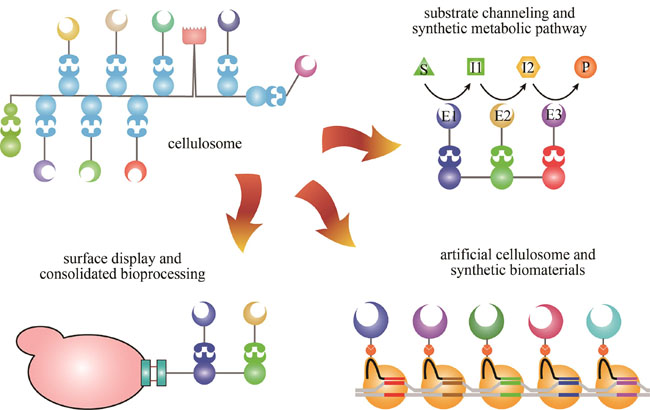
Cellulosomes are multi-enzyme complexes secreted by some anaerobic bacteria which can efficiently degrade lignocellulose. All known cellulosome-producing bacteria belong to Clostridia, including both mesophilic and thermophilic species. Cellulosomes contain non-covalent combinations of scaffolding subunits (scaffoldins) and catalytic subunits. Each of these types of subunits is composed of a variety of covalently-linked tandem modules (i.e. structural domains), which can be roughly classified into four categories: assembly modules, catalytic modules, substrate-binding modules, and cell-binding modules. Cellulosomes in different species have different numbers of scaffoldins and catalytic subunits, and the scaffoldins contain different numbers and types of assembly modules. Therefore, cellulosome structures from different species show great diversity. The architecture of cellulosomes results in multi-level synergistic effects, including the synergy between different enzymes, enzymes and substrates, and enzymes and cells. In addition to the structural complexity and multiple levels of complementary synergy, cellulosomes also have a high degree of conformational flexibility to adapt to the complexity of substrate structures. Furthermore, cellulosome-producing bacteria have developed substrate-coupling regulatory mechanisms to achieve adaptability for complex substrates. These characteristics of modularity, diversity, self-assembly, synergy, high efficiency, and adaptability are highly applicable in synthetic biology. Therefore, in recent years, cellulosomes have been widely used in biotechnology, especially in synthetic biotechnology. This article first briefly introduces the structural basis and mechanism of cellulosomes for highly efficient lignocellulose degradation, and then summarizes the progress in their utilization in different synthetic biotechnology applications, including the construction of designer cellulosomes, substrate channeling and synthetic metabolic pathway construction, cell surface display and enzyme immobilization, and artificial cellulosome construction. The designer cellulosomes contain defined catalytic components located in the specific positions of the scaffoldin, which not only facilitates basic research into the mechanism of the cellulosome, but also provides a solid foundation for biotechnology and synthetic biology applications. Designer cellulosomes can be used to construct a variety of different metabolic pathways in a variety of hosts, often resulting in a several-fold increase in the reaction rate or yield. Natural cellulosomes can be immobilized onto the cell surface through the S-Layer Homology module and/or the surface of the substrate through the carbohydrate-binding module. These immobilization characteristics have also been widely used in synthetic biotechnology. These applications mainly include displaying cellulosomes on the surface of microbial cells that cannot degrade cellulose, giving them the ability to degrade cellulose, and functionalizing the surface of cells, cellulose, or other solid materials (such as nanoparticles) by displaying various enzymes. The major advantages of the cellulosome structure lie in the self-assembly of supramolecular systems and the synergy of multi-enzyme systems, which inspire researchers to develop artificial cellulosomes: supramolecular complexes formed by the interaction of various biomolecules to mimic the structure of cellulosomes. The materials and systems used as scaffolds in artificial cellulosomes are complex and diverse, but the core approach is to construct the scaffold to allow the proximity of different enzymes, resulting in efficient molecular nanomachines or functional biomaterials. Finally, this article looks ahead to the upcoming key areas in cellulosome research and future applications of cellulosomes and cellulosome-producing bacteria. Various protein components and engineering strategies revealed in the study of cellulosomes will play an important role in the future development of synthetic biology.