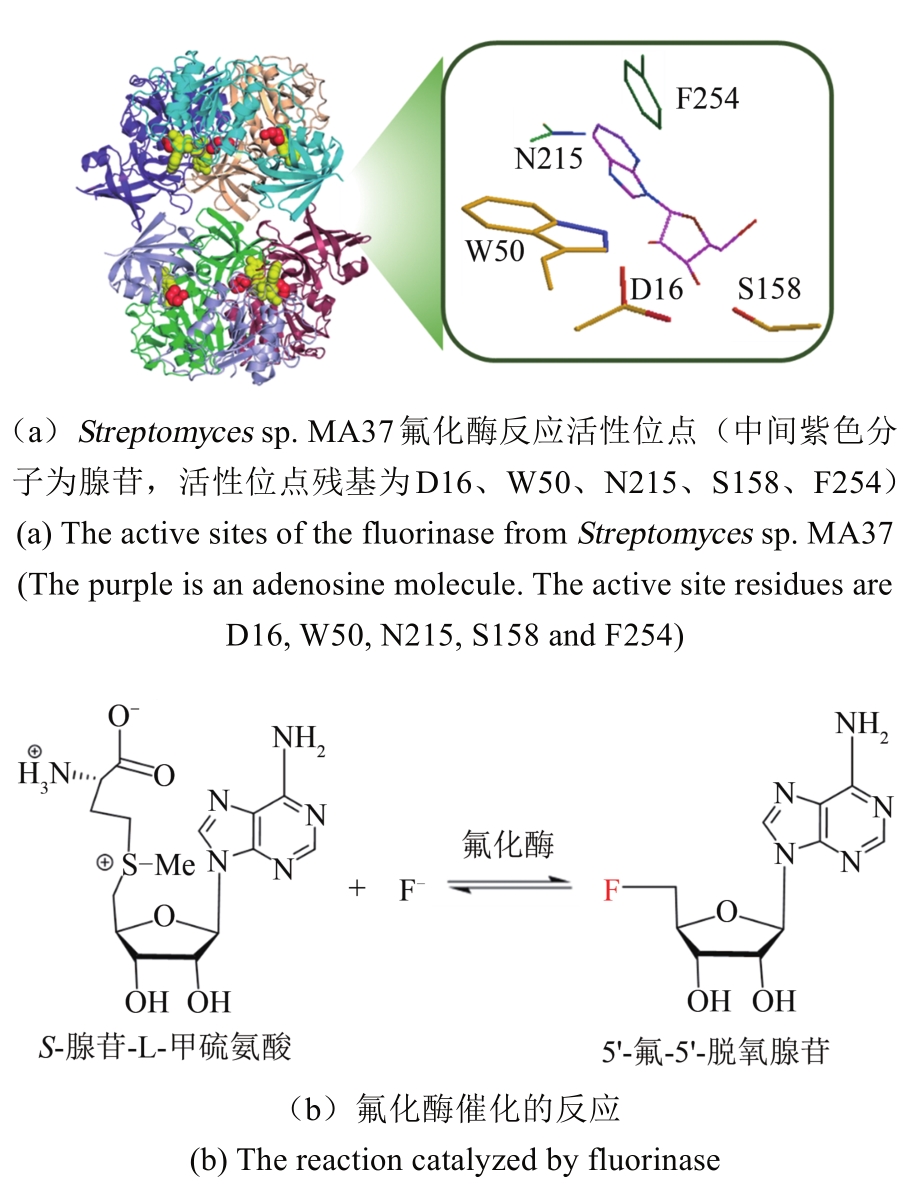
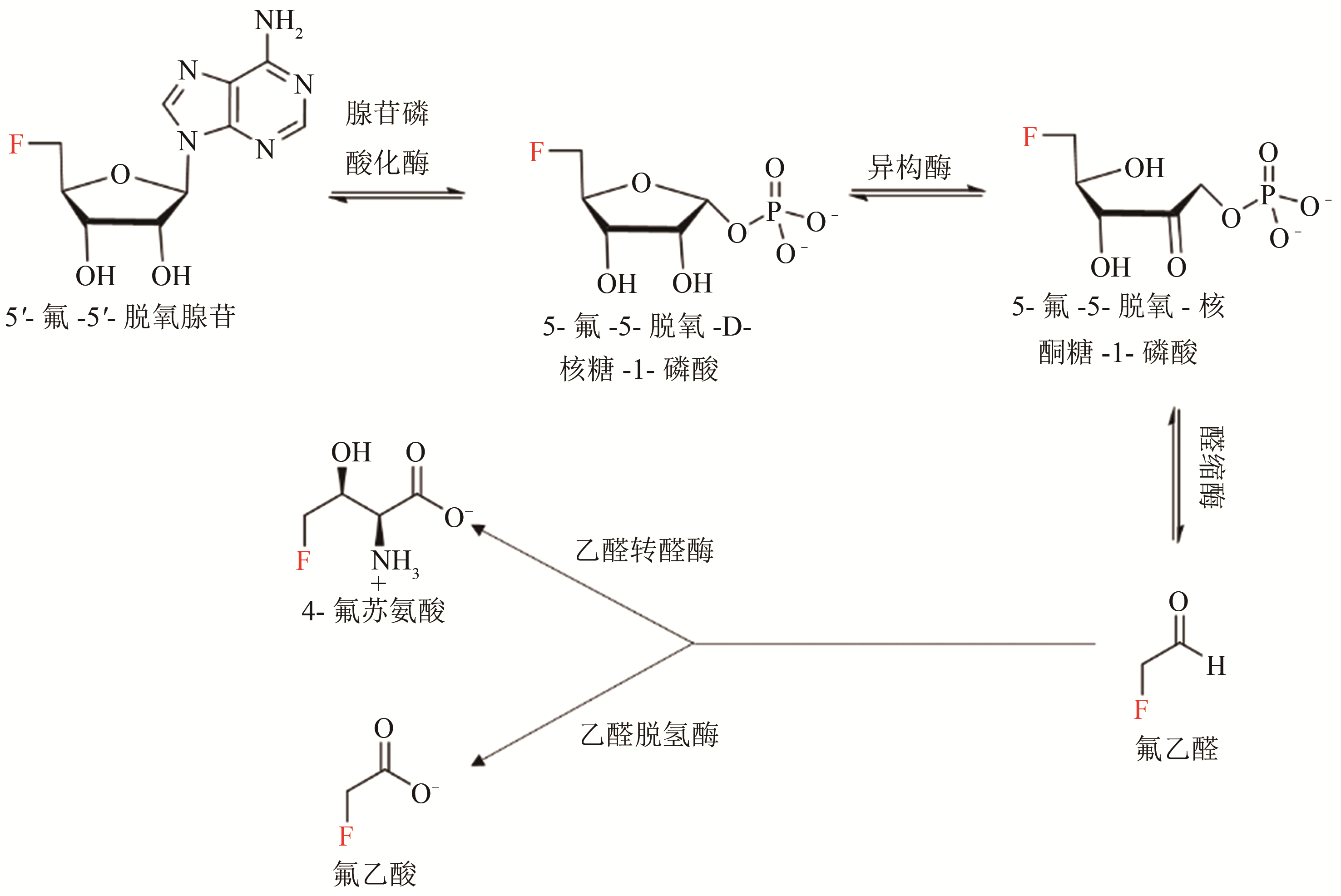
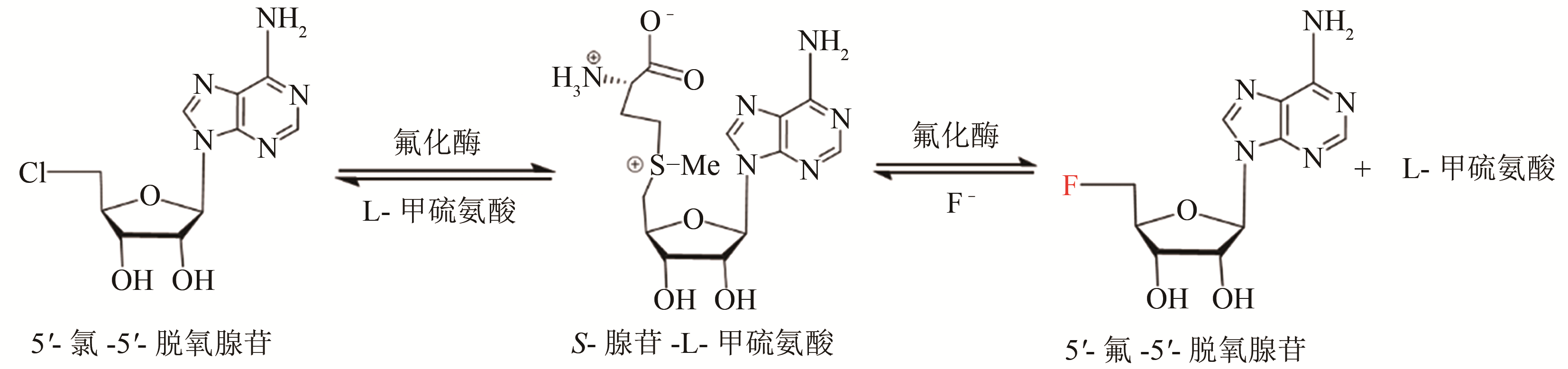
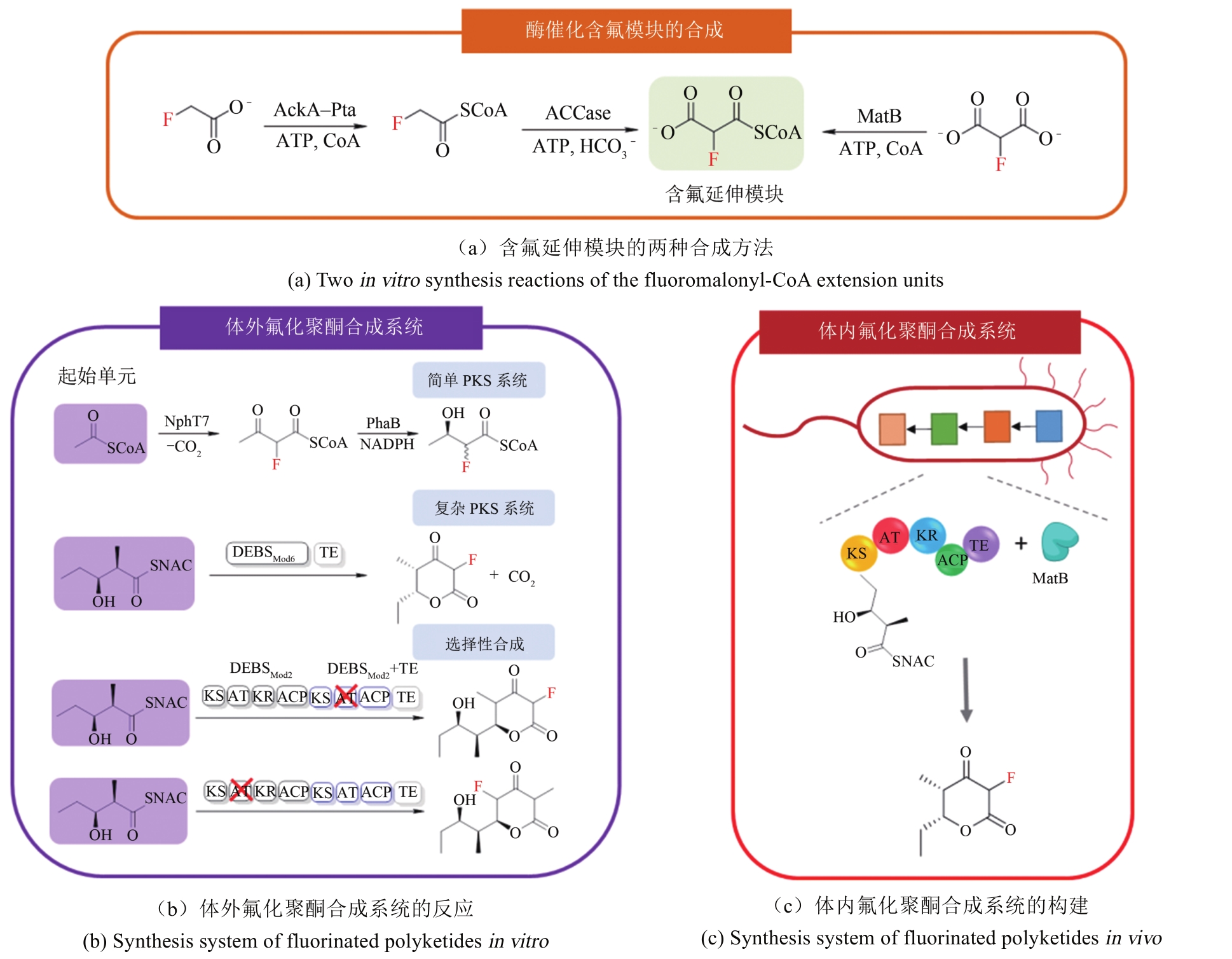
Synthetic Biology Journal ›› 2020, Vol. 1 ›› Issue (3): 358-371.DOI: 10.12211/2096-8280.2020-003
• Invited Review • Previous Articles Next Articles
Applications of synthetic biology in the production of fluorinated compounds
WANG Gaoli1, JIN Xuerui1, LUO Yunzi1,2
- 1.Frontier Science Center for Synthetic Biology and Key Laboratory of Systems Bioengineering (Ministry of Education),School of Chemical Engineering and Technology,Tianjin University,Tianjin 300072,China
2.Collaborative Innovation Center of Chemical Science and Engineering (Tianjin),Tianjin University,Tianjin 300072,China
-
Received:2020-02-27Revised:2020-03-28Online:2020-09-29Published:2020-06-30 -
Contact:LUO Yunzi
合成生物学在含氟化合物生产中的应用
王高丽1, 金雪芮1, 罗云孜1,2
- 1.天津大学化工学院,系统生物工程教育部重点实验室,合成生物学前沿科学中心,天津 300072
2.天津大学化学化工协同创新中心,天津 300072
-
通讯作者:罗云孜 -
作者简介:王高丽(1996—),女,硕士研究生。E-mail:2019207301@tju.edu.cn
金雪芮(1997—),女,硕士研究生。E-mail:2019207303@tju.edu.cn
罗云孜(1985—),女,博士,教授,研究方向为合成生物学。E-mail:luoyunzi827@aliyun.com -
基金资助:天津市自然科学基金(19JCYBJC24200);国家重点研发计划 “合成生物学”重点专项(2018YFA0903300)
CLC Number:
Cite this article
WANG Gaoli, JIN Xuerui, LUO Yunzi. Applications of synthetic biology in the production of fluorinated compounds[J]. Synthetic Biology Journal, 2020, 1(3): 358-371.
王高丽, 金雪芮, 罗云孜. 合成生物学在含氟化合物生产中的应用[J]. 合成生物学, 2020, 1(3): 358-371.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2020-003
| 氟化酶来源 | KM(SAM)/ μmol·L-1 | 转化数kcat/min-1 | (kcat/KM)/×10-3 L·μmol-1·min-1 |
|---|---|---|---|
| S. cattleya[ | 29.2±2.41 | 0.083 | 2.84 |
| Streptomyces sp.MA37[ | 82.4±18.6 | 0.262 | 3.18 |
| N. brasiliensis[ | 27.8±4.23 | 0.122 | 4.40 |
| Actinoplanes sp. N902-109[ | 45.8±7.91 | 0.204 | 4.44 |
| Streptomyces xinghaiensis NRRL B24674[ | 7.04±0.94 | 0.277±0.007 | 39.5±1.51 |
Tab. 1 The summary for the kinetic parameters of fluorinase from different sources
| 氟化酶来源 | KM(SAM)/ μmol·L-1 | 转化数kcat/min-1 | (kcat/KM)/×10-3 L·μmol-1·min-1 |
|---|---|---|---|
| S. cattleya[ | 29.2±2.41 | 0.083 | 2.84 |
| Streptomyces sp.MA37[ | 82.4±18.6 | 0.262 | 3.18 |
| N. brasiliensis[ | 27.8±4.23 | 0.122 | 4.40 |
| Actinoplanes sp. N902-109[ | 45.8±7.91 | 0.204 | 4.44 |
| Streptomyces xinghaiensis NRRL B24674[ | 7.04±0.94 | 0.277±0.007 | 39.5±1.51 |
| 1 | GRIBBLE G W. Occurrence of halogenated alkaloids[J]. The Alkaloids: Chemistry and Biology, 2012, 71:1-165. |
| 2 | O’HAHAN D, PERRY R, LOCK J M, et al. Identification of exceptionally high levels of monofluoroacetate in Dichapetalum braunii from Southeastern Tanzania[J]. Phytochemistry, 1993, 33: 1043–1046. |
| 3 | PAGUIGAN N D, HUNITI M H A, RAJA H A, et al. Chemoselective fluorination and chemoinformatic analysis of griseofulvin: natural vs fluorinated fungal metabolites[J]. Bioorganic and Medicinal Chemistry, 2017, 25(20): 5238-5246. |
| 4 | BOHM H J, BANNER D, BENDELS S, et al. Fluorine in medicinal chemistry[J]. ChemBioChem, 2004, 5(5):637-643. |
| 5 | LIN X X, RONG F, FU D G, et al. Enhanced photocatalytic activity of fluorine doped TiO2 by loaded with Ag for degradation of organic pollutants[J]. Powder Technology, 2012, 219:173-178. |
| 6 | LIANG J, LUO Y Z, ZHAO H M. Synthetic biology: putting synthesis into biology[J]. Wiley Interdisciplinary Reviews Systems Biology & Medicine, 2011, 3(1):7-20. |
| 7 | LUO Y Z, COBB E R, ZHAO H M, Recent advances in natural product discovery [J]. Current Opinion in Biotechnology, 2014, 30: 230-237. |
| 8 | ZHAO Q Y, WANG L P, LUO Y Z. Recent advances in natural products exploitation in Streptomyces via synthetic biology[J]. Engineering in Life Sciences, 2019, 19(6): 452-462. |
| 9 | 王丽苹, 罗云孜. 合成生物学在天然产物研究中的应用[J]. 生物技术通报, 2017, 33(1): 35-47. |
| WANG L P, LUO Y Z. Applications of synthetic biology in the research of natural product[J]. Biotechnology Bulletin, 2017, 33(1): 35-47. | |
| 10 | LUO Y Z, LI B Z, LIU D, et al. Engineered biosynthesis of natural products in heterologous hosts[J]. Chemical Society Reviews, 2015, 44(15): 5265-5290. |
| 11 | PALAZZOTTO E, TONG Y J, LEE S Y, et al. Synthetic biology and metabolic engineering of actinomycetes for natural product discovery[J]. Biotechnology Advances, 2019, 37(6):107366. |
| 12 | TILBURG A Y V, CAO H J, MEULEN S B V D, et al. Metabolic engineering and synthetic biology employing Lactococcus lactis and Bacillus subtilis cell factories[J]. Current Opinion in Biotechnology, 2019, 59: 1-7. |
| 13 | FREY R, HAYASHI T, BULLER R M. Directed evolution of carbon-hydrogen bond activating enzymes[J]. Current Opinion in Biotechnology, 2019, 60: 29-38. |
| 14 | LI F Z, ZHANG X, RENATA H. Enzymatic C—H functionalizations for natural product synthesis[J]. Current Opinion in Chemical Biology, 2019, 49:25-32. |
| 15 | OELSNITZ S D, ELLINGTON A. Continuous directed evolution for strain and protein engineering[J]. Current Opinion in Biotechnology, 2018, 53:158-163. |
| 16 | BELSARE K D, HORN T, RUFF A J, et al. Directed evolution of P450cin for mediated electron transfer[J]. Protein Engineering Design and Selection, 2017, 30(2):119-127. |
| 17 | BALE J B, GONEN S, LIU Y, et al. Accurate design of megadalton-scale two-component icosahedral protein complexes[J]. Science, 2016, 353(6297):389-394. |
| 18 | BAKER D. Computationally designed protein activation[J]. National Science Review, 2019, 6(4): 609-610. |
| 19 | WU Q, PENG Z L, ANISHCHENKO I, et al. Protein contact prediction using metagenome sequence data and residual neural networks[J]. Bioinformatics, 2020, 36(1):41-48. |
| 20 | CONG Q, ANISHCHENKO I, OVCHINNIKOV S, et al. Protein interaction networks revealed by proteome coevolution[J]. Science, 2019, 365(6449):185-189. |
| 21 | NIELAEN J. Cell factory engineering for improved production of natural products[J]. Natural Product Reports, 2019, 36(9):1233-1236. |
| 22 | CHEVRETTE M G, GARCIA K G, MOJJCA N S, et al. Evolutionary dynamics of natural product biosynthesis in bacteria[J]. Natural Product Reports, 2020, 37: 566-599. |
| 23 | JENSEN P E, SCHARFF L B. Engineering of plastids to optimize the production of high-value metabolites and proteins[J]. Current Opinion in Biotechnology, 2019, 59:8-15. |
| 24 | WANG W, LI S, LI Z, et al. Harnessing the intracellular triacylglycerols for titer improvement of polyketides in Streptomyces [J]. Nature Biotechnology, 2020, 38: 76-83. |
| 25 | SHAO Z, RAO G, LI C, et al. Refactoring the silent spectinabilin gene cluster using a plug-and-play scaffold[J]. ACS Synthetic Biology, 2013, 2(11):662-669. |
| 26 | LI L, JIANG W, LU Y. New strategies and approaches for engineering biosynthetic gene clusters of microbial natural products[J]. Biotechnology Advances, 2017, 35(8):936-949. |
| 27 | SPRAKER J E, LUU G T, SANCHEZ L M. Imaging mass spectrometry for natural products discovery: a review of ionization methods[J]. Natural Product Reports,2019,2: |
| 28 | LUO Y Z, HUANG H, LIANG J, et al. Activation and characterization of a cryptic polycyclic tetramate macrolactam biosynthetic gene cluster[J]. Nature Communications, 2013, 4:2894. |
| 29 | CABRERA V L, LOPEZ R O, GONZALEZ R E, et al. Complete genome sequence of Nocardia brasiliensis HUJEG-1[J]. Journal of Bacteriology, 2012, 194(10): 2761-2762. |
| 30 | SUN H H, YEO W L, LIM Y H, et al. Directed evolution of a fluorinase for improved fluorination efficiency with a non-native substrate[J]. Angewandte Chemie-International Edition, 2016, 128(46):14489-14492. |
| 31 | WALKER M C, THURONYI B. W, CHARKOUDIAN L K, et al . Expanding the fluorine chemistry of living systems using engineered polyketide synthase pathways[J]. Science, 2013, 341(6150): 1089-1094. |
| 32 | FRALRY A E, SHERMAN D H. Halogenase engineering and its utility in medicinal chemistry[J]. Bioorganic and Medicinal Chemistry Letters, 2018, 11: 1992-1999. |
| 33 | SANADA M, MIYANO T, IWADARE S W, et al. Biosynthesis of fluorothreonine and fluoroacetic acid by the thienamycin producer, Streptomyces cattleya [J]. The Journal of Antibiotics, 1986, 39: 259-265. |
| 34 | ZHAO C H, LI P, DENG Z X, et al. Insights into fluorometabolite biosynthesis in Streptomyces cattleya DSM46488 through genome sequence and knockout mutants[J]. Bioorganic Chemistry, 2012, 44: 1-7. |
| 35 | O' HAGAN D, SCHAFFRATH C, COBB S L, et al. Biochemistry: biosynthesis of an organofluorine molecule [J]. Nature, 2002, 416(6878): 279-279. |
| 36 | DENG H, MA L, BANDARANAYAKA N, et al. Identification of fluorinases from Streptomyces sp MA37, Norcardia brasiliensis, and Actinoplanes sp N902-109 by genome mining[J]. ChemBioChem, 2014, 15(3): 364-368. |
| 37 | WANG Y Y, DENG Z X, QU X D. Characterization of a SAM-dependent fluorinase from a latent biosynthetic pathway for fluoroacetate and 4-fluorothreonine formation in Nocardia brasiliensis [J]. F1000Research, 2014, 3: 61. |
| 38 | HUANG S, MA L, TONG M H, et al. Fluoroacetate biosynthesis from the marine-derived bacterium Streptomyces xinghaiensis NRRL B-24674[J]. Organic & Biomolecular Chemistry,2014,12(27):4828-4831. |
| 39 | MA L, LI Y F, MENG L P, et al. Biological fluorination from the sea: discovery of a SAM-dependent nucleophilic fluorinating enzyme from the marine-derived bacterium Streptomyces xinghaiensis NRRL B24674[J]. RSC Advances, 2016, 6: 27047-27051. |
| 40 | MEKLAT A, BOURAS N, ZITOUNI A, et al. Actinopolyspora mzabensis sp. nov., a halophilic actinomycete isolated from an Algerian Saharan soil[J]. International Journal of Systematic and Evolutionary Microbiology, 2013, 63(10):3787-3792. |
| 41 | SOOKLAL S A, KONING C D, DEAN B, et al. Identification and characterisation of a fluorinase from Actinopolyspora mzabensis [J]. Protein Expression and Purification, 2020, 166: 105508. |
| 42 | DONG C J, HUANG F L, DENG H, et al. Crystal structure and mechanism of a bacterial fluorinating enzyme[J]. Nature, 2004, 427(6974): 561-565. |
| 43 | PITEL S B R, ZHAO H M. Recent advances in biocatalysis by directed enzyme evolution[J]. Comb. Chem. High Throughput Screen., 2006, 9:247-257. |
| 44 | YEO W L, CHEW X, SMITH D J, et al. Probing the molecular determinants of fluorinase specificity[J]. Chemical Communication, 2017, 53(17):2559-2562. |
| 45 | DENG H, COBB S L, MCEWAN A R, et al. McEwan the fluorinase from Streptomyces cattleya is also a chlorinase[J]. Angewandte Chemie International Edition, 2006, 45(5):759-762. |
| 46 | LOWE P T, COBB S L, O'HAGAN D. An enzymatic Finkelstein reaction: fluorinase catalyses direct halogen exchange[J]. Organic and Biomolecular Chemistry, 2019, 17(32): 7493-7496. |
| 47 | SERGEEV M E, MORGIA F, JAVED M R, et al. Enzymatic radiofluorination: fluorinase accepts methylaza-analog of SAM as substrate for FDA synthesis[J]. Journal of Molecular Catalysis B: Enzymatic, 2013, 97:74-79. |
| 48 | SUN H H, ZHAO H M, ANG E L. A coupled chlorinase-fluorinase system with a high efficiency of trans-halogenation and a shared substrate tolerance[J]. Chemical Communications, 2018, 54(68): 9458-9461. |
| 49 | ZHAO W X, DU G C, LIU S. An efficient thermostabilization strategy based on self-assembling amphipathic peptides for fusion tags[J]. Enzyme and Microbial Technology, 2018,121:68-77. |
| 50 | TU C H, ZHOU J, PENG L. Self-assembled nano-aggregates of fluorinases demonstrate enhanced enzymatic activity, thermostability and reusability[J]. Biomaterials Science, 2019: 2047-4849. |
| 51 | O' HAGAN D. Fluorine in health care: organofluorine containing blockbuster drugs[J]. J. Fluor. Chem., 2010, 131: 1071-1081. |
| 52 | 刘栓栓,王晶,许斌,等.近5年美国FDA批准上市的含氟药物研究进展[J].药学进展,2016,40(10):783-794. |
| LIU S S, WANG J, XU B, et al. Recent advances in R&D of fluorinated drugs approved by FDA in the past five years[J]. Progress in Pharmaceutical Sciences, 2016, 40(10):783-794 | |
| 53 | 赵方诺.氟原子在药物设计中的主要应用以及引入方法[J].国际公关,2019(7):247-248. |
| ZHAO F N. The main applications and introduction methods of fluorine atom in drug design[J]. PR Magazine, 2019(7):247-248. | |
| 54 | 张霁,金传飞,张英俊.含氟药物研究进展和芳(杂)环氟化及N(n=1,2,3)氟甲基化新方法[J].有机化学,2014,34(4):662-680. |
| ZHANG J, JIN C F, ZHANG Y J. Recent advances in research and development of fluorinated drugs and new methods for fluorination, mono-, di-and tri-fluoromethylation[J]. Chinese Journal of Organic Chemistry, 2014, 34(4):662-680. | |
| 55 | 弓添添,肖美娟,陈樱,等.克唑替尼药物关键手性中间体合成进展[J].浙江化工,2018,49(10):10-14. |
| GONG T T, XIAO M J, CHEN Y, et al. Research progress in synthesis of a key chiral intermediate of drug crizotinib[J]. Zhejiang Chemical Industry, 2018, 49(10):10-14. | |
| 56 | 孙冰,赵会,王玉军,等.马来酸阿法替尼的合成工艺研究[J].中国药物化学杂志,2019,29(1):44-48. |
| SUN B, ZHAO H, WANG Y J, et al. Study on synthetic process of afatinib dimaleate[J]. Chinese Journal of Medicinal Chemistry, 2019, 29(1):44-48. | |
| 57 | ODAR C, WINKLER M, WILTSCHI B. Fluoro amino acids: a rarity in nature, yet a prospect for protein engineering [J]. Biotechnology Journal, 2015, 10(3):427-446. |
| 58 | CHAN K K J, O'HAGAN D. The rare fluorinated natural products and biotechnological prospects for fluorine enzymology[J]. Methods in Enzymology, 2012, 516: 219-233. |
| 59 | ZHU X M, HACKL S, THAKER M N, et al. Biosynthesis of the fluorinated natural product nucleocidin in Streptomyces calvus is dependent on the bldA-specified Leu-tRNAUUA molecule[J]. ChemBioChem, 2015,16(17): 2498-2506. |
| 60 | WEISSLEDER R, MAHMOOD U. Molecular imaging[J]. Radiology, 2001, 219 (2): 316-333 |
| 61 | JACOBSON O, KIESEWETTER D O, CHEN X Y. Fluorine-18 radiochemistry, labeling strategies and synthetic routes[J]. Bioconjugate Chemistry, 2015, 26(1):1-18. |
| 62 | THOMPSON S, FLEMING I N, O' HAGAN D, et al. Enzymatic transhalogenation of dendritic RGD peptide constructs with the fluorinase[J]. Organic and Biomolecular Chemistry, 2016, 14(11): 3120-3129. |
| 63 | THOMPSON S, ONEGA M, ASHWORTH S, et al. A two-step fluorinase enzyme mediated 18F labelling of an RGD peptide for positron emission tomography[J]. Chemical Communications, 2015, 51(70): 13542-13545. |
| 64 | ZHANG Q Z, DALL'ANGELO S, FLEMING I N, et al. Last-step enzymatic [18F]-fluorination of cysteine-tethered RGD peptides using modified barbas linkers[J]. Chemistry, 2016, 22(31): 10998-11004. |
| 65 | LOWE P T, DALL'ANGELO S, KRIEGER T M, et al. A new class of fluorinated A2A adenosine receptor agonist with application to last-step enzymatic [18F]fluorination for PET imaging[J]. ChemBioChem, 2017, 18(21): 2156-2164. |
| 66 | LOWE P T, DALL'ANGELO S, DEVINE A, et al. Enzymatic fluorination of biotin and tetrazine conjugates for pretargeting approaches to positron emission tomography imaging[J]. ChemBioChem, 2019, 19(18): 1969-1978. |
| 67 | LOWE P T, DALL'ANGELO S, FLEMING I N, et al. Enzymatic radiosynthesis of a 18F-Glu-Ureido-Lys ligand for the prostate-specific membrane antigen (PSMA) [J]. Organic and Biomolecular Chemistry, 2019, 17(6): 1480-1486. |
| 68 | EUSTAQUIO A S, O'HAGAN D, MOORE B S. Engineering fuorometabolite production: fluorinase expression in Salinispora tropica yields fluorosalinosporamide[J]. Journal of Natural Products, 2010, 73: 378-382. |
| 69 | LUO Y Z, LEE J K, ZHAO H M. Challenges and opportunities in synthetic biology for chemical engineers[J]. Chemical Engineering Science. 2013, 103: 115-119. |
| 70 | WEISSMAN K J. Genetic engineering of modular PKSs: from combinatorial biosynthesis to synthetic biology[J]. Natural Product Reports, 2016, 33(2): 203-230 |
| 71 | WU L R, MAGLABGIT F, DENG H. Fluorine biocatalysis[J]. Current Opinion in Chemical Biology, 2020, 55:119-126 |
| 72 | KLOPEIRS S, KOOPMANS K R M, GARCIA E S, et al. Biosynthesis with fluorine[J]. ChemBioChem, 2014, 15(4): 495-497. |
| 73 | THURONYI B. W, CHANG M C Y. Synthetic biology approaches to fluorinated polyketides[J]. Accounts of Chemical Research, 2015, 48(3): 584-592. |
| 74 | THURONYI B. W, PRIVALSKY T M, CHANG, M C Y. Engineered fluorine metabolism and fluoropolymer production in living cell[J]. Angewandte Chemie-International Edition, 2017, 56(44): 13637-13640. |
| 75 | AD O, THURONYI B. W, CHANG M C Y. Elucidating the mechanism of fluorinated extender unit loading for improved production of fluorine-containing polyketides[J]. Proceedings of the National Academy of Sciences of the United States of America, 2017, 114(5): E660-E668. |
| 76 | JOSE R C, RAJA H A, GRAF T N. Biosynthesis of fluorinated peptaibols using a site-directed building block incorporation approach[J]. Journal of Natural Products, 2017, 80(6): 1883-1892. |
| 77 | FANG J, HAIT D, GORDON M H, et al. Chemoenzymatic platform for synthesis of chiral organofluorines based on type II aldolases[J]. Angewandte Chemie-International Edition, 2019, 131(34): 11967-11971. |
| 78 | ZHANG J, HUANG X Y, ZHANG R K, et al. Enantiodivergent α‑amino C—H fluoroalkylation catalyzed by engineered cytochrome P450s[J]. Journal of the American Chemical Society, 2019, 141: 9798-9802. |
| 79 | GILLIS E P, EASTMAN K J, HILL M D, et al. Applications of fluorine in medicinal chemistry [J]. Journal of Medicinal Chemistry, 2015, 58: 8315-8359. |
| [1] | GAO Ge, BIAN Qi, WANG Baojun. Synthetic genetic circuit engineering: principles, advances and prospects [J]. Synthetic Biology Journal, 2025, 6(1): 45-64. |
| [2] | LI Jiyuan, WU Guosheng. Two hypothesises for the origins of organisms from the synthetic biology perspective [J]. Synthetic Biology Journal, 2025, 6(1): 190-202. |
| [3] | JIAO Hongtao, QI Meng, SHAO Bin, JIANG Jinsong. Legal issues for the storage of DNA data [J]. Synthetic Biology Journal, 2025, 6(1): 177-189. |
| [4] | TANG Xinghua, LU Qianneng, HU Yilin. Philosophical reflections on synthetic biology in the Anthropocene [J]. Synthetic Biology Journal, 2025, 6(1): 203-212. |
| [5] | XU Huaisheng, SHI Xiaolong, LIU Xiaoguang, XU Miaomiao. Key technologies for DNA storage: encoding, error correction, random access, and security [J]. Synthetic Biology Journal, 2025, 6(1): 157-176. |
| [6] | ZHONG Quanzhou, SHAN Yiyi, PEI Qingyun, JIN Yanyun, WANG Yihan, MENG Luyuan, WANG Xinyun, ZHANG Yuxin, LIU Kunyuan, WANG Huizhong, FENG Shangguo. Research progress in the production of α-arbutin through biosynthesis [J]. Synthetic Biology Journal, 2025, 6(1): 118-135. |
| [7] | SHI Ting, SONG Zhan, SONG Shiyi, ZHANG Yi-Heng P. Job. In vitro BioTransformation (ivBT): a new frontier of industrial biomanufacturing [J]. Synthetic Biology Journal, 2024, 5(6): 1437-1460. |
| [8] | CHAI Meng, WANG Fengqing, WEI Dongzhi. Synthesis of organic acids from lignocellulose by biotransformation [J]. Synthetic Biology Journal, 2024, 5(6): 1242-1263. |
| [9] | SHAO Mingwei, SUN Simian, YANG Shimao, CHEN Guoqiang. Bioproduction based on extremophiles [J]. Synthetic Biology Journal, 2024, 5(6): 1419-1436. |
| [10] | ZHU Fanghuan, CEN Xuecong, CHEN Zhen. Research progress of diols production by microbes [J]. Synthetic Biology Journal, 2024, 5(6): 1367-1385. |
| [11] | LIU Yining, PU Wei, YANG Jinxing, WANG Yu. Recent advances in the biosynthesis of ω-amino acids and lactams [J]. Synthetic Biology Journal, 2024, 5(6): 1350-1366. |
| [12] | CHEN Yu, ZHANG Kang, QIU Yijing, CHENG Caiyun, YIN Jingjing, SONG Tianshun, XIE Jingjing. Progress of microbial electrosynthesis for conversion of CO2 [J]. Synthetic Biology Journal, 2024, 5(5): 1142-1168. |
| [13] | ZHENG Haotian, LI Chaofeng, LIU Liangxu, WANG Jiawei, LI Hengrun, NI Jun. Design, optimization and application of synthetic carbon-negative phototrophic community [J]. Synthetic Biology Journal, 2024, 5(5): 1189-1210. |
| [14] | CHENG Xiaolei, LIU Tiangang, TAO Hui. Recent research progress in non-canonical biosynthesis of terpenoids [J]. Synthetic Biology Journal, 2024, 5(5): 1050-1071. |
| [15] | LIU Zijian, MU Baiyang, DUAN Zhiqiang, WANG Xuan, LU Xiaojie. Advances in the development of DNA-compatible chemistries [J]. Synthetic Biology Journal, 2024, 5(5): 1102-1124. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||