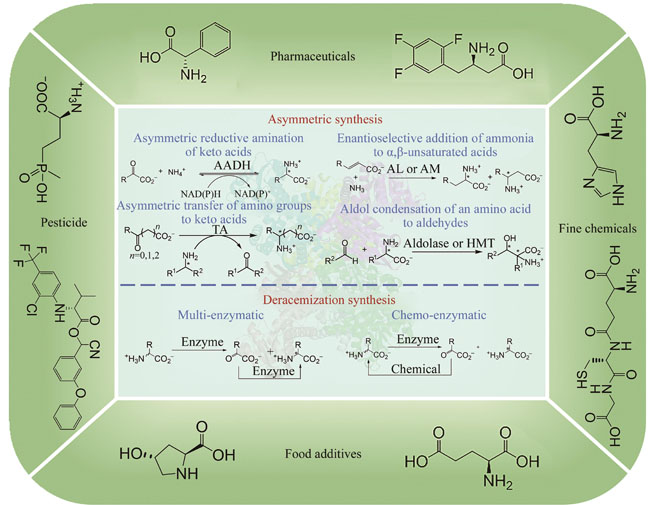
Chiral amino acids represent a crucial class of chiral building blocks with significant value in food, medicine, chemical industry, and agriculture. The market scale of pharmaceuticals, pesticides, food, and chemical industries relying on chiral amino acids is substantial and has been attracting increasing attention. The pursuit of efficient, environmentally friendly, and cost-effective synthesis of chiral amino acids has long been a goal for scientists. Commonly used preparation methods for chiral amino acids fall into four following categories: protein hydrolysis, fermentation, chemical synthesis, and enzyme-catalyzed synthesis. Among these, enzyme-catalyzed synthesis has demonstrated great potential due to its mild reaction conditions, high stereo-selectivity, simplicity of steps, and wide application range. In recent years, with the rapid development of bioinformatics, protein engineering, and computational biology, there has been an increasing number of high-performance enzyme preparations developed, leading to a steady increase in the diversity of enzymes and the gradual diversification of catalyzed reactions, further promoting the wide application of enzyme-catalyzed synthesis of chiral amino acids. The enzyme-catalyzed synthesis of chiral amino acids can be categorized into three groups: asymmetric synthesis, deracemization synthesis, and kinetic resolution. Kinetic resolution, due to its theoretical yield of only 50% and low atom economy, is not suitable for industrial applications. In contrast, asymmetric synthesis and deracemization synthesis with theoretical yield of 100% find wider industrial application. This article reviews the application of enzymatic asymmetric synthesis and deracemization synthesis in the synthesis of chiral amino acids. It includes the development and modification of key enzyme such as amino acid dehydrogenase, transaminase, ammonia lyase, aldolase, amino acid oxidase, and amino acid deaminase, as well as their application in the synthesis of high-value chiral amino acids such as phosphinothricin, tert-leucine, and intermediate of sitagliptin. Additionally, it summarizes the main challenges faced in the field of enzymatic synthesis of chiral amino acids, such as the lack of key enzyme components, and low enantioselectivity, narrow substrate spectra, low catalytic activity, poor stability, limited reaction conditions of wild-type enzymes. Finally, it looks ahead to the application of cutting-edge technologies such as automated experimental devices, machine learning, and artificial intelligence in the field of enzyme modification, as well as the development of more efficient and environmentally friendly catalytic processes through reactor design and reaction process control. These endeavors collectively aim to facilitate the broader industrial application of enzymatic synthesis for chiral amino acids.