR T-cell therapy in B-cell NHL[J]. Current Opinion in Oncology, 2020, 32(5): 408-417. |
| 6 | MULLARD A. FDA approves fourth CAR-T cell therapy[J]. Nature Reviews Drug Discovery, 2021, 20(3): 166. |
| 7 | ANDERSON, A K, STROMNES I M, GREENBERG P D. Obstacles posed by the tumor microenvironment to T cell activity: a case for synergistic therapies[J]. Cancer Cell, 2017, 31(3): 311-325. |
| 8 | LARSON R C, MAUS M V. Recent advances and discoveries in the mechanisms and functions of CAR T cells[J]. Nature Reviews Cancer, 2021, 21(3): 145-161. |
| 9 | NISHIKAWA H, MAEDA Y, ISHIDA T, et al. Cancer/testis antigens are novel targets of immunotherapy for adult T-cell leukemia/lymphoma[J]. Blood, 2012, 119(13): 3097-3104. |
| 10 | JUNGBLUTH A A, ANTONESCU C R, BUSAM K J, et al. Monophasic and biphasic synovial sarcomas abundantly express cancer/testis antigen NY-ESO-1 but not MAGE-A1 or CT7[J]. International Journal of Cancer, 2001, 94(2): 252-256. |
| 11 | D'ANGELO S P, MELCHIORI L, MERCHANT M S, et al. Antitumor activity associated with prolonged persistence of adoptively transferred NY-ESO-1 c259T cells in synovial sarcoma[J]. Cancer Discovery, 2018, 8(8): 944-957. |
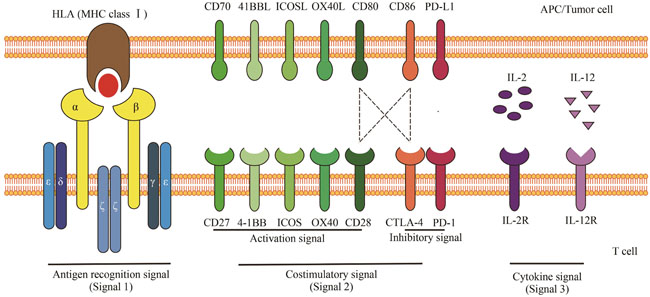
| 12 | CHEN L P, FLIES D B. Molecular mechanisms of T cell co-stimulation and co-inhibition[J]. Nature Reviews Immunology, 2013, 13(4): 227-242. |
| 13 | RAEBER M E, ZURBUCHEN Y, IMPELLIZZIERI D, et al. The role of cytokines in T-cell memory in health and disease[J]. Immunological Reviews, 2018, 283(1): 176-193. |
| 14 | DRIESSENS G, KLINE J, GAJEWSKI T F. Costimulatory and coinhibitory receptors in anti-tumor immunity[J]. Immunological Reviews, 2009, 229(1): 126-144. |
| 15 | GLOBERSON LEVIN A, RIVIÈRE I, ESHHAR Z, et al. CAR T cells: building on the CD19 paradigm[J]. European Journal of Immunology, 2021, 51(9): 2151-2163. |
| 16 | STERNER R C, STERNER R M. CAR-T cell therapy: current limitations and potential strategies[J]. Blood Cancer Journal, 2021, 11: 69. |
| 17 | JAYARAMAN J, MELLODY M P, HOU A J, et al. CAR-T design: elements and their synergistic function[J]. EBioMedicine, 2020, 58: 102931. |
| 18 | CHEEVER M A, ALLISON J P, FERRIS A S, et al. The prioritization of cancer antigens: a national cancer institute pilot project for the acceleration of translational research[J]. Clinical Cancer Research, 2009, 15(17): 5323-5337. |
| 19 | COULIE P G, VAN DEN EYNDE B J, VAN DER BRUGGEN P, et al. Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy[J]. Nature Reviews Cancer, 2014, 14(2): 135-146. |
| 20 | GUBIN M M, ARTYOMOV M N, MARDIS E R, et al. Tumor neoantigens: building a framework for personalized cancer immunotherapy[J]. The Journal of Clinical Investigation, 2015, 125(9): 3413-3421. |
| 21 | SMITH C C, SELITSKY S R, CHAI S J, et al. Alternative tumour-specific antigens[J]. Nature Reviews Cancer, 2019, 19(8): 465-478. |
| 22 | RAPOPORT A P, STADTMAUER E A, BINDER-SCHOLL G K, et al. NY-ESO-1-specific TCR-engineered T cells mediate sustained antigen-specific antitumor effects in myeloma[J]. Nature Medicine, 2015, 21(8): 914-921. |
| 23 | KAGEYAMA S, IKEDA H, MIYAHARA Y, et al. Adoptive transfer of MAGE-A4 T-cell receptor gene-transduced lymphocytes in patients with recurrent esophageal cancer[J]. Clinical Cancer Research, 2015, 21(10): 2268-2277. |
| 24 | MORGAN R A, CHINNASAMY N, ABATE-DAGA D, et al. Cancer regression and neurological toxicity following anti-MAGE-A3 TCR gene therapy[J]. Journal of Immunotherapy, 2013, 36(2): 133-151. |
| 25 | MORGAN R A, DUDLEY M E, WUNDERLICH J R, et al. Cancer regression in patients after transfer of genetically engineered lymphocytes[J]. Science, 2006, 314(5796): 126-129. |
| 26 | HE J J, XIONG X X, YANG H, et al. Defined tumor antigen-specific T cells potentiate personalized TCR-T cell therapy and prediction of immunotherapy response[J]. Cell Research, 2022, 32(6): 530-542. |
| 27 | PARKHURST M R, YANG J C, LANGAN R C, et al. T cells targeting carcinoembryonic antigen can mediate regression of metastatic colorectal cancer but induce severe transient colitis[J]. Molecular Therapy, 2011, 19(3): 620-626. |
| 28 | SHAH N, CHARI A, SCOTT E, et al. B-cell maturation antigen (BCMA) in multiple myeloma: rationale for targeting and current therapeutic approaches[J]. Leukemia, 2020, 34(4): 985-1005. |
| 29 | HAN Z W, LYV Z W, CUI B, et al. The old CEACAMs find their new role in tumor immunotherapy[J]. Investigational New Drugs, 2020, 38(6): 1888-1898. |
| 30 | YAMASHITA N, KUFE D. Addiction of cancer stem cells to MUC1-C in triple-negative breast cancer progression[J]. International Journal of Molecular Sciences, 2022, 23(15): 8219. |
| 31 | KARSCHNIA P, TESKE N, THON N, et al. Chimeric antigen receptor T cells for glioblastoma: current concepts, challenges, and future perspectives[J]. Neurology, 2021, 97(5): 218-230. |
| 32 | CAO W J, XING H Z, LI Y M, et al. Claudin18.2 is a novel molecular biomarker for tumor-targeted immunotherapy[J]. Biomarker Research, 2022, 10(1): 38. |
| 33 | HONG M H, CLUBB J D, CHEN Y Y. Engineering CAR-T cells for next-generation cancer therapy[J]. Cancer Cell, 2020, 38(4): 473-488. |
| 34 | STONE J D, CHERVIN A S, KRANZ D M. T-cell receptor binding affinities and kinetics: impact on T-cell activity and specificity[J]. Immunology, 2009, 126(2): 165-176. |
| 35 | ALEKSIC M, LIDDY N, MOLLOY P E, et al. Different affinity windows for virus and cancer-specific T-cell receptors: implications for therapeutic strategies[J]. European Journal of Immunology, 2012, 42(12): 3174-3179. |
| 36 | SCHUMACHER T N, SCHREIBER R D. Neoantigens in cancer immunotherapy[J]. Science, 2015, 348(6230): 69-74. |
| 37 | STONE J D, KRANZ D M. Role of T cell receptor affinity in the efficacy and specificity of adoptive T cell therapies[J]. Frontiers in Immunology, 2013, 4: 244. |
| 38 | WILLIAMS C M, SCHONNESEN A A, ZHANG S Q, et al. Normalized synergy predicts that CD8 co-receptor contribution to T cell receptor (TCR) and pMHC binding decreases as TCR affinity increases in human viral-specific T cells[J]. Frontiers in Immunology, 2017, 8: 894. |
| 39 | SHARMA P, HARRIS D T, STONE J D, et al. T-cell receptors engineered de novo for peptide specificity can mediate optimal T-cell activity without self cross-reactivity[J]. Cancer Immunology Research, 2019, 7(12): 2025-2035. |
| 40 | TAN M P, GERRY A B, BREWER J E, et al. T cell receptor binding affinity governs the functional profile of cancer-specific CD8+ T cells[J]. Clinical and Experimental Immunology, 2015, 180(2): 255-270. |
| 41 | SCHMID D A, IRVING M B, POSEVITZ V, et al. Evidence for a TCR affinity threshold delimiting maximal CD8 T cell function[J]. Journal of Immunology, 2010, 184(9): 4936-4946. |
| 42 | HOLLER P D, HOLMAN P O, SHUSTA E V, et al. In vitro evolution of a T cell receptor with high affinity for peptide/MHC[J]. Proceedings of the National Academy of Sciences of the United States of America, 2000, 97(10): 5387-5392. |
| 43 | LI Y, MOYSEY R, MOLLOY P E, et al. Directed evolution of human T-cell receptors with picomolar affinities by phage display[J]. Nature Biotechnology, 2005, 23(3): 349-354. |
| 44 | DUNN S M, RIZKALLAH P J, BASTON E, et al. Directed evolution of human T cell receptor CDR2 residues by phage display dramatically enhances affinity for cognate peptide-MHC without increasing apparent cross-reactivity[J]. Protein Science, 2006, 15(4): 710-721. |
| 45 | ZOETE V, IRVING M, FERBER M, et al. Structure-based, rational design of T cell receptors[J]. Frontiers in Immunology, 2013, 4: 268. |
| 46 | PIERCE B G, HELLMAN L M, HOSSAIN M, et al. Computational design of the affinity and specificity of a therapeutic T cell receptor[J]. PLoS Computational Biology, 2014, 10(2): e1003478. |
| 47 | HARRIS D T, WANG N Y, RILEY T P, et al. Deep mutational scans as a guide to engineering high affinity T cell receptor interactions with peptide-bound major histocompatibility complex[J]. The Journal of Biological Chemistry, 2016, 291(47): 24566-24578. |
| 48 | LINETTE G P, STADTMAUER E A, MAUS M V, et al. Cardiovascular toxicity and titin cross-reactivity of affinity-enhanced T cells in myeloma and melanoma[J]. Blood, 2013, 122(6): 863-871. |
| 49 | SANDERSON J P, CROWLEY D J, WIEDERMANN G E, et al. Preclinical evaluation of an affinity-enhanced MAGE-A4-specific T-cell receptor for adoptive T-cell therapy[J]. OncoImmunology, 2020, 9(1): 1682381. |
| 50 | THOMAS S, MOHAMMED F, REIJMERS R M, et al. Framework engineering to produce dominant T cell receptors with enhanced antigen-specific function[J]. Nature Communications, 2019, 10: 4451. |
| 51 | WATSON H A, DURAIRAJ R R P, OHME J, et al. L-selectin enhanced T cells improve the efficacy of cancer immunotherapy[J]. Frontiers in Immunology, 2019, 10: 1321. |
| 52 | AHMADI M, KING J W, XUE S A, et al. CD3 limits the efficacy of TCR gene therapy in vivo [J]. Blood, 2011, 118(13): 3528-3537. |
| 53 | WATANABE K, TERAKURA S, MARTENS A C, et al. Target antigen density governs the efficacy of anti-CD20-CD28-CD3 ζ chimeric antigen receptor-modified effector CD8+ T cells[J]. Journal of Immunology, 2015, 194(3): 911-920. |
| 54 | IRVINE D J, PURBHOO M A, KROGSGAARD M, et al. Direct observation of ligand recognition by T cells[J]. Nature, 2002, 419(6909): 845-849. |
| 55 | HUANG J, BRAMESHUBER M, ZENG X, et al. A single peptide-major histocompatibility complex ligand triggers digital cytokine secretion in CD4+ T cells[J]. Immunity, 2013, 39(5): 846-857. |
| 56 | LIU X J, JIANG S G, FANG C Y, et al. Affinity-tuned ErbB2 or EGFR chimeric antigen receptor T cells exhibit an increased therapeutic index against tumors in mice[J]. Cancer Research, 2015, 75(17): 3596-3607. |
| 57 | CASTELLARIN M, SANDS C, DA T, et al. A rational mouse model to detect on-target, off-tumor CAR T cell toxicity[J]. JCI Insight, 2020, 5(14): e136012. |
| 58 | CARUSO H G, HURTON L V, NAJJAR A, et al. Tuning sensitivity of CAR to EGFR density limits recognition of normal tissue while maintaining potent antitumor activity[J]. Cancer Research, 2015, 75(17): 3505-3518. |
| 59 | AHMED N, SALSMAN V S, KEW Y, et al. HER2-specific T cells target primary glioblastoma stem cells and induce regression of autologous experimental tumors[J]. Clinical Cancer Research, 2010, 16(2): 474-485. |
| 60 | SZÖŐR, Á, TÓTH G, ZSEBIK B, et al. Trastuzumab derived HER2-specific CARs for the treatment of trastuzumab-resistant breast cancer: CAR T cells penetrate and eradicate tumors that are not accessible to antibodies[J]. Cancer Letters, 2020, 484: 1-8. |
| 61 | AHMED N, SALSMAN V S, YVON E, et al. Immunotherapy for osteosarcoma: genetic modification of T cells overcomes low levels of tumor antigen expression[J]. Molecular, 2009, 17(10): 1779-1787 |
| 62 | HUSZNO J, LEŚ D, SARZYCZNY-SŁOTA D, et al. Cardiac side effects of trastuzumab in breast cancer patients-single centere experiences[J]. Contemporary Oncology, 2013, 17(2): 190-195. |
| 63 | SUJJITJOON J, SAYOUR E, TSAO S T, et al. GD2-specific chimeric antigen receptor-modified T cells targeting retinoblastoma-assessing tumor and T cell interaction[J]. Translational Oncology, 2021, 14(2): 100971. |
| 64 | GOLINELLI G, GRISENDI G, PRAPA M, et al. Targeting GD2-positive glioblastoma by chimeric antigen receptor empowered mesenchymal progenitors[J]. Cancer Gene Therapy, 2020, 27(7): 558-570. |
| 65 | YU A L, GILMAN A L, OZKAYNAK F M, et al. Anti-GD2 antibody with GM-CSF, interleukin-2, and isotretinoin for neuroblastoma[J]. The New England Journal of Medicine, 2010, 363(14): 1324-1334. |
| 66 | ARI P, KARS M, MEANY H, et al. Treatment of transient peripheral neuropathy during chimeric 14.18 antibody therapy in children with neuroblastoma: a case series[J]. Journal of Pediatric Hematology/Oncology, 2018, 40(2): e113-e116. |
| 67 | NAZHA B, INAL C, OWONIKOKO T K. Disialoganglioside GD2 expression in solid tumors and role as a target for cancer therapy[J]. Frontiers in Oncology, 2020, 10: 1000. |
| 68 | RICHMAN S A, NUNEZ-CRUZ S, MOGHIMI B, et al. High-affinity GD2-specific CAR T cells induce fatal encephalitis in a preclinical neuroblastoma model[J]. Cancer Immunology Research, 2018, 6(1): 36-46. |
| 69 | KE E E, WU Y L. EGFR as a pharmacological target in EGFR-mutant non-small-cell lung cancer: where do we stand now?[J]. Trends in Pharmacological Sciences, 2016, 37(11): 887-903. |
| 70 | LI H, HUANG Y, JIANG D Q, et al. Antitumor activity of EGFR-specific CAR T cells against non-small-cell lung cancer cells in vitro and in mice[J]. Cell Death & Disease, 2018, 9: 177. |
| 71 | GUO Y L, FENG K C, LIU Y, et al. Phase I study of chimeric antigen receptor-modified T cells in patients with EGFR-positive advanced biliary tract cancers[J]. Clinical Cancer Research, 2018, 24(6): 1277-1286. |
| 72 | LIU Y, GUO Y L, WU Z Q, et al. Anti-EGFR chimeric antigen receptor-modified T cells in metastatic pancreatic carcinoma: a phase I clinical trial[J]. Cytotherapy, 2020, 22(10): 573-580. |
| 73 | PEREZ R, MORENO E, GARRIDO G, et al. EGFR-targeting as a biological therapy: understanding nimotuzumab's clinical effects[J]. Cancers, 2011, 3(2): 2014-2031. |
| 74 | HERNANDEZ-LOPEZ R A, YU W, CABRAL K A, et al. T cell circuits that sense antigen density with an ultrasensitive threshold[J]. Science, 2021, 371(6534): 1166-1171. |
| 75 | HYRENIUS-WITTSTEN A, SU Y, PARK M, et al. SynNotch CAR circuits enhance solid tumor recognition and promote persistent antitumor activity in mouse models[J]. Science Translational Medicine, 2021, 13(591): eabd8836. |
| 76 | BONINI C, FERRARI G, VERZELETTI S, et al. HSV-TK gene transfer into donor lymphocytes for control of allogeneic graft-versus-leukemia[J]. Science, 1997, 276(5319): 1719-1724. |
| 77 | TRAVERSARI C, MARKTEL S, MAGNANI Z, et al. The potential immunogenicity of the TK suicide gene does not prevent full clinical benefit associated with the use of TK-transduced donor lymphocytes in HSCT for hematologic malignancies[J]. Blood, 2007, 109(11): 4708-4715. |
| 78 | GRECO R, OLIVEIRA G, STANGHELLINI M T L, et al. Improving the safety of cell therapy with the TK-suicide gene[J]. Frontiers in Pharmacology, 2015, 6: 95. |
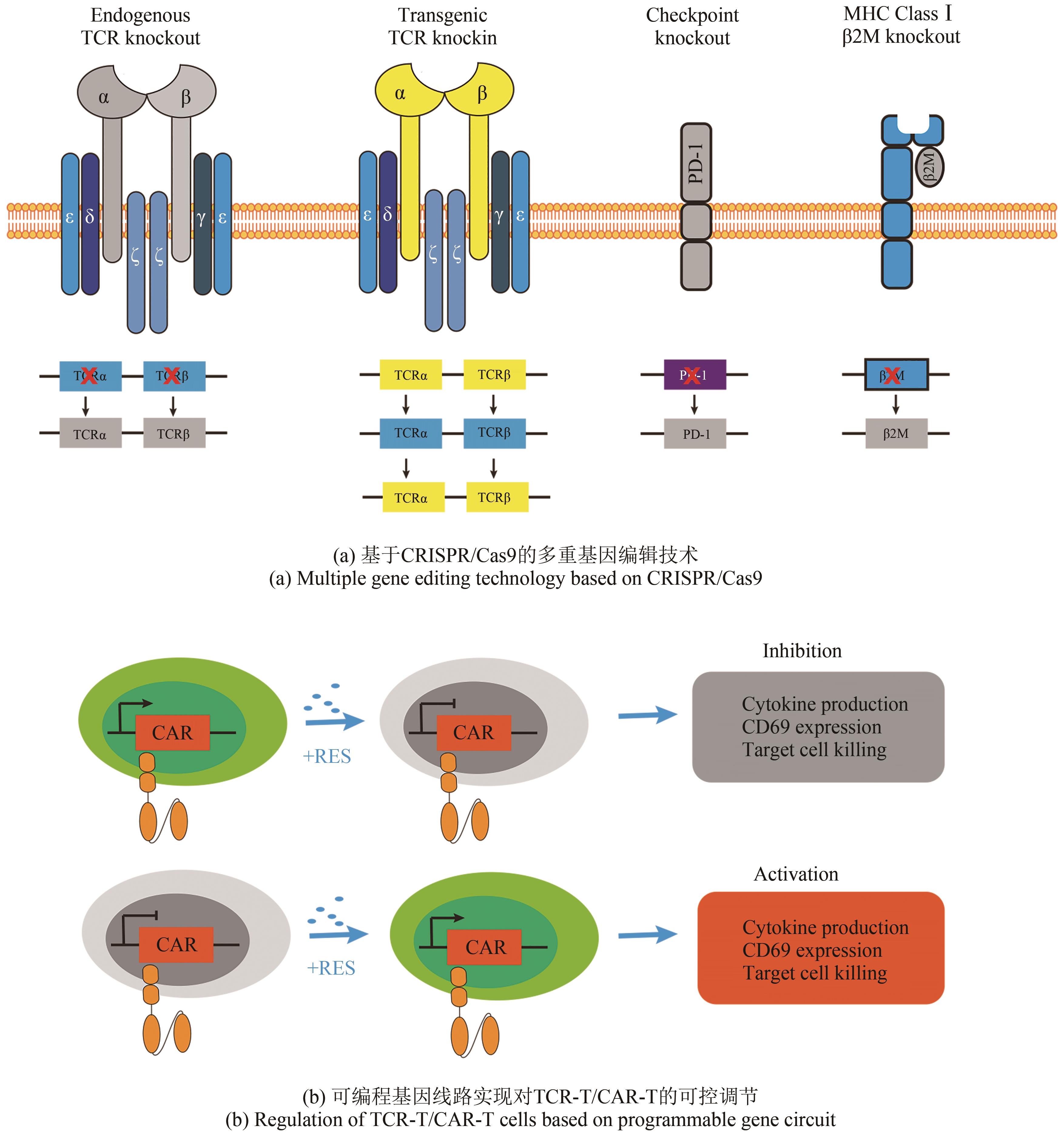
| 79 | STRAATHOF K C, PULÈ M A, YOTNDA P, et al. An inducible caspase 9 safety switch for T-cell therapy[J]. Blood, 2005, 105(11): 4247-4254. |
| 80 | DI STASI A, TEY S K, DOTTI G, et al. Inducible apoptosis as a safety switch for adoptive cell therapy[J]. The New England Journal of Medicine, 2011, 365(18): 1673-1683. |
| 81 | ZHOU X O, DOTTI G, KRANCE R A, et al. Inducible caspase-9 suicide gene controls adverse effects from alloreplete T cells after haploidentical stem cell transplantation[J]. Blood, 2015, 125(26): 4103-4113. |
| 82 | VOGLER I, NEWRZELA S, HARTMANN S, et al. An improved bicistronic CD20/tCD34 vector for efficient purification and in vivo depletion of gene-modified T cells for adoptive immunotherapy[J]. Molecular Therapy, 2010, 18(7): 1330-1338. |
| 83 | PHILIP B, KOKALAKI E, MEKKAOUI L, et al. A highly compact epitope-based marker/suicide gene for easier and safer T-cell therapy[J]. Blood, 2014, 124(8): 1277-1287. |
| 84 | BINNEWIES M, ROBERTS E W, KERSTEN K, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy[J]. Nature Medicine, 2018, 24(5): 541-550. |
| 85 | D'ALOIA M M, ZIZZARI I G, SACCHETTI B, et al. CAR-T cells: the long and winding road to solid tumors[J]. Cell Death & Disease, 2018, 9: 282. |
| 86 | LANITIS E, DANGAJ D, IRVING M, et al. Mechanisms regulating T-cell infiltration and activity in solid tumors[J]. Annals of Oncology, 2017, 28(): xii18-xii32. |
| 87 | NAGARSHETH N, WICHA M S, ZOU W P. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy[J]. Nature Reviews Immunology, 2017, 17(9): 559-572. |
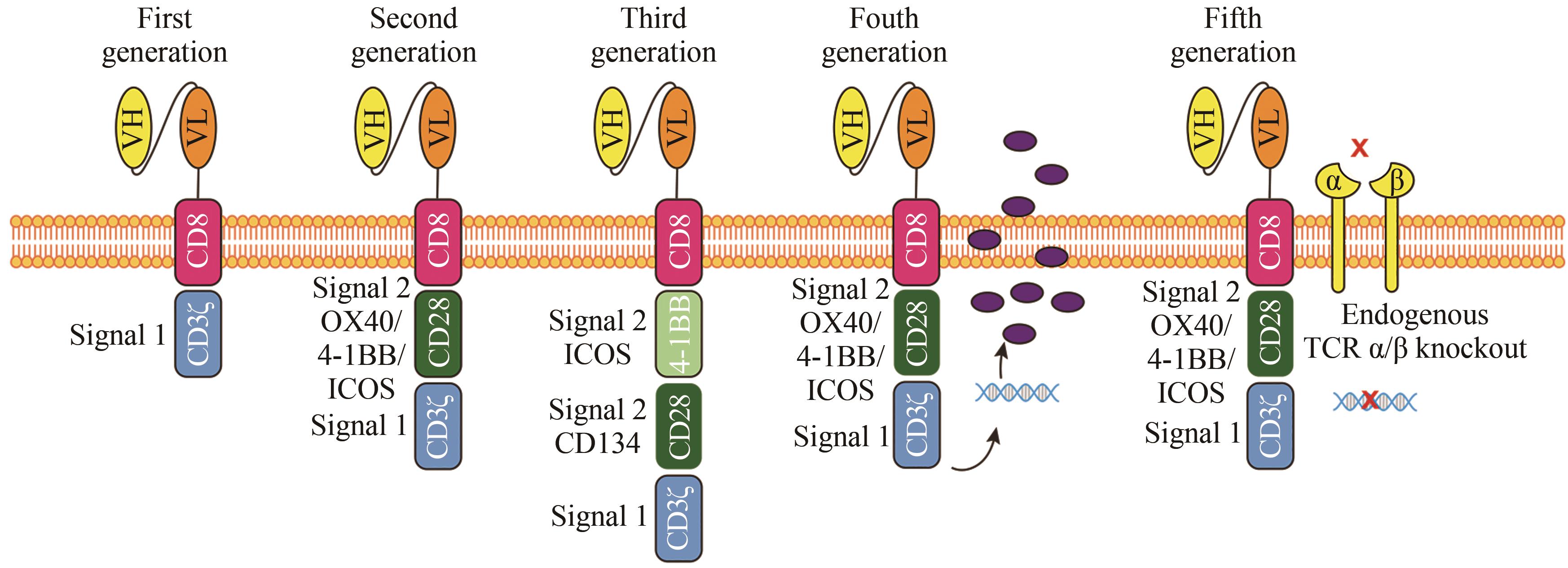
| 88 | MAHER J, BRENTJENS R J, GUNSET G, et al. Human T-lymphocyte cytotoxicity and proliferation directed by a single chimeric TCRζ/CD28 receptor[J]. Nature Biotechnology, 2002, 20(1): 70-75. |
| 89 | KAWALEKAR O U, O'CONNER R S, FRAIETTA J A, et al. Distinct signaling of coreceptors regulates specific metabolism pathways and impacts memory development in CAR T cells[J]. Immunity, 2016, 44(2): 380-390. |
| 90 | WEINKOVE R, GEORGE P, DASYAM N, et al. Selecting costimulatory domains for chimeric antigen receptors: functional and clinical considerations[J]. Clinical & Translational Immunology, 2019, 8(5): e1049. |
| 91 | GROSSER R, CHERKASSKY L, CHINTALAN, et al. Combination immunotherapy with CAR T cells and checkpoint blockade for the treatment of solid tumors[J]. Cancer Cell, 2019, 36(5): 471-482. |
| 92 | HARLIN H, MENG Y R, PETERSON A C, et al. Chemokine expression in melanoma metastases associated with CD8+ T-cell recruitment[J]. Cancer Research, 2009, 69(7): 3077-3085. |
| 93 | IDORN M, SKADBORG S K, KELLERMANN L, et al. Chemokine receptor engineering of T cells with CXCR2 improves homing towards subcutaneous human melanomas in xenograft mouse model[J]. OncoImmunology, 2018, 7(8): e1450715. |
| 94 | DI STASI A, DE ANGELIS B, ROONEY C M, et al. T lymphocytes coexpressing CCR4 and a chimeric antigen receptor targeting CD30 have improved homing and antitumor activity in a Hodgkin tumor model[J]. Blood, 2009, 113(25): 6392-6402. |
| 95 | CRADDOCK J A, LU A, BEAR A, et al. Enhanced tumor trafficking of GD2 chimeric antigen receptor T cells by expression of the chemokine receptor CCR2b[J]. Journal of Immunotherapy, 2010, 33(8): 780-788. |
| 96 | MOON E K, CARPENITO C, SUN J, et al. Expression of a functional CCR2 receptor enhances tumor localization and tumor eradication by retargeted human T cells expressing a mesothelin-specific chimeric antibody receptor[J]. Clinical Cancer Research, 2011, 17(14): 4719-4730. |
| 97 | CARUANA I, SAVOLDO B, HOYOS V, et al. Heparanase promotes tumor infiltration and antitumor activity of CAR-redirected T lymphocytes[J]. Nature Medicine, 2015, 21(5): 524-529. |
| 98 | SCHUBERTH P C, HAGEDORN C, JENSEN S M, et al. Treatment of malignant pleural mesothelioma by fibroblast activation protein-specific re-directed T cells[J]. Journal of Translational Medicine, 2013, 11: 187. |
| 99 | WANG L C S, LO A, SCHOLLER J, et al. Targeting fibroblast activation protein in tumor stroma with chimeric antigen receptor T cells can inhibit tumor growth and augment host immunity without severe toxicity[J]. Cancer Immunology Research, 2014, 2(2): 154-166. |
| 100 | GOVERS C, SEBESTYÉN Z, ROSZIK J, et al. TCRs genetically linked to CD28 and CD3ε do not mispair with endogenous TCR chains and mediate enhanced T cell persistence and anti-melanoma activity[J]. Journal of Immunology, 2014, 193(10): 5315-5326. |
| 101 | STONE J D, HARRIS D T, SOTO C M, et al. A novel T cell receptor single-chain signaling complex mediates antigen-specific T cell activity and tumor control[J]. Cancer Immunology, Immunotherapy, 2014, 63(11): 1163-1176. |
| 102 | KLOSS C C, CONDOMINES M, CARTELLIERI M, et al. Combinatorial antigen recognition with balanced signaling promotes selective tumor eradication by engineered T cells[J]. Nature Biotechnology, 2013, 31(1): 71-75. |
| 103 | LANITIS E, POUSSIN M, KLATTENHOFF A W, et al. Chimeric antigen receptor T Cells with dissociated signaling domains exhibit focused antitumor activity with reduced potential for toxicity in vivo [J]. Cancer Immunology Research, 2013, 1(1): 43-53. |
| 104 | OMER B, CARDENAS M G, PFEIFFER T, et al. A costimulatory CAR improves TCR-based cancer immunotherapy[J]. Cancer Immunology Research, 2022, 10(4): 512-524. |
| 105 | DANIEL-MESHULAM I, HOROVITZ-FRIED M, COHEN C J. Enhanced antitumor activity mediated by human 4-1BB-engineered T cells[J]. International Journal of Cancer, 2013, 133(12): 2903-2913. |
| 106 | NISHIMURA C D, BRENNER D A, MUKHERJEEM, et al. c-MPL provides tumor-targeted T-cell receptor-transgenic T cells with costimulation and cytokine signals[J]. Blood, 2017, 130(25): 2739-2749. |
| 107 | MÉNDEZ-FERRER S, BONNET D, STEENSMA D P, et al. Bone marrow niches in haematological malignancies[J]. Nature Reviews Cancer, 2020, 20(5): 285-298. |
| 108 | WAGNER H J, BOLLARD C M, VIGOUROUX S, et al. A strategy for treatment of Epstein-Barr virus-positive Hodgkin's disease by targeting interleukin 12 to the tumor environment using tumor antigen-specific T cells[J]. Cancer Gene Therapy, 2004, 11(2): 81-91. |
| 109 | KERKAR S P, MURANSKI P, KAISER A, et al. Tumor-specific CD8+ T cells expressing interleukin-12 eradicate established cancers in lymphodepleted hosts[J]. Cancer Research, 2010, 70(17): 6725-6734. |
| 110 | ZHANG L, MORGAN R A, BEANE J D, et al. Tumor-infiltrating lymphocytes genetically engineered with an inducible gene encoding interleukin-12 for the immunotherapy of metastatic melanoma[J]. Clinical Cancer Research, 2015, 21(10): 2278-2288. |
| 111 | DRAKES D J, RAFIQ S, PURDON T J, et al. Optimization of T-cell receptor-modified T cells for cancer therapy[J]. Cancer Immunology Research, 2020, 8(6): 743-755. |
| 112 | BATRA S A, RATHI P, GUO L J, et al. Glypican-3-specific CAR T cells coexpressing IL15 and IL21 have superior expansion and antitumor activity against hepatocellular carcinoma[J]. Cancer Immunology Research, 2020, 8(3): 309-320. |
| 113 | SHUM T, OMER B, TASHIRO H, et al. Constitutive signaling from an engineered IL7 receptor promotes durable tumor elimination by tumor-redirected T cells[J]. Cancer Discovery, 2017, 7(11): 1238-1247. |
| 114 | BOLLARD C M, RÖSSIG C, CALONGE M J, et al. Adapting a transforming growth factor beta-related tumor protection strategy to enhance antitumor immunity[J]. Blood, 2002, 99(9): 3179-3187. |
| 115 | BENDLE G M, LINNEMANN C, BIES L, et al. Blockade of TGF-β signaling greatly enhances the efficacy of TCR gene therapy of cancer[J]. Journal of Immunology, 2013, 191(6): 3232-3239. |
| 116 | BOLLARD C M, TRIPIC T, CRUZ C R, et al. Tumor-specific T-cells engineered to overcome tumor immune evasion induce clinical responses in patients with relapsed Hodgkin lymphoma[J]. Journal of Clinical Oncology, 2018, 36(11): 1128-1139. |
| 117 | YAMAMOTO T N, LEE P H, VODNALA S K, et al. T cells genetically engineered to overcome death signaling enhance adoptive cancer immunotherapy[J]. The Journal of Clinical Investigation, 2019, 129(4): 1551-1565. |
| 118 | ARBER C, YOUNG M, BARTH P. Reprogramming cellular functions with engineered membrane proteins[J]. Current Opinion in Biotechnology, 2017, 47: 92-101. |
| 119 | SHIN J H, PARK H B, OH Y M, et al. Positive conversion of negative signaling of CTLA4 potentiates antitumor efficacy of adoptive T-cell therapy in murine tumor models[J]. Blood, 2012, 119(24): 5678-5687. |
| 120 | ANKRI C, SHAMALOV K, HOROVITZ-FRIED M, et al. Human T cells engineered to express a programmed death 1/28 costimulatory retargeting molecule display enhanced antitumor activity[J]. Journal of Immunology, 2013, 191(8): 4121-4129. |
| 121 | SCHLENKER R, OLGUÍN-CONTRERAS L F, LEISEGANG M, et al. Chimeric PD-1: 28 receptor upgrades low-avidity T cells and restores effector function of tumor-infiltrating lymphocytes for adoptive cell therapy[J]. Cancer Research, 2017, 77(13): 3577-3590. |
| 122 | HOOGI S, EISENBERG V, MAYER S, et al. A TIGIT-based chimeric co-stimulatory switch receptor improves T-cell anti-tumor function[J]. Journal for Immunotherapy of Cancer, 2019, 7(1): 243. |
| 123 | ODA S K, DAMAN A W, GARCIA N M, et al. A CD200R-CD28 fusion protein appropriates an inhibitory signal to enhance T-cell function and therapy of murine leukemia[J]. Blood, 2017, 130(22): 2410-2419. |
| 124 | THEODORE L, ROTH, ROTH T L, LI P J, BLAESCHKE F, et al. Pooled knockin targeting for genome engineering of cellular immunotherapies[J]. Cell, 2020, 181(3): 728-744.e21. |
| 125 | WILKIE S, BURBRIDGE S E, CHIAPERO-STANKE L, et al. Selective expansion of chimeric antigen receptor-targeted T-cells with potent effector function using interleukin-4[J]. The Journal of Biological Chemistry, 2010, 285(33): 25538-25544. |
| 126 | LEEN A M, SUKUMARAN S, WATANABE N, et al. Reversal of tumor immune inhibition using a chimeric cytokine receptor[J]. Molecular Therapy, 2014, 22(6): 1211-1220. |
| 127 | SUKUMARAN S, WATANABE N, BAJGAIN P, et al. Enhancing the potency and specificity of engineered T cells for cancer treatment[J]. Cancer Discovery, 2018, 8(8): 972-987. |
| 128 | SINGH N, SHI J W, JUNE C H, et al. Genome-editing technologies in adoptive T cell immunotherapy for cancer[J]. Current Hematologic Malignancy Reports, 2017, 12(6): 522-529. |
| 129 | PICKAR-OLIVER A, GERSBACH C A. The next generation of CRISPR-Cas technologies and applications[J]. Nature Reviews Molecular Cell Biology, 2019, 20(8): 490-507. |
| 130 | QASIM W, ZHAN H, SAMARASINGHE S, et al. Molecular remission of infant B-ALL after infusion of universal TALEN gene-edited CAR T cells[J]. Science Translational Medicine, 2017, 9(374): eaaj2013. |
| 131 | PROVASI E, GENOVESE P, LOMBARDO A, et al. Editing T cell specificity towards leukemia by zinc finger nucleases and lentiviral gene transfer[J]. Nature Medicine, 2012, 18(5): 807-815. |
| 132 | RASAIYAAH J, GEORGIADIS C, PREECE R, et al. TCRαβ/CD3 disruption enables CD3-specific antileukemic T cell immunotherapy[J]. JCI Insight, 2018, 3(13): e99442. |
| [1] | GAO Ge, BIAN Qi, WANG Baojun. Synthetic genetic circuit engineering: principles, advances and prospects [J]. Synthetic Biology Journal, 2025, 6(1): 45-64. |
| [2] | LI Jiyuan, WU Guosheng. Two hypothesises for the origins of organisms from the synthetic biology perspective [J]. Synthetic Biology Journal, 2025, 6(1): 190-202. |
| [3] | JIAO Hongtao, QI Meng, SHAO Bin, JIANG Jinsong. Legal issues for the storage of DNA data [J]. Synthetic Biology Journal, 2025, 6(1): 177-189. |
| [4] | TANG Xinghua, LU Qianneng, HU Yilin. Philosophical reflections on synthetic biology in the Anthropocene [J]. Synthetic Biology Journal, 2025, 6(1): 203-212. |
| [5] | XU Huaisheng, SHI Xiaolong, LIU Xiaoguang, XU Miaomiao. Key technologies for DNA storage: encoding, error correction, random access, and security [J]. Synthetic Biology Journal, 2025, 6(1): 157-176. |
| [6] | SHI Ting, SONG Zhan, SONG Shiyi, ZHANG Yi-Heng P. Job. In vitro BioTransformation (ivBT): a new frontier of industrial biomanufacturing [J]. Synthetic Biology Journal, 2024, 5(6): 1437-1460. |
| [7] | CHAI Meng, WANG Fengqing, WEI Dongzhi. Synthesis of organic acids from lignocellulose by biotransformation [J]. Synthetic Biology Journal, 2024, 5(6): 1242-1263. |
| [8] | SHAO Mingwei, SUN Simian, YANG Shimao, CHEN Guoqiang. Bioproduction based on extremophiles [J]. Synthetic Biology Journal, 2024, 5(6): 1419-1436. |
| [9] | CHEN Yu, ZHANG Kang, QIU Yijing, CHENG Caiyun, YIN Jingjing, SONG Tianshun, XIE Jingjing. Progress of microbial electrosynthesis for conversion of CO2 [J]. Synthetic Biology Journal, 2024, 5(5): 1142-1168. |
| [10] | ZHENG Haotian, LI Chaofeng, LIU Liangxu, WANG Jiawei, LI Hengrun, NI Jun. Design, optimization and application of synthetic carbon-negative phototrophic community [J]. Synthetic Biology Journal, 2024, 5(5): 1189-1210. |
| [11] | CHEN Ziling, XIANG Yangfei. Integrated development of organoid technology and synthetic biology [J]. Synthetic Biology Journal, 2024, 5(4): 795-812. |
| [12] | CAI Bingyu, TAN Xiangtian, LI Wei. Advances in synthetic biology for engineering stem cell [J]. Synthetic Biology Journal, 2024, 5(4): 782-794. |
| [13] | XIE Huang, ZHENG Yilei, SU Yiting, RUAN Jingyi, LI Yongquan. An overview on reconstructing the biosynthetic system of actinomycetes for polyketides production [J]. Synthetic Biology Journal, 2024, 5(3): 612-630. |
| [14] | ZHA Wenlong, BU Lan, ZI Jiachen. Advances in synthetic biology for producing potent pharmaceutical ingredients of traditional Chinese medicine [J]. Synthetic Biology Journal, 2024, 5(3): 631-657. |
| [15] | HUI Zhen, TANG Xiaoyu. Applications of the CRISPR/Cas9 editing system in the study of microbial natural products [J]. Synthetic Biology Journal, 2024, 5(3): 658-671. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||