合成生物学 ›› 2020, Vol. 1 ›› Issue (6): 674-684.DOI: 10.12211/2096-8280.2020-049
大宗化学品细胞工厂的构建与应用
于勇, 朱欣娜, 张学礼
- 中国科学院天津工业生物技术研究所,天津 300308
-
收稿日期:2020-04-16修回日期:2020-09-25出版日期:2020-12-31发布日期:2021-01-19 -
通讯作者:张学礼 -
作者简介:于勇(1992—),男,博士,博士后。研究方向为代谢工程、合成生物学。E-mail:yu_yong@tib.cas.cn
朱欣娜(1975—),女,博士,副研究员。研究方向为代谢工程、合成生物学。E-mail:zhu_xn@tib.cas.cn
张学礼(1981—),男,博士,研究员。研究方向为代谢工程、合成生物学。E-mail:zhang_xl@tib.cas.cn
第一联系人:共同第一作者 -
基金资助:国家重点研发计划(2019YFA0904900)
Construction and application of microbial cell factories for production of bulk chemicals
YU Yong, ZHU Xinna, ZHANG Xueli
- Tianjin Institute of Industrial Biotechnology,Chinese Academy of Science,Tianjin 300308,China
-
Received:2020-04-16Revised:2020-09-25Online:2020-12-31Published:2021-01-19 -
Contact:ZHANG Xueli
摘要:
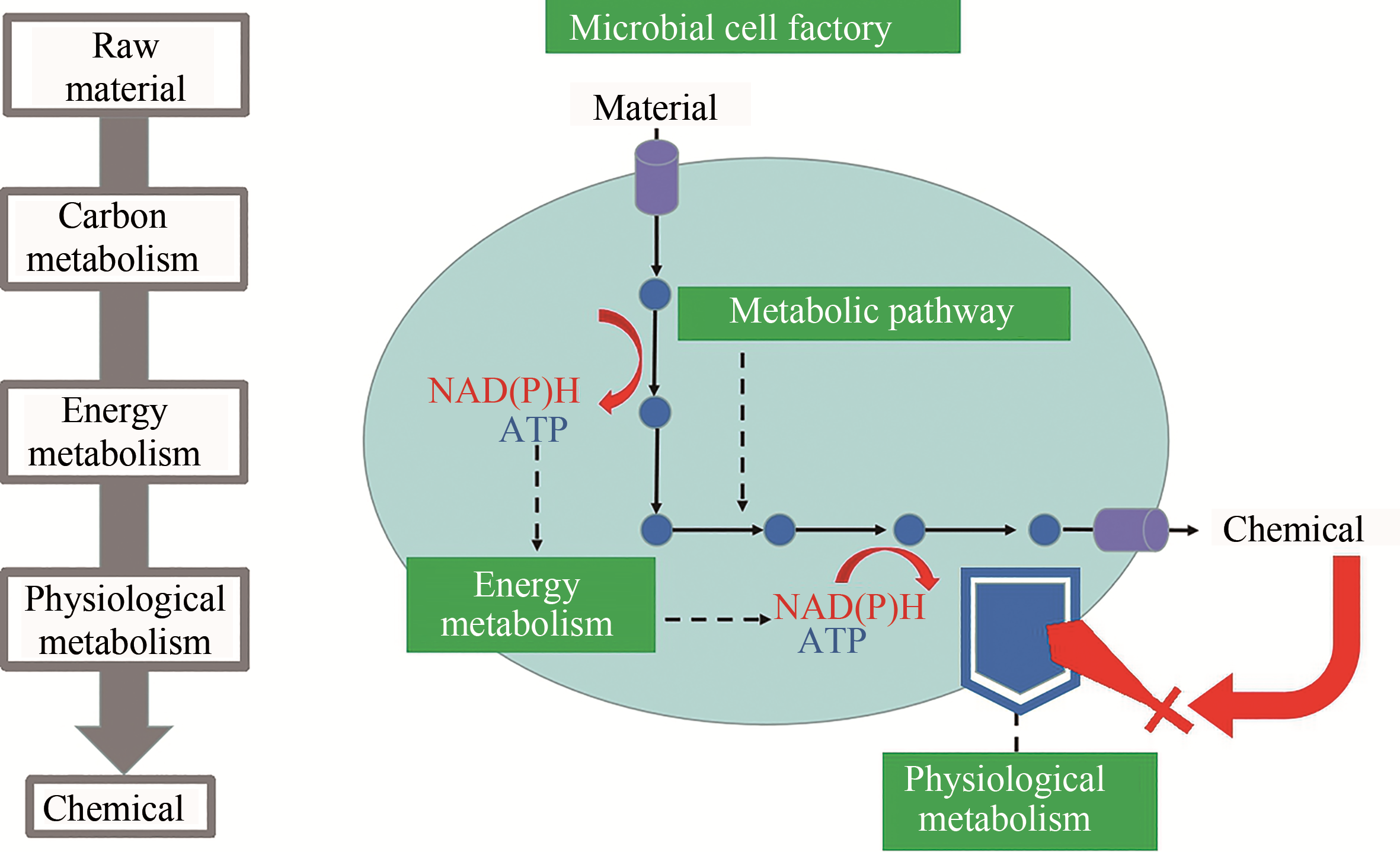
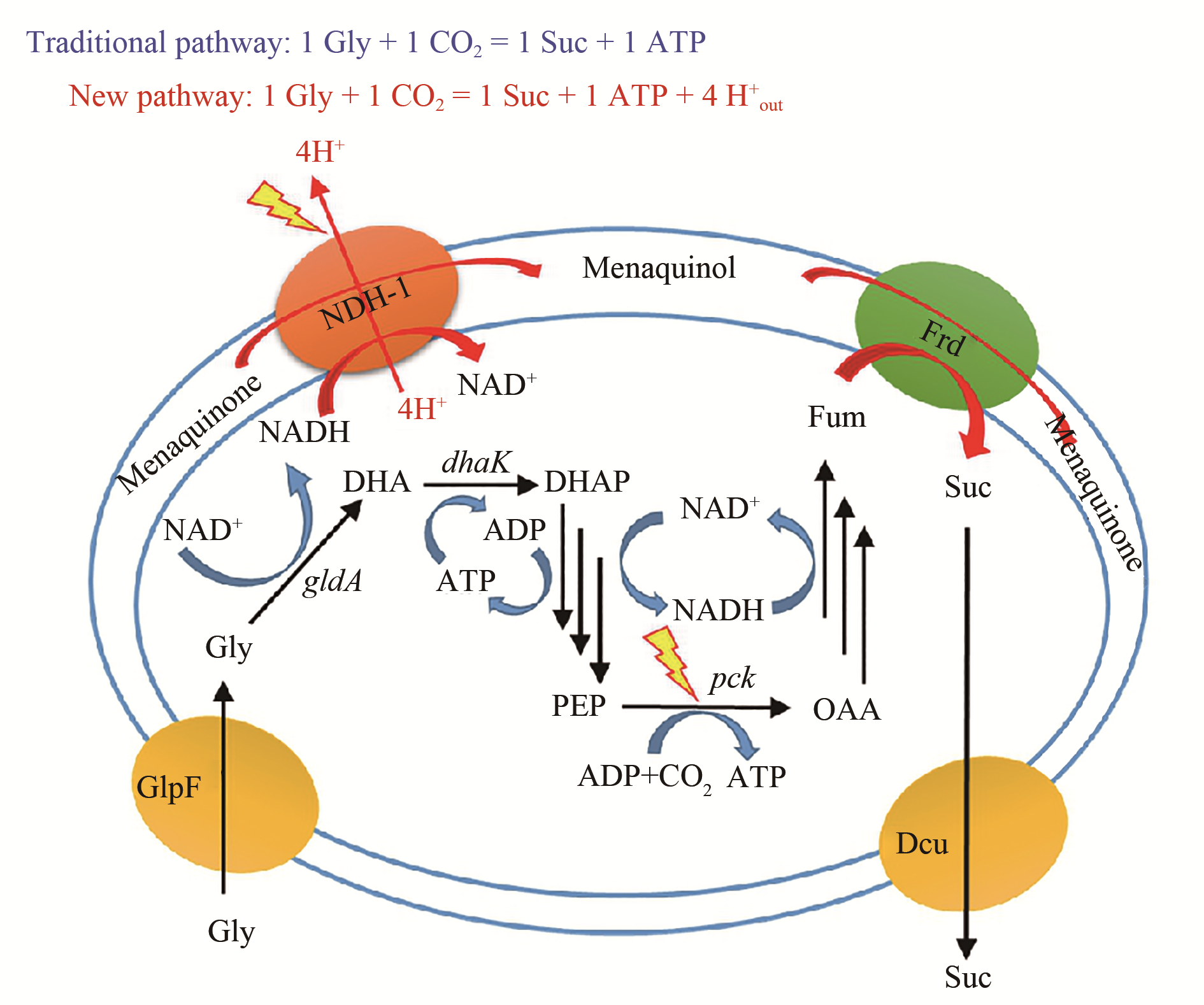
随着合成生物学技术的发展,越来越多的大宗化学品可通过微生物细胞工厂发酵生产,为摆脱石油资源依赖、节能减排提供了可能。本文首先介绍了细胞工厂构建所需的关键技术,包括基因组编辑技术、多基因同时调控技术、蛋白骨架技术、基因动态调控技术、高通量筛选技术。随后结合丁二酸细胞工厂这一具体案例,从物质代谢、能量代谢及细胞生理代谢三方面阐述了如何解析微生物高效合成化学品的代谢调控机制,为高效细胞工厂创建奠定理论基础。并对近年来成功构建的大宗化学品细胞工厂作了举例介绍,包括L-丙氨酸、L-甲硫氨酸、丁二酸、D-乳酸、丙二酸、L-苹果酸、戊二酸、己二酸、1,3-丙二醇、1,4-丁二醇和异丁醇等。未来,进一步增加原料利用效率和拓宽产物范围是微生物细胞工厂的发展方向,但新酶设计与改造是限制代谢途径设计的主要瓶颈。相信随着研究的深入,微生物细胞工厂将在替代化学法生产大宗化学品方面有更广泛的应用。
中图分类号:
引用本文
于勇, 朱欣娜, 张学礼. 大宗化学品细胞工厂的构建与应用[J]. 合成生物学, 2020, 1(6): 674-684.
YU Yong, ZHU Xinna, ZHANG Xueli. Construction and application of microbial cell factories for production of bulk chemicals[J]. Synthetic Biology Journal, 2020, 1(6): 674-684.
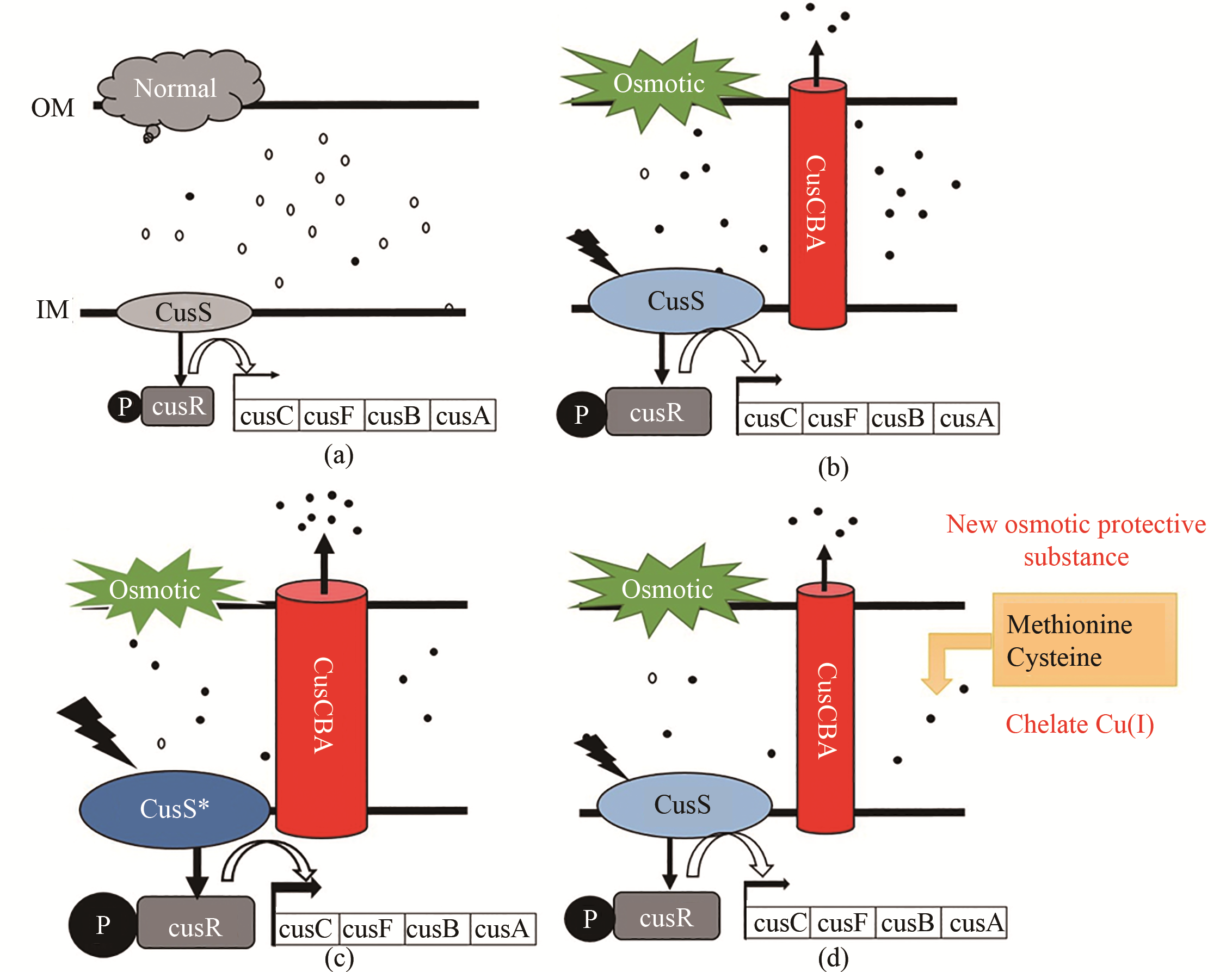
图5 大肠杆菌细胞耐渗透胁迫的新机制(a)在正常渗透压条件下(5%葡萄糖),铜离子大多以低毒的二价离子形式存在,Cus系统不激活;(b)在高渗透压条件下(12%葡萄糖),周质空间中的一价铜离子含量升高,激活了copA、cusS、cusR和cusCFAB表达量提高,转运一价铜离子的能力增强;(c)在高渗透压条件下(12%葡萄糖),突变后的cusS及cusCFBA表达量提高,转运一价铜离子的能力增强;(d)甲硫氨酸、半胱氨酸等渗透保护物种可螯合一价铜离子
Fig. 5 New osmotolerance mechanism of Escherichia coli(a) Under normal conditions (5% glucose), copper exists mostly in the form of less-toxic Cu(Ⅱ), and the CopA and Cus system was not induced; (b) Under high osmotic pressure (12% glucose), increasing free Cu(Ⅰ) in the periplasmic space activates the expression of copA, cusS, cusR and cusCFAB to expel toxic Cu(Ⅰ); (c) Under high osmotic pressure (12% glucose), expression levels of cusS and cusCFAB are increased to expel more Cu(Ⅰ) from the periplasmic space; (d) Osmotic protective substances such as methionine and cysteine can chelate valence copper ions.
| 1 | 于勇, 朱欣娜, 刘萍萍, 等. 微生物细胞工厂生产大宗化学品及其产业化进展[J]. 生物产业技术, 2019(1): 13-18. |
| YU Y, ZHU X N, LIU P P, et al. Production of bulk chemicals through microbial cell factories and its Industrialization[J]. Biotechnology & Business, 2019(1):13-18. | |
| 2 | MURPHY K C. Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli [J]. Journal of Bacteriology, 1998, 180(8): 2063-2071. |
| 3 | ZHANG Y M, BUCHHOLZ F, MUYRERS J P P, et al. A new logic for DNA engineering using recombination in Escherichia coli [J]. Nature Genetics, 1998, 20(2): 123-128. |
| 4 | URNOV F D, REBAR E J, HOLMES M C, et al. Genome editing with engineered zinc finger nucleases[J]. Nature Reviews Genetics, 2010, 11(9): 636-646. |
| 5 | MILLER J C, TAN S, QIAO G, et al. A TALE nuclease architecture for efficient genome editing[J]. Nature Biotechnology, 2011, 29(2): 143-148. |
| 6 | 何庆, 王茜, 朱清华, 等. 基因组工程在微生物细胞工厂构建中的应用进展 [J]. 生物产业技术, 2017(1): 31-36. |
| HE Q, WANG Q, ZHU Q H, et al. Application progress of genome engineering for construction of microbial cell factories[J]. Biotechnology & Business, 2017(1): 31-36. | |
| 7 | SHI S, LIANG Y, ZHANG M M, et al. A highly efficient single-step, markerless strategy for multi-copy chromosomal integration of large biochemical pathways in Saccharomyces cerevisiae [J]. Metabolic Engineering, 2016, 33: 19-27. |
| 8 | HEO M J, JUNG H M, UM J, et al. Controlling citrate synthase expression by CRISPR/Cas9 genome editing for n-butanol production in Escherichia coli [J]. ACS Synthetic Biology, 2017, 6(2): 182-189. |
| 9 | BASSALO M C, GARST A D, HALWEG-EDWARDS A L, et al. Rapid and efficient one-step metabolic pathway integration in E. coli [J]. ACS Synthetic Biology, 2016, 5(7): 561-568. |
| 10 | WASELS F, JEAN-MARIE J, COLLAS F, et al. A two-plasmid inducible CRISPR/Cas9 genome editing tool for Clostridium acetobutylicum [J]. Journal of Microbiol Methods, 2017, 140: 5-11. |
| 11 | CHO J S, CHOI K R, PRABOWO C P S, et al. CRISPR/Cas9-coupled recombineering for metabolic engineering of Corynebacterium glutamicum [J]. Metabolic Engineering, 2017, 42: 157-167. |
| 12 | ANZALONE A V, RANDOLPH P B, DAVIS J R, et al. Search-and-replace genome editing without double-strand breaks or donor DNA[J]. Nature, 2019, 576(7785): 149-157. |
| 13 | WANG H H, ISAACS F J, CARR P A, et al. Programming cells by multiplex genome engineering and accelerated evolution[J]. Nature, 2009, 460(7257): 894-898. |
| 14 | PFLEGER B F, PITERA D J, SMOLKE C D, et al. Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes[J]. Nature Biotechnology, 2006, 24(8): 1027-1032. |
| 15 | WARNER J R, REEDER P J, KARIMPOUR-FARD A, et al. Rapid profiling of a microbial genome using mixtures of barcoded oligonucleotides[J]. Nature Biotechnology, 2010, 28(8): 856-862. |
| 16 | ISAACS F J, CARR P A, WANG H H, et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement[J]. Science, 2011, 333(6040): 348-353. |
| 17 | ZHU X N, ZHAO D D, QIU H N, et al. The CRISPR/Cas9-facilitated multiplex pathway optimization (CFPO) technique and its application to improve the Escherichia coli xylose utilization pathway[J]. Metabolic Engineering, 2017, 43: 37-45. |
| 18 | LEE M J, MANTELL J, HODGSON L, et al. Engineered synthetic scaffolds for organizing proteins within the bacterial cytoplasm[J]. Nature Chemical Biology, 2018, 14(2): 142. |
| 19 | DUEBER J E, WU G C, MALMIRCHEGINI G R, et al. Synthetic protein scaffolds provide modular control over metabolic flux[J]. Nature Biotechnology, 2009, 27(8): 753. |
| 20 | KANG W, MA T, LIU M, et al. Modular enzyme assembly for enhanced cascade biocatalysis and metabolic flux[J]. Nature Communications, 2019, 10(1): 4248. |
| 21 | ZHANG F Z, CAROTHERS J M, KEASLING J D. Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids[J]. Nature Biotechnology, 2012, 30(4): 354. |
| 22 | ZHANG X D. Optimal high-throughput screening: practical experimental design and data analysis for genome-scale RNAi research[M]. New York: Cambridge University Press, 2011. |
| 23 | SALIS H M, MIRSKY E A, VOIGT C A. Automated design of synthetic ribosome binding sites to control protein expression[J]. Nature Biotechnology, 2009, 27(10): 946-950. |
| 24 | KAN S B, LEWIS R D, CHEN K, et al. Directed evolution of cytochrome c for carbon-silicon bond formation: bringing silicon to life[J]. Science, 2016, 354(6315): 1048-1051. |
| 25 | SCHMIDL S R, EKNESS F, SOFJAN K, et al. Rewiring bacterial two-component systems by modular DNA-binding domain swapping[J]. Nature Chemical Biology, 2019, 15(7): 690-698. |
| 26 | SIKORA A E, TEHAN R, MCPHAIL K. Utilization of Vibrio cholerae as a model organism to screen natural product libraries for identification of new antibiotics[J]. Methods in Molecular Biology, 2018, 1839: 135-146. |
| 27 | LU J, TANG J L, LIU Y, et al. Combinatorial modulation of galP and glk gene expression for improved alternative glucose utilization[J]. Applied Microbiology and Biotechnology, 2012, 93(6): 2455-2462. |
| 28 | ZHANG X L, JANTAMA K, MOORE J C, et al. Metabolic evolution of energy-conserving pathways for succinate production in Escherichia coli [J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(48): 20180-20185. |
| 29 | CHEN J, ZHU X N, TAN Z G, et al. Activating C-4-dicarboxylate transporters DcuB and DcuC for improving succinate production[J]. Applied Microbiology and Biotechnology, 2014, 98(5): 2197-2205. |
| 30 | ZHU X N, TAN Z G, XU H T, et al. Metabolic evolution of two reducing equivalent-conserving pathways for high-yield succinate production in Escherichia coli [J]. Metabolic Engineering, 2014, 24: 87-96. |
| 31 | TAN Z G, CHEN J, ZHANG X L. Systematic engineering of pentose phosphate pathway improves Escherichia coli succinate production[J]. Biotechnology for Biofuels, 2016, 9: 13. |
| 32 | YU Y, ZHU X N, XU H T, et al. Construction of an energy-conserving glycerol utilization pathways for improving anaerobic succinate production in Escherichia coli [J]. Metabolic Engineering, 2019, 56: 181-189. |
| 33 | XIAO M Y, ZHU X N, FAN F Y, et al. Osmotolerance in Escherichia coli is improved by activation of copper efflux genes or supplementation with sulfur-containing amino acids[J]. Applied and Environmental Microbiology, 2017, 83(7): 11. |
| 34 | 张学礼, 张冬竹. 一种高产L-丙氨酸的XZ-A26菌株及构建方法与应用: CN102329765A[P]. 2011-08-17. |
| ZHANG X L, ZHANG D Z. High-yield L-alanine XZ-A26 and construction method and application thereof: CN102329765A[P]. 2011-08-17. | |
| 35 | 马桂燕. 2015年蛋氨酸市场回顾及2016年趋势展望[J]. 中国畜牧业, 2016(10): 50-51. |
| MA G Y. Review of methionine market in 2015 and prospect of trend in 2016[J]. China Animal Industry, 2016(10): 50-51. | |
| 36 | 希杰L-蛋氨酸生物利用率高出 40 %[J]. 中国食品工业, 2015(2): 27. |
| The bioavailability of CJ L-methionine was 40 % higher[J]. China Food, 2015(2): 27. | |
| 37 | HUANG J F, SHEN Z Y, MAO Q L, et al. Systematic analysis of bottlenecks in a multibranched and multilevel regulated pathway: the molecular fundamentals of L-methionine biosynthesis in Escherichia coli [J]. ACS Synthetic Biology, 2018, 7(11): 2577-2589. |
| 38 | DIETRICH J A, FORTMAN J L, STEEN E J. Recombinant host cells for the production of malonate: US 10472653[P]. 2019-11-12. |
| 39 | KNUF C, NOOKAEW I, REMMERS I, et al. Physiological characterization of the high malic acid-producing Aspergillus oryzae strain 2103a-68[J]. Applied Microbiology and Biotechnology, 2014, 98(8): 3517-3527. |
| 40 | LI W N, MA L, SHEN X L, et al. Targeting metabolic driving and intermediate influx in lysine catabolism for high-level glutarate production[J]. Nature Communications, 2019, 10: 8. |
| 41 | ZHAO M, HUANG D X, ZHANG X J, et al. Metabolic engineering of Escherichia coli for producing adipic acid through the reverse adipate-degradation pathway[J]. Metabolic Engineering, 2018, 47: 254-262. |
| 42 | NAKAMURA C E, WHITED G M. Metabolic engineering for the microbial production of 1,3-propanediol[J]. Current Opinion in Biotechnology, 2003, 14(5): 454-459. |
| 43 | YIM H, HASELBECK R, NIU W, et al. Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol[J]. Nature Chemical Biology, 2011, 7(7): 445-452. |
| 44 | DIEN S J VAN, BURGARD A P, HASELBECK R, et al. Microorganisms for the production of 1,4 -butanediol and related methods: US 10273508[P]. 2019-04-30. |
| 45 | ATSUMI S, HANAI T, LIAO J C. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels[J]. Nature, 2008, 451(7174): 86-89. |
| 46 | SHELDON R A, PEREIRA P C. Biocatalysis engineering: the big picture[J]. Chemical Society Reviews, 2017, 46(10): 2678-2691. |
| 47 | 曲戈, 朱彤, 蒋迎迎, 等. 蛋白质工程:从定向进化到计算设计[J]. 生物工程学报, 2019, 35(10): 1843-1856. |
| QU G, ZHU T, JIANG Y Y, et al. Protein engineering: from directed evolution to computational design[J]. Chinese Journal of Biotechnology, 2019, 35(10): 1843-1856. | |
| 48 | YU D, WANG J B, REETZ M T. Exploiting designed oxidase-peroxygenase mutual benefit system for asymmetric cascade reactions[J]. Journal of the American Chemical Society, 2019, 141(14): 5655-5658. |
| 49 | The runners-up[J]. Science, 2016, 354(6319): 1518-1523. |
| 50 | LEAVER-FAY A, TYKA M, LEWIS S M, et al. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules[J]. Methods in Enzymology, 2011, 487: 545-574. |
| 51 | SIEGEL J B, SMITH A L, POUST S, et al. Computational protein design enables a novel one-carbon assimilation pathway[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(12): 3704-3709. |
| 52 | JIANG L, ALTHOFF E A, CLEMENTE F R, et al. De novo computational design of retro-aldol enzymes[J]. Science, 2008, 319(5868): 1387-1391. |
| 53 | SIEGEL J B, ZANGHELLINI A, LOVICK H M, et al. Computational design of an enzyme catalyst for a stereoselective bimolecular Diels-Alder reaction[J]. Science, 2010, 329(5989): 309-313. |
| 54 | LU X Y, LIU Y W, YANG Y Q, et al. Constructing a synthetic pathway for acetylcoenzyme A from one-carbon through enzyme design[J]. Nature Communications, 2019, 10(1): 1-10. |
| 55 | LI R, WIJMA H J, SONG L, et al. Computational redesign of enzymes for regio- and enantioselective hydroamination[J]. Nature Chemical Biology, 2018, 14(7): 664-670. |
| [1] | 郭姝媛, 张倩楠, 姑丽克孜·买买提热夏提, 杨一群, 于涛. 液体生物燃料合成与炼制的研究进展[J]. 合成生物学, 2025, 6(1): 18-44. |
| [2] | 应汉杰, 柳东, 王振宇, 沈涛, 庄伟, 朱晨杰. 工业生物制造与“碳中和”目标探讨[J]. 合成生物学, 2025, 6(1): 1-7. |
| [3] | 高歌, 边旗, 王宝俊. 合成基因线路的工程化设计研究进展与展望[J]. 合成生物学, 2025, 6(1): 45-64. |
| [4] | 李冀渊, 吴国盛. 合成生物学视域下有机体的两种隐喻[J]. 合成生物学, 2025, 6(1): 190-202. |
| [5] | 焦洪涛, 齐蒙, 邵滨, 蒋劲松. DNA数据存储技术的法律治理议题[J]. 合成生物学, 2025, 6(1): 177-189. |
| [6] | 唐兴华, 陆钱能, 胡翌霖. 人类世中对合成生物学的哲学反思[J]. 合成生物学, 2025, 6(1): 203-212. |
| [7] | 徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176. |
| [8] | 石婷, 宋展, 宋世怡, 张以恒. 体外生物转化(ivBT):生物制造的新前沿[J]. 合成生物学, 2024, 5(6): 1437-1460. |
| [9] | 柴猛, 王风清, 魏东芝. 综合利用木质纤维素生物转化合成有机酸[J]. 合成生物学, 2024, 5(6): 1242-1263. |
| [10] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [11] | 赵亮, 李振帅, 付丽平, 吕明, 王士安, 张全, 刘立成, 李福利, 刘自勇. 生物转化一碳化合物原料产油脂与单细胞蛋白研究进展[J]. 合成生物学, 2024, 5(6): 1300-1318. |
| [12] | 竺方欢, 岑雪聪, 陈振. 微生物合成二元醇研究进展[J]. 合成生物学, 2024, 5(6): 1367-1385. |
| [13] | 陈雨, 张康, 邱以婧, 程彩云, 殷晶晶, 宋天顺, 谢婧婧. 微生物电合成技术转化二氧化碳研究进展[J]. 合成生物学, 2024, 5(5): 1142-1168. |
| [14] | 郑皓天, 李朝风, 刘良叙, 王嘉伟, 李恒润, 倪俊. 负碳人工光合群落的设计、优化与应用[J]. 合成生物学, 2024, 5(5): 1189-1210. |
| [15] | 夏孔晨, 徐维华, 吴起. 光酶催化混乱性反应的研究进展[J]. 合成生物学, 2024, 5(5): 997-1020. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||