合成生物学 ›› 2023, Vol. 4 ›› Issue (2): 244-262.DOI: 10.12211/2096-8280.2022-059
合成生物学在疾病诊疗中的应用
吴晓昊1, 廖荣东2, 李飞云1, 欧阳中天1, 冉怡1, 公维远1, 曲明灏1, 陈明珏1, 林荔军2, 肖国芝1
- 1.南方科技大学医学院,广东 深圳 518055
2.南方医科大学珠江医院关节骨病外科,广东 广州 510280
-
收稿日期:2022-10-31修回日期:2023-02-01出版日期:2023-04-30发布日期:2023-04-27 -
通讯作者:林荔军,肖国芝 -
作者简介:吴晓昊 (1991—),男,博士研究生。研究方向为骨关节稳态和疾病发生的分子机制。E-mail:wxho0606@163.com廖荣东 (1996—),男,博士研究生。研究方向为骨科疾病的发病机制。E-mail:lrdyxs12138@163.com林荔军 (1976—),男,博士,主任医师,博士生导师。主要从事合成生物学与骨科疾病诊治领域方面的研究,利用合成生物学方法构建新型材料用于骨关节炎、骨折不愈合等骨科多发疾病的治疗,研究方向为骨关节炎发病分子机制研究、骨肉瘤侵袭和转移的分子机制研究、骨组织3D打印和生物力学研究学。E-mail:gost1@smu.edu.cn肖国芝 (1963—),男,教授,乌克兰国家工程院外籍院士。主要研究方向是骨骼发育和疾病的相关分子基础。E-mail:xiaogz@sustech.edu.cn -
基金资助:国家重点研发计划(2019YFA0906004);国家自然科学基金(82230081)
Applications of synthetic biology in disease diagnosis and treatment
WU Xiaohao1, LIAO Rongdong2, LI Feiyun1, OUYANG Zhongtian1, RAN Yi1, GONG Weiyuan1, QU Minghao1, CHEN Mingjue1, LIN Lijun2, XIAO Guozhi1
- 1.School of Medicine,Southern University of Science and Technology,Shenzhen 518055,Guangdong,China
2.Department of Orthopedics,Zhujiang Hospital,Southern Medical University,Guangzhou 510280,Guangdong,China
-
Received:2022-10-31Revised:2023-02-01Online:2023-04-30Published:2023-04-27 -
Contact:LIN Lijun, XIAO Guozhi
摘要:
合成生物学是一门新兴学科。从广义上讲,合成生物学是通过将基因工程、系统生物学、计算机工程等多学科作为工具,根据特定需求进行设计,乃至重新合成生物体系。近20年来,合成生物学领域的相关研究不断取得突破,并已在针对人类疾病的诊断、临床治疗、药物研发等诸多方面获得重要应用。合成生物学不仅为疾病的早期、精确诊断提供新的思路和技术手段,还发展出多种新型的疾病治疗手段,包括基于合成生物学原理设计的细胞疗法、细菌疗法、疫苗、生物医学材料等。利用合成生物学方法,我们可以精确诊断早期疾病、精准改造细胞或细菌、进行疾病机制研究和药物筛选、快速生产新型疫苗和生物医学材料。基于合成生物学的疾病诊疗方法将是科研领域重要的发展方向之一,并将在未来彻底改变临床疾病的诊疗方式。本文综述合成生物学原理和技术在疾病诊断和治疗中的应用,并进一步探讨合成生物学在疫苗生产、生物医学材料、新药研发等方面的应用。
中图分类号:
引用本文
吴晓昊, 廖荣东, 李飞云, 欧阳中天, 冉怡, 公维远, 曲明灏, 陈明珏, 林荔军, 肖国芝. 合成生物学在疾病诊疗中的应用[J]. 合成生物学, 2023, 4(2): 244-262.
WU Xiaohao, LIAO Rongdong, LI Feiyun, OUYANG Zhongtian, RAN Yi, GONG Weiyuan, QU Minghao, CHEN Mingjue, LIN Lijun, XIAO Guozhi. Applications of synthetic biology in disease diagnosis and treatment[J]. Synthetic Biology Journal, 2023, 4(2): 244-262.
| 项目 | 酶传感器 | 质粒传感器 | 哺乳动物细胞传感器 | 细菌传感器 |
|---|---|---|---|---|
| 原始信号 | 酶活性失调 | 转录活性改变 | 转录或代谢活性改变 | 生化特性改变(如营养物质,pH等) |
| 放大方式 | 催化放大 尿液浓缩 | 分泌物增加 | 细胞增殖 分泌物增加 | 细菌增殖 催化放大 尿液浓缩 |
| 信号特征 | 底物信号扩大 | 特异性转录 | 肿瘤微环境驱动信号激活 | 特异性肿瘤靶向浸润 |
| 检测方式 | 尿液,血液,呼吸 | 尿液,血液 | 尿液,血液 | 尿液,血液 |
表1 合成生物学在癌症诊断中的生物传感器特点
Table 1 Biosensors developed through synthetic biology for cancer diagnosis
| 项目 | 酶传感器 | 质粒传感器 | 哺乳动物细胞传感器 | 细菌传感器 |
|---|---|---|---|---|
| 原始信号 | 酶活性失调 | 转录活性改变 | 转录或代谢活性改变 | 生化特性改变(如营养物质,pH等) |
| 放大方式 | 催化放大 尿液浓缩 | 分泌物增加 | 细胞增殖 分泌物增加 | 细菌增殖 催化放大 尿液浓缩 |
| 信号特征 | 底物信号扩大 | 特异性转录 | 肿瘤微环境驱动信号激活 | 特异性肿瘤靶向浸润 |
| 检测方式 | 尿液,血液,呼吸 | 尿液,血液 | 尿液,血液 | 尿液,血液 |

图1 在CAR-T细胞中引入的“逻辑门”系统“与门”:当synthetic notch(SynNotch)受体识别并结合肿瘤抗原时,激活CAR的表达,由CAR识别另一肿瘤抗原后激活T细胞并介导其对肿瘤的杀伤功能(如图左上所示)。另一种“与门”:弱活化信号的CAR和共刺激受体(co-stimulatory receptor, CCR)均不足以单独提供激活T细胞的信号。当两个受体共同识别出对应肿瘤抗原时,才足以激活T细胞功能(如图右上所示)。“或门”:串联表达不同CAR的单链可变区片段(single-chain variable fragment, ScFv),任一ScFv片段识别到对应肿瘤抗原都可以激活T细胞功能(如图左下所示)。“非门”:当CAR识别到肿瘤抗原后激活T细胞,同时当inhibitory CAR(iCAR)识别到正常细胞抗原时,iCAR发出抑制信号抑制CAR的激活信号从而保护正常细胞(如图右下所示)
Fig. 1 Diagram for the logic gate to CAR-T cellsAND gate: When the synthetic notch (SynNotch) receptor recognizes and binds to a tumor antigen, it activates the expression of CAR to recognizes another tumor antigen for activating T cells to mediate its killing function against tumors (as shown in the upper left of the figure). Another AND gate: both CAR and CCR with weak activation signals are not sufficient to activate T cells. When the two receptors jointly recognize the corresponding tumor antigen, the combined signal is sufficient to activate T cell for function (as shown in the upper right of the figure). OR gate: any ScFv fragment expressing different CARs in tandem can activate T cell for function when it recognizes the corresponding tumor antigen (as shown in the lower left of the figure). Not gate: CAR recognizes tumor antigens to activate T cells, but when iCAR recognizes normal cell antigens, it sends out an inhibitory signal to inhibit the activation signal of CAR for the protection of normal cells (as shown in the lower right of the figure)
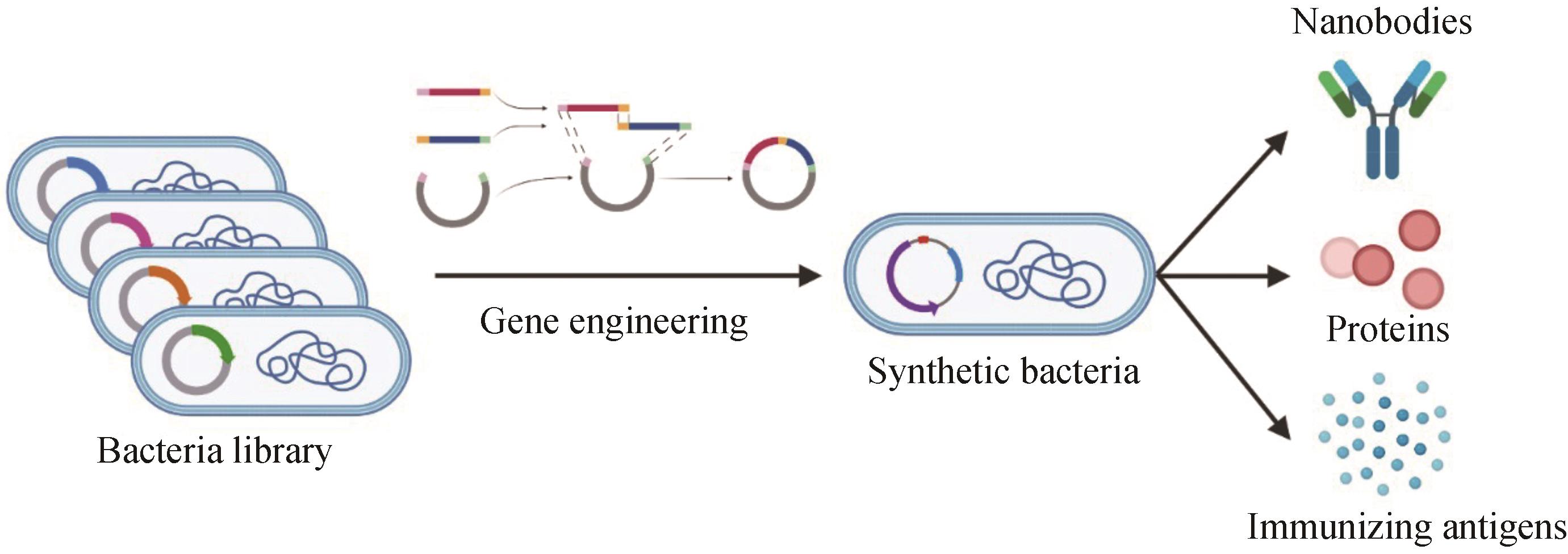
图2 工程细菌生物合成和细菌疗法原理示意图运用合成生物学应用工程原理,将特化和标准化的DNA序列或基因编码产物等基序混合匹配构建为功能齐全的遗传装置并将其集成到底盘细菌中,最终构建出一个能够产生纳米抗体、分泌特定蛋白或表达免疫抗原刺激免疫通路激活的工程细菌
Fig. 2 Schematic diagram for engineering bacterial biosynthesis and corresponding therapyWith the engineering principles of synthetic biology, specialized and standardized DNA sequences or gene coding products and other sequence motifs are mixed and matched to construct fully functional genetic devices for integrating them into chassis bacteria to finally develop an engineered bacteria that can produce nanobodies, secrete specific proteins or express immune antigens to stimulate the activation of the immune pathways
| 材料种类 | 材料名称 | 材料特性 | 参考文献 |
|---|---|---|---|
| 多糖材料 | 细菌纤维素 | 高物理强度和高保水能力,可生产人造血管支架 | [ |
| 纤维素-几丁质共聚物 | 可作为静脉假体,在动物体内能被降解 | [ | |
| 核酸材料 | ssDNA纳米材料 | 制作病原体传感器,对动物体内的病原体进行探测 | [ |
| RNA纳米材料 | 控制体内细胞代谢过程 | [ | |
| 蛋白质材料 | 弹性蛋白样多肽 | 用于组织工程和药物递送 | [ |
| 弹性蛋白样重组体 | 制作基于ELR的水凝胶,用于传递药物和疫苗 | [ | |
| 活细胞材料 | curli纳米纤维 | 与生物膜融合后可结合在特定表面,用于治疗小鼠胃肠道炎症 | [ |
| 大肠杆菌ECM材料 | 生产抗肿瘤药物——脱氧紫罗兰素 | [ | |
| E.coli Nissle材料 | 一种无毒性的大肠杆菌材料,对患者进行持续给药 | [ | |
| 枯草芽孢杆菌水凝膜 | 具有多功能再生和可调性,可作为潜在的医疗生物材料 | [ |
表2 合成生物医学材料总结
Table 2 Summary of synthetic biomedical materials
| 材料种类 | 材料名称 | 材料特性 | 参考文献 |
|---|---|---|---|
| 多糖材料 | 细菌纤维素 | 高物理强度和高保水能力,可生产人造血管支架 | [ |
| 纤维素-几丁质共聚物 | 可作为静脉假体,在动物体内能被降解 | [ | |
| 核酸材料 | ssDNA纳米材料 | 制作病原体传感器,对动物体内的病原体进行探测 | [ |
| RNA纳米材料 | 控制体内细胞代谢过程 | [ | |
| 蛋白质材料 | 弹性蛋白样多肽 | 用于组织工程和药物递送 | [ |
| 弹性蛋白样重组体 | 制作基于ELR的水凝胶,用于传递药物和疫苗 | [ | |
| 活细胞材料 | curli纳米纤维 | 与生物膜融合后可结合在特定表面,用于治疗小鼠胃肠道炎症 | [ |
| 大肠杆菌ECM材料 | 生产抗肿瘤药物——脱氧紫罗兰素 | [ | |
| E.coli Nissle材料 | 一种无毒性的大肠杆菌材料,对患者进行持续给药 | [ | |
| 枯草芽孢杆菌水凝膜 | 具有多功能再生和可调性,可作为潜在的医疗生物材料 | [ |
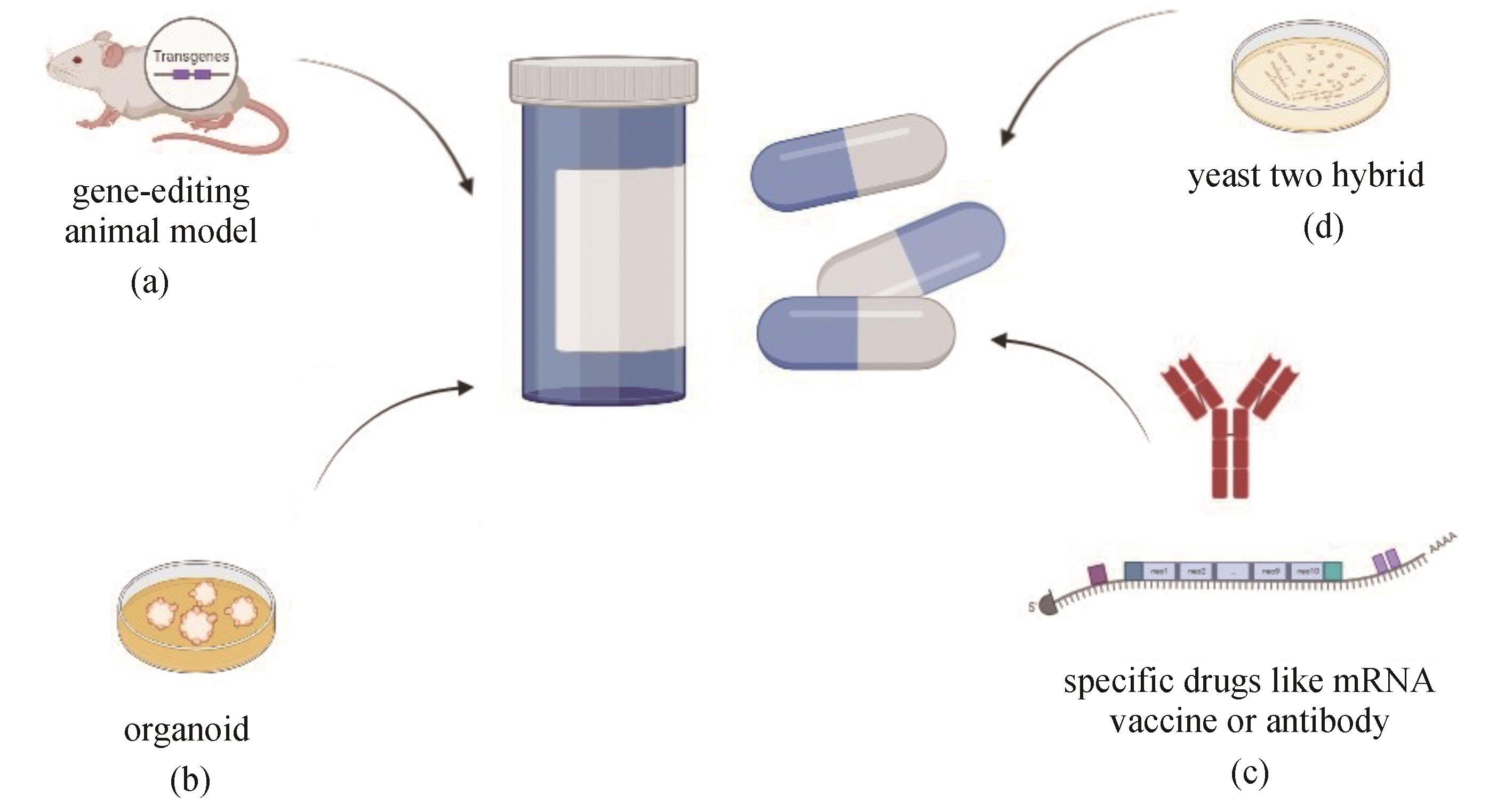
图3 合成生物学在新药研发中的应用(a)在药物机制研究中构建基因编辑动物模型;(b)在药物筛选与机制研究中使用类器官模型;(c)生产特异性药物,如mRNA疫苗和特异性抗体;(d)在抗体生产和筛选过程中应用广泛的酵母双杂交技术
Fig. 3 Applications of synthetic biology in drug discovery(a) Building animal models for gene-editing in drug mechanism research; (b) Building organoid models for drug screening and mechanism discovery; (c) Expressing specific drugs like mRNA vaccines or antibodies; (d) Yeast two-hybrid technology for screening and producing antibodies
| 1 | YE H F, FUSSENEGGER M. Synthetic therapeutic gene circuits in mammalian cells[J]. FEBS Letters, 2014, 588(15): 2537-2544. |
| 2 | ELOWITZ M B, LEIBLER S. A synthetic oscillatory network of transcriptional regulators[J]. Nature, 2000, 403(6767): 335-338. |
| 3 | GARDNER T S, CANTOR C R, COLLINS J J. Construction of a genetic toggle switch in Escherichia coli [J]. Nature, 2000, 403(6767): 339-342. |
| 4 | KRAMER B P, FISCHER C, FUSSENEGGER M. BioLogic gates enable logical transcription control in mammalian cells[J]. Biotechnology and Bioengineering, 2004, 87(4): 478-484. |
| 5 | BECSKEI A, SERRANO L. Engineering stability in gene networks by autoregulation[J]. Nature, 2000, 405(6786): 590-593. |
| 6 | KITADA T, DIANDRETH B, TEAGUE B, et al. Programming gene and engineered-cell therapies with synthetic biology[J]. Science, 2018, 359(6376): eaad1067. |
| 7 | BATEY R T, GILBERT S D, MONTANGE R K. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine[J]. Nature, 2004, 432(7015): 411-415. |
| 8 | BADORREK C S, GHERGHE C M, WEEKS K M. Structure of an RNA switch that enforces stringent retroviral genomic RNA dimerization[J]. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(37): 13640-13645. |
| 9 | SIUTI P, YAZBEK J, LU T K. Synthetic circuits integrating logic and memory in living cells[J]. Nature Biotechnology, 2013, 31(5): 448-452. |
| 10 | KEMMER C, GITZINGER M, DAOUD-EL BABA M, et al. Self-sufficient control of urate homeostasis in mice by a synthetic circuit[J]. Nature Biotechnology, 2010, 28(4): 355-360. |
| 11 | RICHARDS R M, ZHAO F F, FREITAS K A, et al. NOT-gated CD93 CAR T cells effectively target AML with minimized endothelial cross-reactivity[J]. Blood Cancer Discovery, 2021, 2(6): 648-665. |
| 12 | GIBSON D G, YOUNG L, CHUANG R Y, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases[J]. Nature Methods, 2009, 6(5): 343-345. |
| 13 | MCNERNEY M P, DOIRON K E, NG T L, et al. Theranostic cells: emerging clinical applications of synthetic biology[J]. Nature Reviews Genetics, 2021, 22(11): 730-746. |
| 14 | ENDY D. Foundations for engineering biology[J]. Nature, 2005, 438(7067): 449-453. |
| 15 | SLUSARCZYK A L, LIN A, WEISS R. Foundations for the design and implementation of synthetic genetic circuits[J]. Nature Reviews Genetics, 2012, 13(6): 406-420. |
| 16 | WANG Y H, WEI K Y, SMOLKE C D. Synthetic biology: advancing the design of diverse genetic systems[J]. Annual Review of Chemical and Biomolecular Engineering, 2013, 4: 69-102. |
| 17 | ISHINO Y, SHINAGAWA H, MAKINO K, et al. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product[J]. Journal of Bacteriology, 1987, 169(12): 5429-5433. |
| 18 | YIN H, XUE W, CHEN S D, et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype[J]. Nature Biotechnology, 2014, 32(6): 551-553. |
| 19 | BAKONDI B, LV W J, LU B, et al. In vivo CRISPR/Cas9 gene editing corrects retinal dystrophy in the S334ter-3 rat model of autosomal dominant retinitis pigmentosa[J]. Molecular Therapy, 2016, 24(3): 556-563. |
| 20 | LIANG C, LI F F, WANG L Y, et al. Tumor cell-targeted delivery of CRISPR/Cas9 by aptamer-functionalized lipopolymer for therapeutic genome editing of VEGFA in osteosarcoma[J]. Biomaterials, 2017, 147: 68-85. |
| 21 | KAN M J, DOUDNA J A. Treatment of genetic diseases with CRISPR genome editing[J]. JAMA, 2022, 328(10): 980-981. |
| 22 | SOLEIMANY A P, BHATIA S N. Activity-based diagnostics: an emerging paradigm for disease detection and monitoring[J]. Trends in Molecular Medicine, 2020, 26(5): 450-468. |
| 23 | SLOMOVIC S, PARDEE K, COLLINS J J. Synthetic biology devices for in vitro and in vivo diagnostics[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(47): 14429-14435. |
| 24 | TIAN Q, PRICE N D, HOOD L. Systems cancer medicine: towards realization of predictive, preventive, personalized and participatory (P4) medicine[J]. Journal of Internal Medicine, 2012, 271(2): 111-121. |
| 25 | SIU A L. Screening for breast cancer: U.S. preventive services task force recommendation statement[J]. Annals of Internal Medicine, 2016, 164(4): 279-296. |
| 26 | US Preventive Services Task Force. Screening for colorectal cancer: US preventive services task force recommendation statement[J]. JAMA, 2016, 315(23): 2564-2575. |
| 27 | SIEGEL R L, MILLER K D, JEMAL A. Cancer statistics, 2020[J]. CA: A Cancer Journal for Clinicians, 2020, 70(1): 7-30. |
| 28 | LUTZ A M, WILLMANN J K, COCHRAN F V, et al. Cancer screening: a mathematical model relating secreted blood biomarker levels to tumor sizes[J]. PLoS Medicine, 2008, 5(8): e170. |
| 29 | KWONG G A, GHOSH S, GAMBOA L, et al. Synthetic biomarkers: a twenty-first century path to early cancer detection[J]. Nature Reviews Cancer, 2021, 21(10): 655-668. |
| 30 | HANAHAN D, WEINBERG R A. Hallmarks of cancer: the next generation[J]. Cell, 2011, 144(5): 646-674. |
| 31 | KWON E J, DUDANI J S, BHATIA S N. Ultrasensitive tumour-penetrating nanosensors of protease activity[J]. Nature Biomedical Engineering, 2017, 1: 54. |
| 32 | KESSENBROCK K, PLAKS V, WERB Z. Matrix metalloproteinases: regulators of the tumor microenvironment[J]. Cell, 2010, 141(1): 52-67. |
| 33 | DUDANI J S, IBRAHIM M, KIRKPATRICK J, et al. Classification of prostate cancer using a protease activity nanosensor library[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(36): 8954-8959. |
| 34 | KWONG G A, VON MALTZAHN G, MURUGAPPAN G, et al. Mass-encoded synthetic biomarkers for multiplexed urinary monitoring of disease[J]. Nature Biotechnology, 2013, 31(1): 63-70. |
| 35 | DUDANI J S, WARREN A D, BHATIA S N. Harnessing protease activity to improve cancer care[J]. Annual Review of Cancer Biology, 2018, 2: 353-376. |
| 36 | BHANG H E C, GABRIELSON K L, LATERRA J, et al. Tumor-specific imaging through progression elevated gene-3 promoter-driven gene expression[J]. Nature Medicine, 2011, 17(1): 123-129. |
| 37 | AALIPOUR A, CHUANG H Y, MURTY S, et al. Engineered immune cells as highly sensitive cancer diagnostics[J]. Nature Biotechnology, 2019, 37(5): 531-539. |
| 38 | NIU G, CHEN X Y. Molecular imaging with activatable reporter systems[J]. Theranostics, 2012, 2(4): 413-423. |
| 39 | REAGAN M R, KAPLAN D L. Concise review: mesenchymal stem cell tumor-homing: detection methods in disease model systems[J]. Stem Cells, 2011, 29(6): 920-927. |
| 40 | ZHOU S B, GRAVEKAMP C, BERMUDES D, et al. Tumour-targeting bacteria engineered to fight cancer[J]. Nature Reviews Cancer, 2018, 18(12): 727-743. |
| 41 | WEI T Y, CHENG C M. Synthetic biology-based point-of-care diagnostics for infectious disease[J]. Cell Chemical Biology, 2016, 23(9): 1056-1066. |
| 42 | SHORR A F, MICEK S T, JACKSON W L, et al. Economic implications of an evidence-based sepsis protocol: can we improve outcomes and lower costs?[J]. Critical Care Medicine, 2007, 35(5): 1257-1262. |
| 43 | MANI V, WANG S Q, INCI F, et al. Emerging technologies for monitoring drug-resistant tuberculosis at the point-of-care[J]. Advanced Drug Delivery Reviews, 2014, 78: 105-117. |
| 44 | COURBET A, ENDY D, RENARD E, et al. Detection of pathological biomarkers in human clinical samples via amplifying genetic switches and logic gates[J]. Science Translational Medicine, 2015, 7(289): 289ra83. |
| 45 | DONALDSON T, DATTELBAUM J D. Designing a thermostable switch-based biosensor[J]. Biophysical Journal, 2014, 106(2): 810a. |
| 46 | SILVERMAN A D, KARIM A S, JEWETT M C. Cell-free gene expression: an expanded repertoire of applications[J]. Nature Reviews Genetics, 2020, 21(3): 151-170. |
| 47 | TAN X, LETENDRE J H, COLLINS J J, et al. Synthetic biology in the clinic: engineering vaccines, diagnostics, and therapeutics[J]. Cell, 2021, 184(4): 881-898. |
| 48 | GREEN A A, SILVER P A, COLLINS J J, et al. Toehold switches: de-novo-designed regulators of gene expression[J]. Cell, 2014, 159(4): 925-939. |
| 49 | MAKAROVA K S, WOLF Y I, IRANZO J, et al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants[J]. Nature Reviews Microbiology, 2020, 18(2): 67-83. |
| 50 | LESLIE H H, SPIEGELMAN D, ZHOU X, et al. Service readiness of health facilities in Bangladesh, Haiti, Kenya, Malawi, Namibia, Nepal, Rwanda, Senegal, Uganda and the United Republic of Tanzania[J]. Bulletin of the World Health Organization, 2017, 95(11): 738-748. |
| 51 | JUNE C H, SADELAIN M. Chimeric antigen receptor therapy[J]. New England Journal of Medicine, 2018, 379(1): 64-73. |
| 52 | LANITIS E, DANGAJ D, IRVING M, et al. Mechanisms regulating T-cell infiltration and activity in solid tumors[J]. Annals of Oncology, 2017, 28(): xii18-xii32. |
| 53 | LANITIS E, IRVING M, COUKOS G. Targeting the tumor vasculature to enhance T cell activity[J]. Current Opinion in Immunology, 2015, 33: 55-63. |
| 54 | KLOSS C C, CONDOMINES M, CARTELLIERI M, et al. Combinatorial antigen recognition with balanced signaling promotes selective tumor eradication by engineered T cells[J]. Nature Biotechnology, 2013, 31(1): 71-75. |
| 55 | WU M R, JUSIAK B, LU T K. Engineering advanced cancer therapies with synthetic biology[J]. Nature Reviews Cancer, 2019, 19(4): 187-195. |
| 56 | FEDOROV V D, THEMELI M, SADELAIN M. PD-1- and CTLA-4-based inhibitory chimeric antigen receptors (iCARs) divert off-target immunotherapy responses[J]. Science Translational Medicine, 2013, 5(215): 215ra172. |
| 57 | SADELAIN M. Chimeric antigen receptors: a paradigm shift in immunotherapy[J]. Annual Review of Cancer Biology, 2017, 1: 447-466. |
| 58 | XU N, PALMER D C, ROBESON A C, et al. STING agonist promotes CAR T cell trafficking and persistence in breast cancer[J]. The Journal of Experimental Medicine, 2021, 218(2): e20200844. |
| 59 | LÓPEZ-LÁZARO M. The migration ability of stem cells can explain the existence of cancer of unknown primary site. Rethinking metastasis[J]. Oncoscience, 2015, 2(5): 467-475. |
| 60 | LANDSKRON G, DE LA FUENTE M, THUWAJIT P, et al. Chronic inflammation and cytokines in the tumor microenvironment[J]. Journal of Immunology Research, 2014, 2014: 149185. |
| 61 | ABOODY K S, NAJBAUER J, DANKS M K. Stem and progenitor cell-mediated tumor selective gene therapy[J]. Gene Therapy, 2008, 15(10): 739-752. |
| 62 | SPAETH E, KLOPP A, DEMBINSKI J, et al. Inflammation and tumor microenvironments: defining the migratory itinerary of mesenchymal stem cells[J]. Gene Therapy, 2008, 15(10): 730-738. |
| 63 | TAKAYAMA Y, KUSAMORI K, NISHIKAWA M. Click chemistry as a tool for cell engineering and drug delivery[J]. Molecules, 2019, 24(1): 172. |
| 64 | LAYEK B, SADHUKHA T, PRABHA S. Glycoengineered mesenchymal stem cells as an enabling platform for two-step targeting of solid tumors[J]. Biomaterials, 2016, 88: 97-109. |
| 65 | HARGADON K M, JOHNSON C E, WILLIAMS C J. Immune checkpoint blockade therapy for cancer: an overview of FDA-approved immune checkpoint inhibitors[J]. International Immunopharmacology, 2018, 62: 29-39. |
| 66 | DARVIN P, TOOR S M, SASIDHARAN NAIR V, et al. Immune checkpoint inhibitors: recent progress and potential biomarkers[J]. Experimental & Molecular Medicine, 2018, 50(12): 1-11. |
| 67 | HU Q Y, SUN W J, WANG J Q, et al. Conjugation of haematopoietic stem cells and platelets decorated with anti-PD-1 antibodies augments anti-leukaemia efficacy[J]. Nature Biomedical Engineering, 2018, 2(11): 831-840. |
| 68 | STÜDEMANN T, RÖSSINGER J, MANTHEY C, et al. Contractile force of transplanted cardiomyocytes actively supports heart function after injury[J]. Circulation, 2022, 146(15): 1159-1169. |
| 69 | LLOYD-PRICE J, ABU-ALI G, HUTTENHOWER C. The healthy human microbiome[J]. Genome Medicine, 2016, 8(1): 51. |
| 70 | YU H. Bacteria-mediated disease therapy[J]. Applied Microbiology and Biotechnology, 2011, 92(6): 1107-1113. |
| 71 | FLORES BUESO Y, LEHOURITIS P, TANGNEY M. In situ biomolecule production by bacteria; a synthetic biology approach to medicine[J]. Journal of Controlled Release: Official Journal of the Controlled Release Society, 2018, 275: 217-228. |
| 72 | LIM B, YIN Y T, YE H, et al. Reprogramming synthetic cells for targeted cancer therapy[J]. ACS Synthetic Biology, 2022, 11(3): 1349-1360. |
| 73 | CHOWDHURY S, CASTRO S, COKER C, et al. Programmable bacteria induce durable tumor regression and systemic antitumor immunity[J]. Nature Medicine, 2019, 25(7): 1057-1063. |
| 74 | GURBATRI C R, LIA I, VINCENT R, et al. Engineered probiotics for local tumor delivery of checkpoint blockade nanobodies[J]. Science Translational Medicine, 2020, 12(530): eaax0876. |
| 75 | LEVENTHAL D S, SOKOLOVSKA A, LI N, et al. Immunotherapy with engineered bacteria by targeting the STING pathway for anti-tumor immunity[J]. Nature Communications, 2020, 11: 2739. |
| 76 | 高梦学, 王丽娜, 黄鹤. 合成生物学在肠道微生态疗法研发中的应用[J]. 合成生物学, 2022, 3(1): 35-52. |
| GAO M X, WANG L N, HUANG H. Advances in synthetic biology assisted intestinal microecological therapy[J]. Synthetic Biology Journal, 2022, 3(1): 35-52. | |
| 77 | STEIDLER L, HANS W, SCHOTTE L, et al. Treatment of murine colitis by lactococcus lactis secreting interleukin-10[J]. Science, 2000, 289(5483): 1352-1355. |
| 78 | VANDENBROUCKE K, DE HAARD H, BEIRNAERT E, et al. Orally administered L. lactis secreting an anti-TNF nanobody demonstrate efficacy in chronic colitis[J]. Mucosal Immunology, 2010, 3(1): 49-56. |
| 79 | FANG X, ZHOU X T, MIAO Y Q, et al. Therapeutic effect of GLP-1 engineered strain on mice model of Alzheimer's disease and Parkinson's disease[J]. AMB Express, 2020, 10(1): 80. |
| 80 | CHEN T T, TIAN P Y, HUANG Z X, et al. Engineered commensal bacteria prevent systemic inflammation-induced memory impairment and amyloidogenesis via producing GLP-1[J].Applied Microbiology and Biotechnology, 2018, 102(17): 7565-7575. |
| 81 | CECARINI V, BONFILI L, GOGOI O, et al. Neuroprotective effects of p62(SQSTM1)-engineered lactic acid bacteria in Alzheimer's disease: a pre-clinical study[J]. Aging, 2020, 12(16): 15995-16020. |
| 82 | LINDNER F, DIEPOLD A. Optogenetics in bacteria-applications and opportunities[J]. FEMS Microbiology Reviews, 2022, 46(2): fuab055. |
| 83 | BAUMSCHLAGER A, KHAMMASH M. Synthetic biological approaches for optogenetics and tools for transcriptional light-control in bacteria[J]. Advanced Biology, 2021, 5(5): e2000256. |
| 84 | WEI J J, Jin F. Illuminating bacterial behaviors with optogenetics[J]. Current Opinion in Solid State and Materials Science, 2022, 26(6): 101023. |
| 85 | TANDAR S T, SENOO S, TOYA Y, et al. Optogenetic switch for controlling the central metabolic flux of Escherichia coli [J]. Metabolic Engineering, 2019, 55: 68-75. |
| 86 | MIYAKE K, ABE K, FERRI S, et al. A green-light inducible lytic system for cyanobacterial cells[J].Biotechnology for Biofuels, 2014, 7: 56. |
| 87 | LI X, ZHANG C C, XU X P, et al. A single-component light sensor system allows highly tunable and direct activation of gene expression in bacterial cells[J]. Nucleic Acids Research, 2020, 48(6): e33. |
| 88 | ZHANG J Y, LUO Y H, POH C L. Blue light-directed cell migration, aggregation, and patterning[J]. Journal of Molecular Biology, 2020, 432(10): 3137-3148. |
| 89 | CHEN X J, LIU R M, MA Z C, et al. An extraordinary stringent and sensitive light-switchable gene expression system for bacterial cells[J]. Cell Research, 2016, 26(7): 854-857. |
| 90 | SUN R, LIU M Z, LU J P, et al. Bacteria loaded with glucose polymer and photosensitive ICG silicon-nanoparticles for glioblastoma photothermal immunotherapy[J]. Nature Communications, 2022, 13: 5127. |
| 91 | KANEKIYO M, ELLIS D, KING N P. New vaccine design and delivery technologies[J]. The Journal of Infectious Diseases, 2019, 219(S1): S88-S96. |
| 92 | SKWARCZYNSKI M, TOTH I. Peptide-based synthetic vaccines[J]. Chemical Science, 2016, 7(2): 842-854. |
| 93 | ALLEMAN M M, JORBA J, GREENE S A, et al. Update on vaccine-derived poliovirus outbreaks-worldwide, July 2019-February 2020[J]. MMWR Morbidity and Mortality Weekly Report, 2020, 69(16): 489-495. |
| 94 | POLACK F P, HOFFMAN S J, CRUJEIRAS G, et al. A role for nonprotective complement-fixing antibodies with low avidity for measles virus in atypical measles[J]. Nature Medicine, 2003, 9(9): 1209-1213. |
| 95 | LE NOUËN C, COLLINS P L, BUCHHOLZ U J. Attenuation of human respiratory viruses by synonymous genome recoding[J]. Frontiers in Immunology, 2019, 10: 1250. |
| 96 | COLEMAN J R, PAPAMICHAIL D, SKIENA S, et al. Virus attenuation by genome-scale changes in codon pair bias[J]. Science, 2008, 320(5884): 1784-1787. |
| 97 | BURNS C C, SHAW J, CAMPAGNOLI R, et al. Modulation of poliovirus replicative fitness in HeLa cells by deoptimization of synonymous codon usage in the capsid region[J]. Journal of Virology, 2006, 80(7): 3259-3272. |
| 98 | SCHLUB T E, BUCHMANN J P, HOLMES E C. A simple method to detect candidate overlapping genes in viruses using single genome sequences[J]. Molecular Biology and Evolution, 2018, 35(10): 2572-2581. |
| 99 | ATHEY J, ALEXAKI A, OSIPOVA E, et al. A new and updated resource for codon usage tables[J]. BMC Bioinformatics, 2017, 18(1): 391. |
| 100 | TULLOCH F, ATKINSON N J, EVANS D J, et al. RNA virus attenuation by codon pair deoptimisation is an artefact of increases in CpG/UpA dinucleotide frequencies[J]. eLife, 2014, 3: e04531. |
| 101 | JONES K L, DRANE D, GOWANS E J. Long-term storage of DNA-free RNA for use in vaccine studies[J]. BioTechniques, 2007, 43(5): 675-681. |
| 102 | MCMAHON H T, GALLOP J L. Membrane curvature and mechanisms of dynamic cell membrane remodelling[J]. Nature, 2005, 438(7068): 590-596. |
| 103 | GUPTA A, ANDRESEN J L, MANAN R S, et al. Nucleic acid delivery for therapeutic applications[J]. Advanced Drug Delivery Reviews, 2021, 178: 113834. |
| 104 | WHITEHEAD K A, LANGER R, ANDERSON D G. Knocking down barriers: advances in siRNA delivery[J]. Nature Reviews Drug Discovery, 2009, 8(2): 129-138. |
| 105 | ALLEN T M, CULLIS P R. Liposomal drug delivery systems: from concept to clinical applications[J]. Advanced Drug Delivery Reviews, 2013, 65(1): 36-48. |
| 106 | ALLISON S J, MILNER J. RNA interference by single- and double-stranded siRNA with a DNA extension containing a 3′ nuclease-resistant mini-hairpin structure[J]. Molecular Therapy-Nucleic Acids, 2014, 2: e141. |
| 107 | KUDCHODKAR S B, CHOI H, REUSCHEL E L, et al. Rapid response to an emerging infectious disease - lessons learned from development of a synthetic DNA vaccine targeting Zika virus[J]. Microbes and Infection, 2018, 20(11/12): 676-684. |
| 108 | TEBAS P, KRAYNYAK K A, PATEL A, et al. Intradermal SynCon® Ebola GP DNA vaccine is temperature stable and safely demonstrates cellular and humoral immunogenicity advantages in healthy volunteers[J]. The Journal of Infectious Diseases, 2019, 220(3): 400-410. |
| 109 | WOLFF J A, LUDTKE J J, ACSADI G, et al. Long-term persistence of plasmid DNA and foreign gene expression in mouse muscle[J]. Human Molecular Genetics, 1992, 1(6): 363-369. |
| 110 | LI L, PETROVSKY N. Molecular mechanisms for enhanced DNA vaccine immunogenicity[J]. Expert Review of Vaccines, 2016, 15(3): 313-329. |
| 111 | GAUDINSKI M R, HOUSER K V, MORABITO K M, et al. Safety, tolerability, and immunogenicity of two Zika virus DNA vaccine candidates in healthy adults: randomised, open-label, phase 1 clinical trials[J]. The Lancet, 2018, 391(10120): 552-562. |
| 112 | REICHMUTH A M, OBERLI M A, JAKLENEC A, et al. mRNA vaccine delivery using lipid nanoparticles[J]. Therapeutic Delivery, 2016, 7(5): 319-334. |
| 113 | PARDI N, HOGAN M J, WEISSMAN D. Recent advances in mRNA vaccine technology[J]. Current Opinion in Immunology, 2020, 65: 14-20. |
| 114 | CHEN R, WANG S K, BELK J A, et al. Engineering circular RNA for enhanced protein production[J]. Nature Biotechnology, 2023, 41, 262-272. |
| 115 | WANG J, TAVAKOLI J, TANG Y H. Bacterial cellulose production, properties and applications with different culture methods - a review[J]. Carbohydrate Polymers, 2019, 219: 63-76. |
| 116 | SINGH A, WALKER K T, LEDESMA-AMARO R, et al. Engineering bacterial cellulose by synthetic biology[J]. International Journal of Molecular Sciences, 2020, 21(23): 9185. |
| 117 | MATHUR D, MEDINTZ I L. The growing development of DNA nanostructures for potential healthcare-related applications[J]. Advanced Healthcare Materials, 2019, 8(9): e1801546. |
| 118 | LI M, ZHENG M X, WU S Y, et al. In vivo production of RNA nanostructures via programmed folding of single-stranded RNAs[J]. Nature Communications, 2018, 9: 2196. |
| 119 | MCDANIEL J R, RADFORD D C, CHILKOTI A. A unified model for de novo design of elastin-like polypeptides with tunable inverse transition temperatures[J]. Biomacromolecules, 2013, 14(8): 2866-2872. |
| 120 | IBÁÑEZ-FONSECA A, FLORA T, ACOSTA S, et al. Trends in the design and use of elastin-like recombinamers as biomaterials[J]. Matrix Biology, 2019, 84: 111-126. |
| 121 | DURAJ-THATTE A M, DORVAL COURCHESNE N M, PRAVESCHOTINUNT P, et al. Hydrogels: genetically programmable self-regenerating bacterial hydrogels[J]. Advanced Materials, 2019, 31(40): 1901826. |
| 122 | SANKARAN S, BECKER J, WITTMANN C, et al. Optoregulated drug release from an engineered living material: self-replenishing drug depots for long-term, light-regulated delivery[J]. Small, 2019, 15(5): e1804717. |
| 123 | OU B M, YANG Y, THAM W L, et al. Genetic engineering of probiotic Escherichia coli Nissle 1917 for clinical application[J]. Applied Microbiology and Biotechnology, 2016, 100(20): 8693-8699. |
| 124 | XU L J, WANG X Y, SUN F, et al. Harnessing proteins for engineered living materials[J]. Current Opinion in Solid State and Materials Science, 2021, 25(1): 100896. |
| 125 | RYNGAJŁŁO M, JĘDRZEJCZAK-KRZEPKOWSKA M, KUBIAK K, et al. Towards control of cellulose biosynthesis by Komagataeibacter using systems-level and strain engineering strategies: current progress and perspectives[J]. Applied Microbiology and Biotechnology, 2020, 104(15): 6565-6585. |
| 126 | ELBAZ J, YIN P, VOIGT C A. Genetic encoding of DNA nanostructures and their self-assembly in living bacteria[J]. Nature Communications, 2016, 7: 11179. |
| 127 | VARANKO A K, SU J C, CHILKOTI A. Elastin-like polypeptides for biomedical applications[J]. Annual Review of Biomedical Engineering, 2020, 22: 343-369. |
| 128 | WANG Y Y, AN B L, XUE B, et al. Living materials fabricated via gradient mineralization of light-inducible biofilms[J]. Nature Chemical Biology, 2021, 17(3): 351-359. |
| 129 | ZHOU P, YANG X L, WANG X G, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin[J]. Nature, 2020, 579(7798): 270-273. |
| 130 | SEFIK E, ISRAELOW B, MIRZA H, et al. A humanized mouse model of chronic COVID-19[J]. Nature Biotechnology, 2022, 40(6): 906-920. |
| 131 | YE C Y, QI L N, WANG J, et al. COVID-19 pandemic: advances in diagnosis, treatment, organoid applications and impacts on cancer patient management[J]. Frontiers in Medicine, 2021, 8: 606755. |
| 132 | CHEN D, SU X, CHEN H B, et al. Human organoids as a promising platform for fighting COVID-19[J]. International Journal of Biological Sciences, 2022, 18(3): 901-910. |
| 133 | HAN Y L, DUAN X H, YANG L L, et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids[J]. Nature, 2021, 589(7841): 270-275. |
| 134 | EBISUDANI T, SUGIMOTO S, HAGA K, et al. Direct derivation of human alveolospheres for SARS-CoV-2 infection modeling and drug screening[J]. Cell Reports, 2021, 35(10): 109218. |
| 135 | PARK J W, LAGNITON P N P, LIU Y, et al. mRNA vaccines for COVID-19: what, why and how[J]. International Journal of Biological Sciences, 2021, 17(6): 1446-1460. |
| 136 | JAHANSHAHLU L, REZAEI N. Monoclonal antibody as a potential anti-COVID-19[J]. Biomedicine & Pharmacotherapy, 2020, 129: 110337. |
| 137 | MOOSAVI B, MOUSAVI B, YANG W C, et al. Yeast-based assays for detecting protein-protein/drug interactions and their inhibitors[J]. European Journal of Cell Biology, 2017, 96(6): 529-541. |
| 138 | 郑美云, 李妙君, 沈国婴, 等. 用酵母双杂交系统筛选抗人p53单链抗体[J] 细胞与分子免疫学杂志, 2016, 32(1): 112-117. |
| ZHENG M Y, LI M J, SHEN G Y, et al. Screening of special scFv antibody against human p53 protein by yeast two-hybrid system[J]. Chinese Journal of Cellular and Molecular Immunology, 2016, 32(1):112-117. | |
| 139 | ROZENBLATT-ROSEN O, REGEV A, OBERDOERFFER P, et al. The human tumor atlas network: charting tumor transitions across space and time at single-cell resolution[J]. Cell, 2020, 181(2): 236-249. |
| [1] | 高歌, 边旗, 王宝俊. 合成基因线路的工程化设计研究进展与展望[J]. 合成生物学, 2025, 6(1): 45-64. |
| [2] | 李冀渊, 吴国盛. 合成生物学视域下有机体的两种隐喻[J]. 合成生物学, 2025, 6(1): 190-202. |
| [3] | 焦洪涛, 齐蒙, 邵滨, 蒋劲松. DNA数据存储技术的法律治理议题[J]. 合成生物学, 2025, 6(1): 177-189. |
| [4] | 唐兴华, 陆钱能, 胡翌霖. 人类世中对合成生物学的哲学反思[J]. 合成生物学, 2025, 6(1): 203-212. |
| [5] | 徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176. |
| [6] | 石婷, 宋展, 宋世怡, 张以恒. 体外生物转化(ivBT):生物制造的新前沿[J]. 合成生物学, 2024, 5(6): 1437-1460. |
| [7] | 柴猛, 王风清, 魏东芝. 综合利用木质纤维素生物转化合成有机酸[J]. 合成生物学, 2024, 5(6): 1242-1263. |
| [8] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [9] | 陈雨, 张康, 邱以婧, 程彩云, 殷晶晶, 宋天顺, 谢婧婧. 微生物电合成技术转化二氧化碳研究进展[J]. 合成生物学, 2024, 5(5): 1142-1168. |
| [10] | 郑皓天, 李朝风, 刘良叙, 王嘉伟, 李恒润, 倪俊. 负碳人工光合群落的设计、优化与应用[J]. 合成生物学, 2024, 5(5): 1189-1210. |
| [11] | 夏孔晨, 徐维华, 吴起. 光酶催化混乱性反应的研究进展[J]. 合成生物学, 2024, 5(5): 997-1020. |
| [12] | 陈子苓, 向阳飞. 类器官技术与合成生物学协同研究进展[J]. 合成生物学, 2024, 5(4): 795-812. |
| [13] | 蔡冰玉, 谭象天, 李伟. 合成生物学在干细胞工程化改造中的研究进展[J]. 合成生物学, 2024, 5(4): 782-794. |
| [14] | 洪源, 刘妍. 脑类器官在再生医学中的研究进展[J]. 合成生物学, 2024, 5(4): 754-769. |
| [15] | 王达庆, 陶婷婷, 张旭, 李洪敬. 骨骼肌芯片及其在生物医学领域的研究进展[J]. 合成生物学, 2024, 5(4): 867-882. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
