合成生物学 ›› 2023, Vol. 4 ›› Issue (6): 1140-1160.DOI: 10.12211/2096-8280.2023-044
真核微藻脂质代谢工程的研究进展和展望
孙翰1, 刘进1,2
- 1.深圳大学高等研究院,广东 深圳 518060
2.北京大学工学院,北京 100871
-
收稿日期:2023-06-29修回日期:2023-08-06出版日期:2023-12-31发布日期:2024-01-19 -
通讯作者:刘进 -
作者简介:孙翰 (1991—),男,博士,副研究员。研究方向为微藻代谢工程和微藻生物质精制。E-mail:shlyg2242@163.com刘进 (1980—),男,博士,研究员,博士生导师。研究方向为微藻代谢与合成生物。E-mail:gjinliu@pku.edu.cn -
基金资助:国家重点研发计划(2019YFA0902500);广东省“珠江人才计划”(2019ZT08H476);深圳市孔雀团队项目(KQTD20180412181334790)
Research progress and prospects in lipid metabolic engineering of eukaryotic microalgae
SUN Han1, LIU Jin1,2
- 1.Institute for Advanced Study,Shenzhen University,Shenzhen 518060,Guangdong,China
2.College of Engineering,Peking University,Beijing 100871,China
-
Received:2023-06-29Revised:2023-08-06Online:2023-12-31Published:2024-01-19 -
Contact:LIU Jin
摘要:
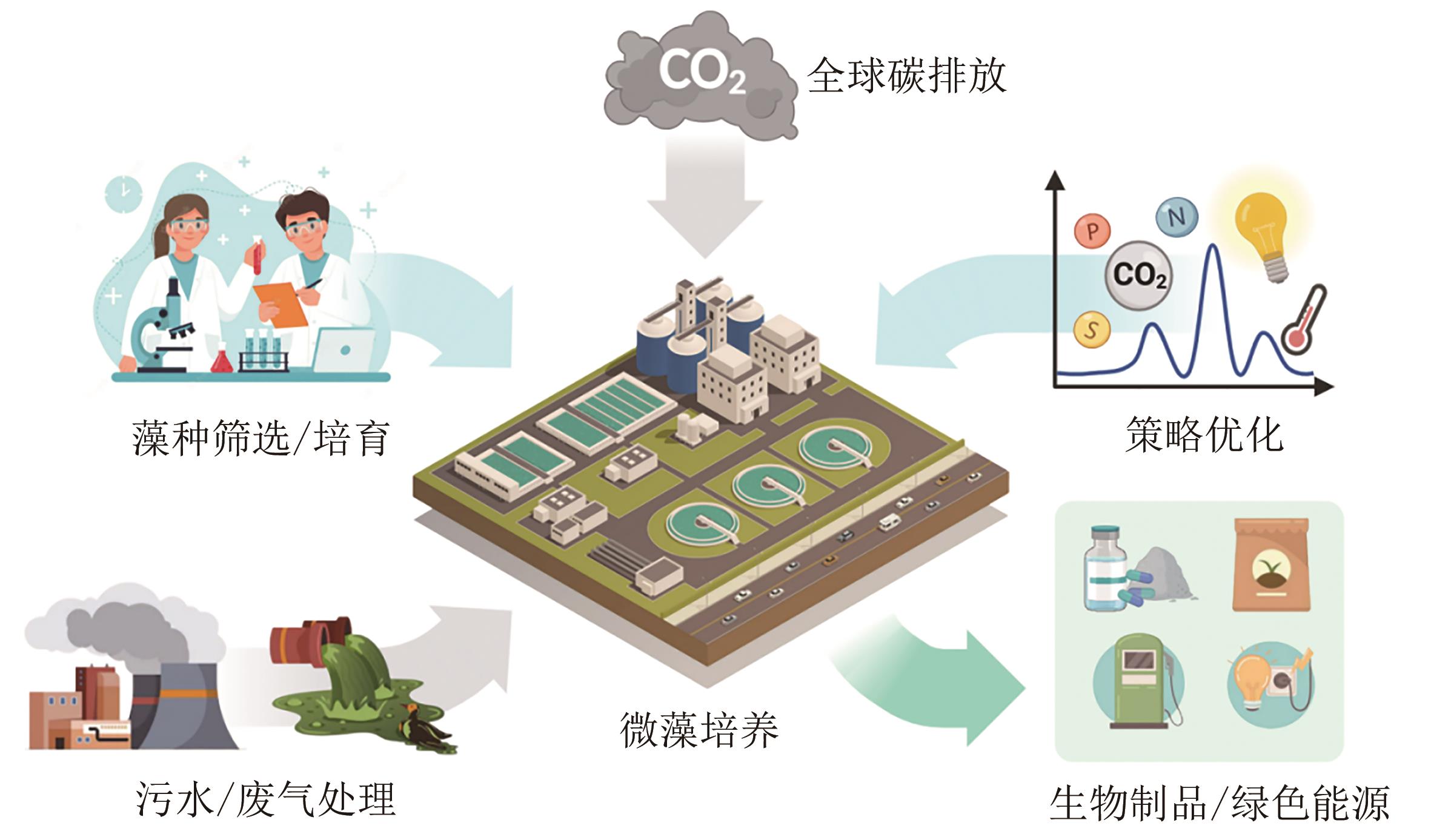
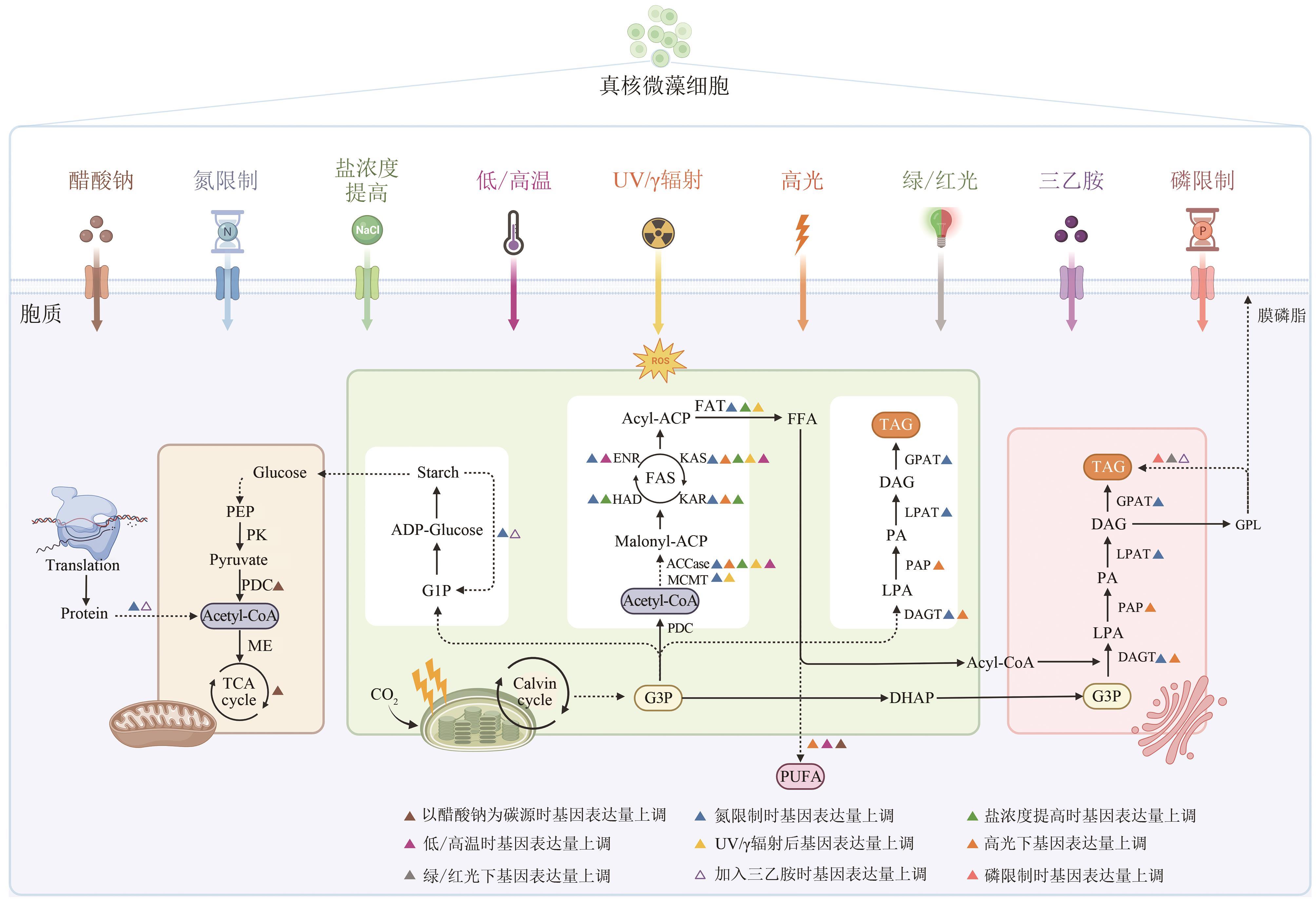
真核微藻作为一类重要的生物资源,脂质含量高,广泛应用于能源、化工和食品等领域。然而,真核微藻生物能源成本偏高,其产业化应用仍然面临着一系列挑战。通过代谢工程手段改造微藻,促进脂质的合成与积累,可提高微藻脂质生产的经济可行性。本文介绍真核微藻脂质代谢途径和关键酶基因,并总结了不同培养条件下代谢途径相关基因在转录水平上的变化。还探讨通过代谢工程调控脂质合成相关酶、转录因子和竞争途径等方法,以提高微藻脂质含量和调整脂肪酸组成。通过基因组学、转录组学和蛋白质组学数据的整合分析可揭示脂质代谢中的关键节点和主效调控因子,有助于确定代谢工程的潜在目标。此外,基因工具和基因编辑技术的开发和拓展可显著提高转化效率,实现对微藻底盘细胞的精准改造。通过重塑能量和碳代谢途径,可设计优化微藻脂质生物合成过程。在微藻遗传工具、基因编辑技术、代谢通路调控和产业化等方面的进一步研究和探索对于推动微藻脂质工程的研究和发展具有重要意义。
中图分类号:
引用本文
孙翰, 刘进. 真核微藻脂质代谢工程的研究进展和展望[J]. 合成生物学, 2023, 4(6): 1140-1160.
SUN Han, LIU Jin. Research progress and prospects in lipid metabolic engineering of eukaryotic microalgae[J]. Synthetic Biology Journal, 2023, 4(6): 1140-1160.
| 门 (分类) | 种 | 基因库 | 参考文献 | ||
|---|---|---|---|---|---|
| 细胞核 | 线粒体 | 叶绿体 | |||
| 绿藻门 | Chromochloris zofingiensis SAG UTEX32 | SRR5310949-5310954 | SRR5310949-5310954 | SRR5310949-5310954 | [ |
| Ostreococcus tauri RCC1115 | NERT00000000 | — | — | [ | |
| Botryococcus braunii | MVGU00000000 | KR057902.1 | KM462884.1 | [ | |
| Chlamydomonas reinhardtii CC503 cw92 mt+ | ABCN00000000 | U03843 | BK000554.2 | [ | |
| Auxenochlorella pyrenoidosa FACHB-9 | ANZC00000000 | — | — | [ | |
| Chlorella vulgaris | LDKB00000000 | — | AB001684.1 | [ | |
| Floydiella terrestris | ADIC01000000 | — | — | [ | |
| Ostreococcus lucimarinus CCE9901 | GCA_000092065.1 | — | — | [ | |
| Micromonas pusilla CCMP1545 | ACCP00000000 | FJ858268.1 | FJ858269 | [ | |
| Micromonas sp. RCC299 | GCA_000090985.2 | FJ859351.1 | FJ858267.1 | [ | |
| Volvox carteri Eve | ACJH00000000 | — | — | [ | |
| Ostreococcus tauri RCC 4221 | CAID00000000 | CR954200.2 | CR954199.2 | [ | |
| Auxenochlorella protothecoides UTEX 2341/0710 | MUYL00000000 APJO00000000 | — | /KC63163 4.1 | [ | |
| 硅藻门 | Phaeodactylum tricornutum CCAP1055/1 | ABQD00000000 | HQ840789.1 | EF067920.1 | [ |
| Thalassiosira pseudonana CCMP1335 | AAFD00000000 | DQ186202.1 | EF067921.1 | [ | |
| 红藻门 | Cyanidioschzon merolae 10D | AP006483 | D89861.1 | AB002583.1 | [ |
| Porphyridium purpureum NIES 2140 | AROW00000000 | — | AP012987.1 | [ | |
| 真眼点藻门 | Nannochloropsis oceanica IMET1 | MPCS00000000.1 | KC598090.1 | KC568462.1 | [ |
| Nannochloropsis gaditana CCMP526 | AGNI00000000 | KC012945.1 | KC012944.1 | [ | |
表1 微藻基因组相关信息
Table 1 The details of microalgae genome
| 门 (分类) | 种 | 基因库 | 参考文献 | ||
|---|---|---|---|---|---|
| 细胞核 | 线粒体 | 叶绿体 | |||
| 绿藻门 | Chromochloris zofingiensis SAG UTEX32 | SRR5310949-5310954 | SRR5310949-5310954 | SRR5310949-5310954 | [ |
| Ostreococcus tauri RCC1115 | NERT00000000 | — | — | [ | |
| Botryococcus braunii | MVGU00000000 | KR057902.1 | KM462884.1 | [ | |
| Chlamydomonas reinhardtii CC503 cw92 mt+ | ABCN00000000 | U03843 | BK000554.2 | [ | |
| Auxenochlorella pyrenoidosa FACHB-9 | ANZC00000000 | — | — | [ | |
| Chlorella vulgaris | LDKB00000000 | — | AB001684.1 | [ | |
| Floydiella terrestris | ADIC01000000 | — | — | [ | |
| Ostreococcus lucimarinus CCE9901 | GCA_000092065.1 | — | — | [ | |
| Micromonas pusilla CCMP1545 | ACCP00000000 | FJ858268.1 | FJ858269 | [ | |
| Micromonas sp. RCC299 | GCA_000090985.2 | FJ859351.1 | FJ858267.1 | [ | |
| Volvox carteri Eve | ACJH00000000 | — | — | [ | |
| Ostreococcus tauri RCC 4221 | CAID00000000 | CR954200.2 | CR954199.2 | [ | |
| Auxenochlorella protothecoides UTEX 2341/0710 | MUYL00000000 APJO00000000 | — | /KC63163 4.1 | [ | |
| 硅藻门 | Phaeodactylum tricornutum CCAP1055/1 | ABQD00000000 | HQ840789.1 | EF067920.1 | [ |
| Thalassiosira pseudonana CCMP1335 | AAFD00000000 | DQ186202.1 | EF067921.1 | [ | |
| 红藻门 | Cyanidioschzon merolae 10D | AP006483 | D89861.1 | AB002583.1 | [ |
| Porphyridium purpureum NIES 2140 | AROW00000000 | — | AP012987.1 | [ | |
| 真眼点藻门 | Nannochloropsis oceanica IMET1 | MPCS00000000.1 | KC598090.1 | KC568462.1 | [ |
| Nannochloropsis gaditana CCMP526 | AGNI00000000 | KC012945.1 | KC012944.1 | [ | |
| 藻种 | 株 | 细胞器 | 转化方法 | 启动子 | 报告基因 (标记) | 参考文献 |
|---|---|---|---|---|---|---|
| Chlamydomonas reinhardtii | dw15.1 | 细胞核 | 电穿孔 | Hsp70A-RBCS2 | ble | [ |
| 137c | 原生质体 | 农杆菌转化 | ChlL | nptⅡ (GUS) | [ | |
| Chlorella vulgaris | UMT-M1 | 细胞核 | 农杆菌转化 | CaMV35S/OlexA-TATA | hpt (GFP) | [ |
| CBS 15-2075 | 叶绿体 | PEG介导 | CaMV35S | aphⅧ (EGFP) | [ | |
| 211/11B | 细胞核 | 基因枪 | CaMV35S | nptⅡ (GUS) | [ | |
| Chlorella ellipsoidea | SD0801 | 细胞核 | PEG介导 | Ubi-1 (Polyubiquitin-1) | ble (GUS) | [ |
| Nrm-4 | 细胞核 | 电穿孔 | Ubi | nptⅡ | [ | |
| Chlorella sp. | DT | 细胞核 | PEG介导 | Actin1/CaMV35S | hpt (GUS) | [ |
| Chromochloris zofingiensis | ATCC 30412 | 细胞核 | 电穿孔/基因枪 | PDS/NIT/RBCS2 | pds | [ |
| Nannochloropsis sp. | PP983 | 细胞核 | 电穿孔 | TUB | ble (GUS) | [ |
| W2J3B | 细胞核 | 电穿孔 | VCP1/2 | ble | [ | |
| Nannochloropsis salina | CCMP1776 | 细胞核 | 基因枪 | TUB | ble | [ |
| Nannochloropsis oceanica | IMET1 | 细胞核 | 电穿孔 | β-tubulin | ble (GUS) | [ |
| Nannochloropsis oculata | NIES-2146 | 细胞核 | 纤维素酶预处理和电穿孔 | Hsp70A-RBCS2 | CP (purple chromoprotein) | [ |
| Nannochloropsis gaditana | CCMP526 | 细胞核 | 电穿孔 | TUB/UEP/Hsp70A | ble | [ |
| Dunaliella salina | — | 细胞核 | 农杆菌转化 | RBCS2/CaMV35S | hptⅡ (GFP) | [ |
| UTEX-1644 | 细胞核 | 玻璃珠法 | Ubi-1 | pat | [ | |
| L1644 | 细胞核 | 基因枪 | Actin | bar | [ | |
| Dunaliella tertiolecta | — | 细胞核 | 玻璃珠法 | CaMV35S | bar (EGFP) | [ |
| SE0045 | 叶绿体 | 基因枪 | PsbD | ereB | [ | |
| Phaeodactylum tricornutum | NRIA-0065 | 细胞核 | 基因枪 | DIV/FCP | ble (EGFP) | [ |
| CCMP2561 | 细胞核 | 电穿孔 | FCPB | ble (GFP/GUS) | [ | |
| Thalassiosira pseudonana | CCMP1335 | 细胞核 | 基因枪 | FCP2 | ble (EGFP) | [ |
| Haematococcus pluvialis | 1844 | 细胞核 | 基因枪 | PsaD | ble/pds | [ |
| CCAP 34/7 | 叶绿体 | PEG介导 | RBCL | aadA | [ | |
| NIES-144 | 细胞核 | 基因枪 | PDS | pds | [ | |
| — | 细胞核 | 农杆菌转化 | CaMV35S | hpt (GFP) | [ | |
| Fistulifera solaris | JPCC DA0580 | 细胞核 | 基因枪 | GAPDH | AphⅧ (G418) (GFP) | [ |
| JPCC DA0580 | 叶绿体 | 基因枪 | CaMV35S/FCPB/RSV | nptⅡ (GFP) | [ | |
| Monoraphidium neglectum | SAG 48.87 | 细胞核 | 电穿孔 | CAB2 (Chlorophyll a/b-binding protein) | AphⅧ (HygromycinB) | [ |
| Neochloris oleoabundans | UTEX 1185 | 细胞核 | 电穿孔 | β2-tubulin | Hyg3 (HygromycinB) | [ |
| Botryococcus braunii | UTEX572 | 细胞核 | 纤维素酶预 处理和电穿孔 | CaMV35S | aphⅧ | [ |
| Tetraselmis chuii | CCAP 66/21B | 细胞核 | 农杆菌转化 | CaMV35S | ble | [ |
| Symbiodinium spp. | Mf11.5b.1 | 细胞核 | 玻璃珠法 | CaMV35S | nptⅡ | [ |
| Lobosphaera incisa | SAG 2468 | 细胞核 | 电穿孔 | RBCS | ble | [ |
| Isochrysis sp. | H-13 | 细胞核 | 农杆菌转化 | LAT | pds | [ |
| Parachlorella kessleri | — | 细胞核 | 农杆菌转化 | CaMV35S | hpt | [ |
| Fistulifera sp. | JPCC DA0580 | 细胞核 | 基因枪 | FCPB | nptⅡ | [ |
| Aurantiochytrium sp. | KRS101 | 细胞核 | 电穿孔 | GAP | CYH | [ |
| Scenedesmus obliquus | FSP-3 | 细胞核 | 电穿孔 | CaMV35S | CAT | [ |
| Schizochytrium sp. | TIO1101 | 细胞核 | 农杆菌转化 | TEF1/CaMV35S | nptⅡ | [ |
| Platymonas subcordiformis | — | 细胞核 | 玻璃珠法 | CaMV35S | — | [ |
| Porphyridium sp. | — | 叶绿体 | 基因枪 | AHAS | AHAS | [ |
| Arthrospira sp. | PCC9438 | 细胞核 | 电穿孔 | CAT | CAT | [ |
表2 真核微藻转化方法和遗传工具相关信息[90]
Table 2 Transformation methods and genetic tools for eukaryotic microalgae[90]
| 藻种 | 株 | 细胞器 | 转化方法 | 启动子 | 报告基因 (标记) | 参考文献 |
|---|---|---|---|---|---|---|
| Chlamydomonas reinhardtii | dw15.1 | 细胞核 | 电穿孔 | Hsp70A-RBCS2 | ble | [ |
| 137c | 原生质体 | 农杆菌转化 | ChlL | nptⅡ (GUS) | [ | |
| Chlorella vulgaris | UMT-M1 | 细胞核 | 农杆菌转化 | CaMV35S/OlexA-TATA | hpt (GFP) | [ |
| CBS 15-2075 | 叶绿体 | PEG介导 | CaMV35S | aphⅧ (EGFP) | [ | |
| 211/11B | 细胞核 | 基因枪 | CaMV35S | nptⅡ (GUS) | [ | |
| Chlorella ellipsoidea | SD0801 | 细胞核 | PEG介导 | Ubi-1 (Polyubiquitin-1) | ble (GUS) | [ |
| Nrm-4 | 细胞核 | 电穿孔 | Ubi | nptⅡ | [ | |
| Chlorella sp. | DT | 细胞核 | PEG介导 | Actin1/CaMV35S | hpt (GUS) | [ |
| Chromochloris zofingiensis | ATCC 30412 | 细胞核 | 电穿孔/基因枪 | PDS/NIT/RBCS2 | pds | [ |
| Nannochloropsis sp. | PP983 | 细胞核 | 电穿孔 | TUB | ble (GUS) | [ |
| W2J3B | 细胞核 | 电穿孔 | VCP1/2 | ble | [ | |
| Nannochloropsis salina | CCMP1776 | 细胞核 | 基因枪 | TUB | ble | [ |
| Nannochloropsis oceanica | IMET1 | 细胞核 | 电穿孔 | β-tubulin | ble (GUS) | [ |
| Nannochloropsis oculata | NIES-2146 | 细胞核 | 纤维素酶预处理和电穿孔 | Hsp70A-RBCS2 | CP (purple chromoprotein) | [ |
| Nannochloropsis gaditana | CCMP526 | 细胞核 | 电穿孔 | TUB/UEP/Hsp70A | ble | [ |
| Dunaliella salina | — | 细胞核 | 农杆菌转化 | RBCS2/CaMV35S | hptⅡ (GFP) | [ |
| UTEX-1644 | 细胞核 | 玻璃珠法 | Ubi-1 | pat | [ | |
| L1644 | 细胞核 | 基因枪 | Actin | bar | [ | |
| Dunaliella tertiolecta | — | 细胞核 | 玻璃珠法 | CaMV35S | bar (EGFP) | [ |
| SE0045 | 叶绿体 | 基因枪 | PsbD | ereB | [ | |
| Phaeodactylum tricornutum | NRIA-0065 | 细胞核 | 基因枪 | DIV/FCP | ble (EGFP) | [ |
| CCMP2561 | 细胞核 | 电穿孔 | FCPB | ble (GFP/GUS) | [ | |
| Thalassiosira pseudonana | CCMP1335 | 细胞核 | 基因枪 | FCP2 | ble (EGFP) | [ |
| Haematococcus pluvialis | 1844 | 细胞核 | 基因枪 | PsaD | ble/pds | [ |
| CCAP 34/7 | 叶绿体 | PEG介导 | RBCL | aadA | [ | |
| NIES-144 | 细胞核 | 基因枪 | PDS | pds | [ | |
| — | 细胞核 | 农杆菌转化 | CaMV35S | hpt (GFP) | [ | |
| Fistulifera solaris | JPCC DA0580 | 细胞核 | 基因枪 | GAPDH | AphⅧ (G418) (GFP) | [ |
| JPCC DA0580 | 叶绿体 | 基因枪 | CaMV35S/FCPB/RSV | nptⅡ (GFP) | [ | |
| Monoraphidium neglectum | SAG 48.87 | 细胞核 | 电穿孔 | CAB2 (Chlorophyll a/b-binding protein) | AphⅧ (HygromycinB) | [ |
| Neochloris oleoabundans | UTEX 1185 | 细胞核 | 电穿孔 | β2-tubulin | Hyg3 (HygromycinB) | [ |
| Botryococcus braunii | UTEX572 | 细胞核 | 纤维素酶预 处理和电穿孔 | CaMV35S | aphⅧ | [ |
| Tetraselmis chuii | CCAP 66/21B | 细胞核 | 农杆菌转化 | CaMV35S | ble | [ |
| Symbiodinium spp. | Mf11.5b.1 | 细胞核 | 玻璃珠法 | CaMV35S | nptⅡ | [ |
| Lobosphaera incisa | SAG 2468 | 细胞核 | 电穿孔 | RBCS | ble | [ |
| Isochrysis sp. | H-13 | 细胞核 | 农杆菌转化 | LAT | pds | [ |
| Parachlorella kessleri | — | 细胞核 | 农杆菌转化 | CaMV35S | hpt | [ |
| Fistulifera sp. | JPCC DA0580 | 细胞核 | 基因枪 | FCPB | nptⅡ | [ |
| Aurantiochytrium sp. | KRS101 | 细胞核 | 电穿孔 | GAP | CYH | [ |
| Scenedesmus obliquus | FSP-3 | 细胞核 | 电穿孔 | CaMV35S | CAT | [ |
| Schizochytrium sp. | TIO1101 | 细胞核 | 农杆菌转化 | TEF1/CaMV35S | nptⅡ | [ |
| Platymonas subcordiformis | — | 细胞核 | 玻璃珠法 | CaMV35S | — | [ |
| Porphyridium sp. | — | 叶绿体 | 基因枪 | AHAS | AHAS | [ |
| Arthrospira sp. | PCC9438 | 细胞核 | 电穿孔 | CAT | CAT | [ |
| 1 | KATIYAR R, GURJAR B R, BISWAS S, et al. Microalgae: an emerging source of energy based bio-products and a solution for environmental issues[J]. Renewable and Sustainable Energy Reviews, 2017, 72: 1083-1093. |
| 2 | CARNEIRO M L N M, PRADELLE F, BRAGA S L, et al. Potential of biofuels from algae: comparison with fossil fuels, ethanol and biodiesel in Europe and Brazil through life cycle assessment (LCA)[J]. Renewable and Sustainable Energy Reviews, 2017, 73: 632-653. |
| 3 | HEPBURN C, ADLEN E, BEDDINGTON J, et al. The technological and economic prospects for CO2 utilization and removal[J]. Nature, 2019, 575(7781): 87-97. |
| 4 | TZACHOR A, RICHARDS C E, HOLT L. Future foods for risk-resilient diets[J]. Nature Food, 2021, 2(5): 326-329. |
| 5 | ADESANYA V O, CADENA E, SCOTT S A, et al. Life cycle assessment on microalgal biodiesel production using a hybrid cultivation system[J]. Bioresource Technology, 2014, 163: 343-355. |
| 6 | KUMAR K, MISHRA S K, SHRIVASTAV A, et al. Recent trends in the mass cultivation of algae in raceway ponds[J]. Renewable and Sustainable Energy Reviews, 2015, 51: 875-885. |
| 7 | TIBOCHA-BONILLA J D, ZUÑIGA C, GODOY-SILVA R D, et al. Advances in metabolic modeling of oleaginous microalgae[J].Biotechnology for Biofuels, 2018, 11(1): 1241. |
| 8 | LI J, ZHU D L, NIU J F, et al. An economic assessment of astaxanthin production by large scale cultivation of Haematococcus pluvialis [J]. Biotechnology Advances, 2011, 29(6): 568-574. |
| 9 | RUIZ J, OLIVIERI G, DE VREE J, et al. Towards industrial products from microalgae[J]. Energy & Environmental Science, 2016, 9(10): 3036-3043. |
| 10 | MASRI M A, GARBE D, MEHLMER N, et al. A sustainable, high-performance process for the economic production of waste-free microbial oils that can replace plant-based equivalents[J]. Energy & Environmental Science, 2019, 12(9): 2717-2732. |
| 11 | SUN H, MAO X M, WU T, et al. Novel insight of carotenoid and lipid biosynthesis and their roles in storage carbon metabolism in Chlamydomonas reinhardtii [J]. Bioresource Technology, 2018, 263: 450-457. |
| 12 | LAURENS L M L, MARKHAM J, TEMPLETON D W, et al. Development of algae biorefinery concepts for biofuels and bioproducts; a perspective on process-compatible products and their impact on cost-reduction[J]. Energy & Environmental Science, 2017, 10(8): 1716-1738. |
| 13 | ROTH M S, COKUS S J, GALLAHER S D, et al. Chromosome-level genome assembly and transcriptome of the green alga Chromochloris zofingiensis illuminates astaxanthin production[J]. Proceedings of the National Academy of Sciences of the United States of America, 2017, 114(21): E4296-E4305. |
| 14 | BLANC-MATHIEU R, SANCHEZ-FERANDIN S, EYRE-WALKER A, et al. Organellar inheritance in the green lineage: insights from Ostreococcus tauri [J]. Genome Biology and Evolution, 2013, 5(8): 1503-1511. |
| 15 | WEISS T L, JOHNSTON J S, FUJISAWA K, et al. Genome size and phylogenetic analysis of the A and L races of Botryococcus braunii [J]. Journal of Applied Phycology, 2011, 23(5): 833-839. |
| 16 | MERCHANT S S, PROCHNIK S E, VALLON O, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions[J]. Science, 2007, 318(5848): 245-250. |
| 17 | FAN J H, NING K, ZENG X W, et al. Genomic foundation of starch-to-lipid switch in oleaginous Chlorella spp[J]. Plant Physiology, 2015, 169(4): 2444-2461. |
| 18 | GUARNIERI M T, BOORE J L, HENARD C A, et al. LDKB00000000.1 Chlorella vulgaris [DB/OL]. NCBI, 2015[2023-06-01]. . |
| 19 | BROUARD J S, OTIS C, LEMIEUX C, et al. The exceptionally large chloroplast genome of the green alga Floydiella terrestris illuminates the evolutionary history of the Chlorophyceae[J]. Genome Biology and Evolution, 2010, 2: 240-256. |
| 20 | PALENIK B, GRIMWOOD J, AERTS A, et al. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation[J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(18): 7705-7710. |
| 21 | WORDEN A Z, LEE J H, MOCK T, et al. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas [J]. Science, 2009, 324(5924): 268-272. |
| 22 | PROCHNIK S E, UMEN J, NEDELCU A M, et al. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri [J]. Science, 2010, 329(5988): 223-226. |
| 23 | BLANC-MATHIEU R, VERHELST B, DERELLE E, et al. An improved genome of the model marine alga Ostreococcus tauri unfolds by assessing Illumina de novo assemblies[J].BMC Genomics, 2014, 15: 1103. |
| 24 | GAO C F, WANG Y, SHEN Y, et al. Oil accumulation mechanisms of the oleaginous microalga Chlorella protothecoides revealed through its genome, transcriptomes, and proteomes[J].BMC Genomics, 2014, 15: 582. |
| 25 | BOWLER C, ALLEN A E, BADGER J H, et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes[J]. Nature, 2008, 456(7219): 239-244. |
| 26 | ARMBRUST E V, BERGES J A, BOWLER C, et al. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism[J]. Science, 2004, 306(5693): 79-86. |
| 27 | NOZAKI H, TAKANO H, MISUMI O, et al. A 100%-complete sequence reveals unusually simple genomic features in the hot-spring red alga Cyanidioschyzon merolae [J].BMC Biology, 2007, 5: 28. |
| 28 | OHTA N, MATSUZAKI M, MISUMI O, et al. Complete sequence and analysis of the plastid genome of the unicellular red alga Cyanidioschyzon merolae [J]. DNA Research, 2003, 10(2): 67-77. |
| 29 | TAJIMA N, SATO S, MARUYAMA F, et al. Analysis of the complete plastid genome of the unicellular red alga Porphyridium purpureum [J]. Journal of Plant Research, 2014, 127(3): 389-397. |
| 30 | WANG D M, NING K, LI J, et al. Nannochloropsis genomes reveal evolution of microalgal oleaginous traits[J]. PLoS Genetics, 2014, 10(1): e1004094. |
| 31 | RADAKOVITS R, JINKERSON R E, FUERSTENBERG S I, et al. Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropsis gaditana [J]. Nature Communications, 2012, 3: 686. |
| 32 | MUÑOZ C F, SÜDFELD C, NADUTHODI M I S, et al. Genetic engineering of microalgae for enhanced lipid production[J]. Biotechnology Advances, 2021, 52: 107836. |
| 33 | JIA J, HAN D X, GERKEN H G, et al. Molecular mechanisms for photosynthetic carbon partitioning into storage neutral lipids in Nannochloropsis oceanica under nitrogen-depletion conditions[J]. Algal Research, 2015, 7: 66-77. |
| 34 | LIU J, SUN Z, MAO X M, et al. Multiomics analysis reveals a distinct mechanism of oleaginousness in the emerging model alga Chromochloris zofingiensis [J]. The Plant Journal, 2019, 98(6): 1060-1077. |
| 35 | KAMALANATHAN M, PIERANGELINI M, SHEARMAN L A, et al. Impacts of nitrogen and phosphorus starvation on the physiology of Chlamydomonas reinhardtii [J]. Journal of Applied Phycology, 2016, 28(3): 1509-1520. |
| 36 | LIANG J B, IQBAL S, WEN F, et al. Phosphorus-induced lipid class alteration revealed by lipidomic and transcriptomic profiling in oleaginous microalga Nannochloropsis sp. PJ12[J]. Marine Drugs, 2019, 17(9): 519. |
| 37 | JIN X J, GONG S Q, YANG B J, et al. Transcriptomic analysis for phosphorus limitation-induced β-glucans accumulation in Chlorella sorokiniana SCSIO 46784 during the early phase of growth[J]. Algal Research, 2021, 54: 102208. |
| 38 | HULATT C J, SMOLINA I, DOWLE A, et al. Proteomic and transcriptomic patterns during lipid remodeling in Nannochloropsis gaditana [J]. International Journal of Molecular Sciences, 2020, 21(18): 6946. |
| 39 | MANDOTRA S K, KUMAR P, SUSEELA M R, et al. Evaluation of fatty acid profile and biodiesel properties of microalga Scenedesmus abundans under the influence of phosphorus, pH and light intensities[J]. Bioresource Technology, 2016, 201: 222-229. |
| 40 | HE Q N, YANG H J, WU L, et al. Effect of light intensity on physiological changes, carbon allocation and neutral lipid accumulation in oleaginous microalgae[J]. Bioresource Technology, 2015, 191: 219-228. |
| 41 | LI Y L, SUN H, WANG Y N, et al. Integrated metabolic tools reveal carbon alternative in Isochrysis zhangjiangensis for fucoxanthin improvement[J]. Bioresource Technology, 2022, 347: 126401. |
| 42 | ATTA M, IDRIS A, BUKHARI A, et al. Intensity of blue LED light: a potential stimulus for biomass and lipid content in fresh water microalgae Chlorella vulgaris [J]. Bioresource Technology, 2013, 148: 373-378. |
| 43 | GAYTÁN-LUNA D E, OCHOA-ALFARO A E, ROCHA-URIBE A, et al. Effect of green and red light in lipid accumulation and transcriptional profile of genes implicated in lipid biosynthesis in Chlamydomonas reinhardtii [J]. Biotechnology Progress, 2016, 32(6): 1404-1411. |
| 44 | VIELER A, WU G X, TSAI C H, et al. Genome, functional gene annotation, and nuclear transformation of the heterokont oleaginous alga Nannochloropsis oceanica CCMP1779[J]. PLoS Genetics, 2012, 8(11): e1003064. |
| 45 | POLINER E, BUSCH A W U, NEWTON L, et al. Aureochromes maintain polyunsaturated fatty acid content in Nannochloropsis oceanica [J]. Plant Physiology, 2022, 189(2): 906-921. |
| 46 | PATELOU M, INFANTE C, DARDELLE F, et al. Transcriptomic and metabolomic adaptation of Nannochloropsis gaditana grown under different light regimes[J]. Algal Research, 2020, 45: 101735. |
| 47 | WU L F, CHEN P C, LEE C M. The effects of nitrogen sources and temperature on cell growth and lipid accumulation of microalgae[J]. International Biodeterioration & Biodegradation, 2013, 85: 506-510. |
| 48 | CALHOUN S, BELL T A S, DAHLIN L R, et al. A multi-omic characterization of temperature stress in a halotolerant Scenedesmus strain for algal biotechnology[J]. Communications Biology, 2021, 4: 333. |
| 49 | XING G L, YUAN H L, YANG J S, et al. Integrated analyses of transcriptome, proteome and fatty acid profilings of the oleaginous microalga Auxenochlorella protothecoides UTEX 2341 reveal differential reprogramming of fatty acid metabolism in response to low and high temperatures[J]. Algal Research, 2018, 33: 16-27. |
| 50 | SU H, FENG J, LV J P, et al. Molecular mechanism of lipid accumulation and metabolism of oleaginous Chlorococcum sphacosum GD from soil under salt stress[J]. International Journal of Molecular Sciences, 2021, 22(3): 1304. |
| 51 | QU D H, MIAO X L. Carbon flow conversion induces alkali resistance and lipid accumulation under alkaline conditions based on transcriptome analysis in Chlorella sp. BLD[J]. Chemosphere, 2021, 265: 129046. |
| 52 | SINGH R D, SETHY S, GHOSH S, et al. UV and γ-radiation induced molecular changes for rapid lipid accumulation in Chlorella sorokiniana [J]. Biomass and Bioenergy, 2022, 163: 106493. |
| 53 | NAJAFABADI H A, MALEKZADEH M, JALILIAN F, et al. Effect of various carbon sources on biomass and lipid production of Chlorella vulgaris during nutrient sufficient and nitrogen starvation conditions[J]. Bioresource Technology, 2015, 180: 311-317. |
| 54 | UDAYAN A, ARUMUGAM M. Selective enrichment of eicosapentaenoic acid (20:5n-3) in N. oceanica CASA CC201 by natural auxin supplementation[J]. Bioresource Technology, 2017, 242: 329-333. |
| 55 | XUE L L, JIANG J G. Cultivation of Dunaliella tertiolecta intervened by triethylamine enhances the lipid content[J]. Algal Research, 2017, 25: 136-141. |
| 56 | ROESSLER P G. Changes in the activities of various lipid and carbohydrate biosynthetic enzymes in the diatom Cyclotella cryptica in response to silicon deficiency[J]. Archives of Biochemistry and Biophysics, 1988, 267(2): 521-528. |
| 57 | DUNAHAY T G, JARVIS E E, DAIS S S, et al. Manipulation of microalgal lipid production using genetic engineering[J]. Applied Biochemistry and Biotechnology, 1996, 57: 223-231. |
| 58 | GOMMA A E, LEE S K, SUN S M, et al. Improvement in oil production by increasing malonyl-CoA and glycerol-3-phosphate pools in Scenedesmus quadricauda [J]. Indian Journal of Microbiology, 2015, 55(4): 447-455. |
| 59 | CHEN J W, LIU W J, HU D X, et al. Identification of a malonyl CoA-acyl carrier protein transacylase and its regulatory role in fatty acid biosynthesis in oleaginous microalga Nannochloropsis oceanica [J]. Biotechnology and Applied Biochemistry, 2017, 64(5): 620-626. |
| 60 | RENGEL R, SMITH R T, HASLAM R P, et al. Overexpression of acetyl-CoA synthetase (ACS) enhances the biosynthesis of neutral lipids and starch in the green microalga Chlamydomonas reinhardtii [J]. Algal Research, 2018, 31: 183-193. |
| 61 | YAN J F, CHENG R B, LIN X Z, et al. Overexpression of acetyl-CoA synthetase increased the biomass and fatty acid proportion in microalga Schizochytrium [J]. Applied Microbiology and Biotechnology, 2013, 97(5): 1933-1939. |
| 62 | OSADA K, MAEDA Y, YOSHINO T, et al. Enhanced NADPH production in the pentose phosphate pathway accelerates lipid accumulation in the oleaginous diatom Fistulifera solaris [J]. Algal Research, 2017, 23: 126-134. |
| 63 | NIU Y F, WANG X, HU D X, et al. Molecular characterization of a glycerol-3-phosphate acyltransferase reveals key features essential for triacylglycerol production in Phaeodactylum tricornutum [J].Biotechnology for Biofuels, 2016, 9: 60. |
| 64 | MUÑOZ C F, WEUSTHUIS R A, D'ADAMO S, et al. Effect of single and combined expression of lysophosphatidic acid acyltransferase, glycerol-3-phosphate acyltransferase, and diacylglycerol acyltransferase on lipid accumulation and composition in Neochloris oleoabundans [J]. Frontiers in Plant Science, 2019, 10: 1573. |
| 65 | MARAVI D K, KUMAR S, SHARMA P K, et al. Ectopic expression of AtDGAT1, encoding diacylglycerol O-acyltransferase exclusively committed to TAG biosynthesis, enhances oil accumulation in seeds and leaves of Jatropha [J]. Biotechnology for Biofuels, 2016, 9: 226. |
| 66 | WANG Z K, HUANG W J, CHANG J M, et al. Overexpression of SiDGAT1, a gene encoding acyl-CoA: diacylglycerol acyltransferase from Sesamum indicum L. increases oil content in transgenic Arabidopsis and soybean[J]. Plant Cell, Tissue and Organ Culture (PCTOC), 2014, 119(2): 399-410. |
| 67 | NIU Y F, ZHANG M H, LI D W, et al. Improvement of neutral lipid and polyunsaturated fatty acid biosynthesis by overexpressing a type 2 diacylglycerol acyltransferase in marine diatom Phaeodactylum tricornutum [J]. Marine Drugs, 2013, 11(11): 4558-4569. |
| 68 | LI D W, CEN S Y, LIU Y H, et al. A type 2 diacylglycerol acyltransferase accelerates the triacylglycerol biosynthesis in heterokont oleaginous microalga Nannochloropsis oceanica [J]. Journal of Biotechnology, 2016, 229: 65-71. |
| 69 | DENG X D, GU B, LI Y J, et al. The roles of acyl-CoA: diacylglycerol acyltransferase 2 genes in the biosynthesis of triacylglycerols by the green algae Chlamydomonas reinhardtii [J]. Molecular Plant, 2012, 5(4): 945-947. |
| 70 | COUSO I, PÉREZ-PÉREZ M E, FORD M M, et al. Phosphorus availability regulates TORC1 signaling via LST8 in Chlamydomonas [J]. The Plant Cell, 2020, 32(1): 69-80. |
| 71 | NGAN C Y, WONG C H, CHOI C, et al. Lineage-specific chromatin signatures reveal a regulator of lipid metabolism in microalgae[J]. Nature Plants, 2015, 1: 15107. |
| 72 | BAJHAIYA A K, DEAN A P, ZEEF L A H, et al. PSR1 is a global transcriptional regulator of phosphorus deficiency responses and carbon storage metabolism in Chlamydomonas reinhardtii [J]. Plant Physiology, 2016, 170(3): 1216-1234. |
| 73 | JIA B, XIE X F, WU M, et al. Understanding the functions of endogenous DOF transcript factor in Chlamydomonas reinhardtii [J]. Biotechnology for Biofuels, 2019, 12: 67. |
| 74 | ZHANG J H, HAO Q, BAI L L, et al. Overexpression of the soybean transcription factor GmDof4 significantly enhances the lipid content of Chlorella ellipsoidea [J]. Biotechnology for Biofuels, 2014, 7: 128. |
| 75 | LI D W, BALAMURUGAN S, YANG Y F, et al. Transcriptional regulation of microalgae for concurrent lipid overproduction and secretion[J]. Science Advances, 2019, 5(1): eaau3795. |
| 76 | YAMAOKA Y, SHIN S, CHOI B Y, et al. The bZIP1 transcription factor regulates lipid remodeling and contributes to ER stress management in Chlamydomonas reinhardtii [J]. The Plant Cell, 2019, 31(5): 1127-1140. |
| 77 | AJJAWI I, VERRUTO J, AQUI M, et al. Lipid production in Nannochloropsis gaditana is doubled by decreasing expression of a single transcriptional regulator[J]. Nature Biotechnology, 2017, 35(7): 647-652. |
| 78 | RADAKOVITS R, EDUAFO P M, POSEWITZ M C. Genetic engineering of fatty acid chain length in Phaeodactylum tricornutum [J]. Metabolic Engineering, 2011, 13: 89-95. |
| 79 | GONG Y M, GUO X J, WAN X, et al. Characterization of a novel thioesterase (PtTE) from Phaeodactylum tricornutum [J]. Journal of Basic Microbiology, 2011, 51(6): 666-672. |
| 80 | PENG K T, ZHENG C N, XUE J, et al. Delta 5 fatty acid desaturase upregulates the synthesis of polyunsaturated fatty acids in the marine diatom Phaeodactylum tricornutum [J]. Journal of Agricultural and Food Chemistry, 2014, 62(35): 8773-8776. |
| 81 | HAMILTON M L, HASLAM R P, NAPIER J A, et al. Metabolic engineering of Phaeodactylum tricornutum for the enhanced accumulation of omega-3 long chain polyunsaturated fatty acids[J]. Metabolic Engineering, 2014, 22: 3-9. |
| 82 | SHI H S, LUO X, WU R N, et al. Production of eicosapentaenoic acid by application of a delta-6 desaturase with the highest ALA catalytic activity in algae[J]. Microbial Cell Factories, 2018, 17: 7. |
| 83 | POLINER E, PULMAN J A, ZIENKIEWICZ K, et al. A toolkit for Nannochloropsis oceanica CCMP1779 enables gene stacking and genetic engineering of the eicosapentaenoic acid pathway for enhanced long-chain polyunsaturated fatty acid production[J]. Plant Biotechnology Journal, 2018, 16: 298-309. |
| 84 | NORASHIKIN M N, LOH S H, AZIZ A, et al. Metabolic engineering of fatty acid biosynthesis in Chlorella vulgaris using an endogenous omega-3 fatty acid desaturase gene with its promoter[J]. Algal Research, 2018, 31: 262-275. |
| 85 | WANG X, DONG H P, WEI W, et al. Dual expression of plastidial GPAT1 and LPAT1 regulates triacylglycerol production and the fatty acid profile in Phaeodactylum tricornutum [J].Biotechnology for Biofuels, 2018, 11: 318. |
| 86 | LIU J, LIU M J, PAN Y F, et al. Metabolic engineering of the oleaginous alga Nannochloropsis for enriching eicosapentaenoic acid in triacylglycerol by combined pulling and pushing strategies[J]. Metabolic Engineering, 2022, 69: 163-174. |
| 87 | XIN Y, LU Y D, LEE Y Y, et al. Producing designer oils in industrial microalgae by rational modulation of Co-evolving type-2 diacylglycerol acyltransferases[J]. Molecular Plant, 2017, 10(12): 1523-1539. |
| 88 | LU N, WEI D, JIANG X L, et al. Regulation of lipid metabolism in the snow alga Chlamydomonas nivalis in response to NaCl stress: an integrated analysis by cytomic and lipidomic approaches[J]. Process Biochemistry, 2012, 47(7): 1163-1170. |
| 89 | SUN H, REN Y Y, MAO X M, et al. Harnessing C/N balance of Chromochloris zofingiensis to overcome the potential conflict in microalgal production[J]. Communications Biology, 2020, 3: 186. |
| 90 | SUN H, WU T, CHEN S H Y, et al. Powerful tools for productivity improvements in microalgal production[J]. Renewable and Sustainable Energy Reviews, 2021, 152: 111609. |
| 91 | KONG F T, LIANG Y X, LÉGERET B, et al. Chlamydomonas carries out fatty acid β-oxidation in ancestral peroxisomes using a bona fide acyl-CoA oxidase[J]. The Plant Journal, 2017, 90(2): 358-371. |
| 92 | CRAIG W, GARGANO D, SCOTTI N, et al. Direct gene transfer in potato: a comparison of particle bombardment of leaf explants and PEG-mediated transformation of protoplasts[J]. Plant Cell Reports, 2005, 24(10): 603-611. |
| 93 | NG S L, HARIKRISHNA J A, BAKAR F ABU, et al. Heterologous expression of the Streptococcus pneumoniae yoeB and pezT toxin genes is lethal in Chlorella vulgaris [J]. Algal Research, 2016, 19: 21-29. |
| 94 | YANG B, LIU J, LIU B, et al. Development of a stable genetic system for Chlorella vulgaris—a promising green alga for CO2 biomitigation[J]. Algal Research, 2015, 12: 134-141. |
| 95 | TALEBI A F, TOHIDFAR M, TABATABAEI M, et al. Genetic manipulation, a feasible tool to enhance unique characteristic of Chlorella vulgaris as a feedstock for biodiesel production[J]. Molecular Biology Reports, 2013, 40(7): 4421-4428. |
| 96 | LIU L L, WANG Y Q, ZHANG Y C, et al. Development of a new method for genetic transformation of the green alga Chlorella ellipsoidea [J]. Molecular Biotechnology, 2013, 54(2): 211-219. |
| 97 | BAI L L, YIN W B, CHEN Y H, et al. A new strategy to produce a defensin: stable production of mutated NP-1 in nitrate reductase-deficient Chlorella ellipsoidea [J]. PLoS One, 2013, 8(1): e54966. |
| 98 | HUANG C C, CHEN M W, HSIEH J L, et al. Expression of mercuric reductase from Bacillus megaterium MB1 in eukaryotic microalga Chlorella sp. DT: an approach for mercury phytoremediation[J]. Applied Microbiology and Biotechnology, 2006, 72(1): 197-205. |
| 99 | LIU J, SUN Z, GERKEN H, et al. Genetic engineering of the green alga Chlorella zofingiensis: a modified norflurazon-resistant phytoene desaturase gene as a dominant selectable marker[J]. Applied Microbiology and Biotechnology, 2014, 98(11): 5069-5079. |
| 100 | LI F J, GAO D W, HU H H. High-efficiency nuclear transformation of the oleaginous marine Nannochloropsis species using PCR product[J]. Bioscience, Biotechnology, and Biochemistry, 2014, 78(5): 812-817. |
| 101 | KILIAN O, BENEMANN C S E, NIYOGI K K, et al. High-efficiency homologous recombination in the oil-producing alga Nannochloropsis sp.[J]. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(52): 21265-21269. |
| 102 | KANG N K, JEON S, KWON S, et al. Effects of overexpression of a bHLH transcription factor on biomass and lipid production in Nannochloropsis salina [J].Biotechnology for Biofuels, 2015, 8: 200. |
| 103 | WEI L, XIN Y, WANG Q T, et al. RNAi-based targeted gene knockdown in the model oleaginous microalgae Nannochloropsis oceanica [J]. The Plant Journal, 2017, 89(6): 1236-1250. |
| 104 | SHIH C H, CHEN H Y, LEE H C, et al. Purple chromoprotein gene serves as a new selection marker for transgenesis of the microalga Nannochloropsis oculata [J]. PLoS One, 2015, 10(3): e0120780. |
| 105 | SIMON D P, ANILA N, GAYATHRI K, et al. Heterologous expression of β-carotene hydroxylase in Dunaliella salina by Agrobacterium-mediated genetic transformation[J]. Algal Research, 2016, 18: 257-265. |
| 106 | FENG S Y, FENG W P, ZHAO L, et al. Preparation of transgenic Dunaliella salina for immunization against white spot syndrome virus in crayfish[J]. Archives of Virology, 2014, 159(3): 519-525. |
| 107 | 姜国忠, 吕玉民, 牛向丽, 等. 盐藻肌动蛋白基因启动子驱动的bar基因表达作为核转化筛选标记[J]. 遗传学报, 2005, 32(4): 424-433. |
| JIANG G Z, LÜ Y M, NIU X L, et al. The actin gene promoter-driven bar as a dominant selectable marker for nuclear transformation of Dunaliella salina [J]. Acta Genetica Sinica, 2005, 32(4): 424-433. | |
| 108 | ZHANG J Y, JEON H, SIM S J, et al. Homologous sense and antisense expression of a gene in Dunaliella tertiolecta [J]. Planta, 2015, 242(4): 1051-1058. |
| 109 | GEORGIANNA D R, HANNON M J, MARCUSCHI M, et al. Production of recombinant enzymes in the marine alga Dunaliella tertiolecta [J]. Algal Research, 2013, 2(1): 2-9. |
| 110 | KADONO T, MIYAGAWA-YAMAGUCHI A, KIRA N, et al. Characterization of marine diatom-infecting virus promoters in the model diatom Phaeodactylum tricornutum [J]. Scientific Reports, 2015, 5: 18708. |
| 111 | ZHANG C Y, HU H H. High-efficiency nuclear transformation of the diatom Phaeodactylum tricornutum by electroporation[J]. Marine Genomics, 2014, 16: 63-66. |
| 112 | POULSEN N, CHESLEY P M, KRÖGER N. Molecular genetic manipulation of the diatom Thalassiosira pseudonana (Bacillariophyceae)[J]. Journal of Phycology, 2006, 42(5): 1059-1065. |
| 113 | SHARON-GOJMAN R, MAIMON E, LEU S, et al. Advanced methods for genetic engineering of Haematococcus pluvialis (Chlorophyceae, Volvocales)[J]. Algal Research, 2015, 10: 8-15. |
| 114 | GUTIÉRREZ C L, GIMPEL J, ESCOBAR C, et al. Chloroplast genetic tool for the green microalgae Haematococcus pluvialis (Chlorophyceae, Volvocales)1 [J]. Journal of Phycology, 2012, 48(4): 976-983. |
| 115 | STEINBRENNER J, SANDMANN G. Transformation of the green alga Haematococcus pluvialis with a phytoene desaturase for accelerated astaxanthin biosynthesis[J]. Applied and Environmental Microbiology, 2006, 72(12): 7477-7484. |
| 116 | KATHIRESAN S, CHANDRASHEKAR A, RAVISHANKAR G A, et al. Agrobacterium-mediated transformation in the green alga Haematococcus pluvialis(Chlorophyceae, Volvocales)[J]. Journal of Phycology, 2009, 45(3): 642-649. |
| 117 | MUTO M, FUKUDA Y, NEMOTO M, et al. Establishment of a genetic transformation system for the marine pennate diatom Fistulifera sp. strain JPCC DA0580—a high triglyceride producer[J]. Marine Biotechnology, 2013, 15(1): 48-55. |
| 118 | JAEGER D, HÜBNER W, HUSER T, et al. Nuclear transformation and functional gene expression in the oleaginous microalga Monoraphidium neglectum [J]. Journal of Biotechnology, 2017, 249: 10-15. |
| 119 | CHUNGJATUPORNCHAI W, KITRAKSA P, FA-AROONSAWAT S. Stable nuclear transformation of the oleaginous microalga Neochloris oleoabundans by electroporation[J].Journal of Applied Phycology, 2016, 28(1): 191-199. |
| 120 | BERRIOS H, ZAPATA M, RIVAS M. A method for genetic transformation of Botryococcus braunii using a cellulase pretreatment[J]. Journal of Applied Phycology, 2016, 28(1): 201-208. |
| 121 | ÚBEDA-MÍNGUEZ P, CHILEH T, DAUTOR Y, et al. Tools for microalgal biotechnology: development of an optimized transformation method for an industrially promising microalga—Tetraselmis chuii [J]. Journal of Applied Phycology, 2015, 27(1): 223-232. |
| 122 | ORTIZ-MATAMOROS M F, VILLANUEVA M A, ISLAS-FLORES T. Transient transformation of cultured photosynthetic dinoflagellates (Symbiodinium spp.) with plant-targeted vectors[J]. Ciencias Marinas, 2015, 41(1): 21-32. |
| 123 | ZORIN B, GRUNDMAN O, KHOZIN-GOLDBERG I, et al. Development of a nuclear transformation system for Oleaginous Green Alga Lobosphaera (Parietochloris) incisa and genetic complementation of a mutant strain, deficient in arachidonic acid biosynthesis[J]. PLoS One, 2014, 9(8): e105223. |
| 124 | PRASAD B, VADAKEDATH N, JEONG H J, et al. Agrobacterium tumefaciens-mediated genetic transformation of haptophytes (Isochrysis species)[J].Applied Microbiology and Biotechnology, 2014, 98(20): 8629-8639. |
| 125 | RATHOD J P, PRAKASH G, PANDIT R, et al. Agrobacterium-mediated transformation of promising oil-bearing marine algae Parachlorella kessleri [J].Photosynthesis Research, 2013, 118(1/2): 141-146. |
| 126 | HONG W K, HEO S Y, OH B R, et al. A transgene expression system for the marine microalgae Aurantiochytrium sp. KRS101 using a mutant allele of the gene encoding ribosomal protein L44 as a selectable transformation marker for cycloheximide resistance[J]. Bioprocess and Biosystems Engineering, 2013, 36(9): 1191-1197. |
| 127 | GUO S L, ZHAO X Q, TANG Y, et al. Establishment of an efficient genetic transformation system in Scenedesmus obliquus [J]. Journal of Biotechnology, 2013, 163(1): 61-68. |
| 128 | CHENG R B, MA R J, LI K, et al. Agrobacterium tumefaciens mediated transformation of marine microalgae Schizochytrium [J]. Microbiological Research, 2012, 167(3): 179-186. |
| 129 | CUI Y L, WANG J F, JIANG P, et al. Transformation of Platymonas (Tetraselmis) subcordiformis (Prasinophyceae, Chlorophyta) by agitation with glass beads[J]. World Journal of Microbiology and Biotechnology, 2010, 26(9): 1653-1657. |
| 130 | LAPIDOT M, RAVEH D, SIVAN A, et al. Stable chloroplast transformation of the unicellular red alga Porphyridium species[J]. Plant Physiology, 2002, 129: 7-12. |
| 131 | KAWATA Y, YANO S, KOJIMA H, et al. Transformation of Spirulina platensis strain C1 (Arthrospira sp. PCC9438) with Tn5 transposase-transposon DNA-cation liposome complex[J]. Marine Biotechnology, 2004, 6(4): 355-363. |
| 132 | JAENICKE L, KUHNE W, SPESSERT R, et al. Cell-wall lytic enzymes (autolysins) of Chlamydomonas reinhardtii are (hydroxy)proline-specific proteases[J]. European Journal of Biochemistry, 1987, 170(1/2): 485-491. |
| 133 | ZHU Z, SUN J, FA Y, et al. Enhancing microalgal lipid accumulation for biofuel production[J]. Frontiers in Microbiology, 2022, 13: 1024441. |
| 134 | WU X G, GIRIDHAR BABU A, KIM B L, et al. Ultrasound-mediated intracellular delivery of fluorescent dyes and DNA into microalgal cells[J]. Algal Research, 2016, 15: 210-216. |
| 135 | KINDLE K L, SCHNELL R A, FERNÁNDEZ E, et al. Stable nuclear transformation of Chlamydomonas using the Chlamydomonas gene for nitrate reductase[J]. The Journal of Cell Biology, 1989, 109(6): 2589-2601. |
| 136 | SØRENSEN I, FEI Z J, ANDREAS A, et al. Stable transformation and reverse genetic analysis of Penium margaritaceum: a platform for studies of charophyte green algae, the immediate ancestors of land plants[J]. The Plant Journal, 2014, 77(3): 339-351. |
| 137 | JIA Y L, XUE L X, LIU H T, et al. Characterization of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene from the halotolerant alga Dunaliella salina and inhibition of its expression by RNAi[J]. Current Microbiology, 2009, 58(5): 426-431. |
| 138 | RANDHAWA S, SENGAR S. The evolution and history of gene editing technologies[M/OL]//Progress in molecular biology and translational science. Amsterdam: Elsevier, 2021, 178: 1-62 [2023-07-01]. . |
| 139 | SIZOVA I, GREINER A, AWASTHI M, et al. Nuclear gene targeting in Chlamydomonas using engineered zinc-finger nucleases[J]. The Plant Journal, 2013, 73(5): 873-882. |
| 140 | DABOUSSI F, LEDUC S, MARÉCHAL A, et al. Genome engineering empowers the diatom Phaeodactylum tricornutum for biotechnology[J]. Nature Communications, 2014, 5: 3831. |
| 141 | WEYMAN P D, BEERI K, LEFEBVRE S C, et al. Inactivation of Phaeodactylum tricornutum urease gene using transcription activator-like effector nuclease-based targeted mutagenesis[J]. Plant Biotechnology Journal, 2015, 13(4): 460-470. |
| 142 | NGUYEN T H T, PARK S, JEONG J, et al. Enhancing lipid productivity by modulating lipid catabolism using the CRISPR-Cas9 system in Chlamydomonas [J]. Journal of Applied Phycology, 2020, 32(5): 2829-2840. |
| 143 | FENG S Y, HU L N, ZHANG Q H, et al. CRISPR/Cas technology promotes the various application of Dunaliella salina system[J].Applied Microbiology and Biotechnology, 2020, 104(20): 8621-8630. |
| 144 | LIN W R, NG I S. Development of CRISPR/Cas9 system in Chlorella vulgaris FSP-E to enhance lipid accumulation[J]. Enzyme and Microbial Technology, 2020, 133: 109458. |
| 145 | VERRUTO J, FRANCIS K, WANG Y J, et al. Unrestrained markerless trait stacking in Nannochloropsis gaditana through combined genome editing and marker recycling technologies[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(30): E7015-E7022. |
| 146 | WANG Q T, GONG Y H, HE Y H, et al. Genome engineering of Nannochloropsis with hundred-kilobase fragment deletions by Cas9 cleavages[J]. The Plant Journal, 2021, 106(4): 1148-1162. |
| 147 | BAEK K, KIM D H, JEONG J, et al. DNA-free two-gene knockout in Chlamydomonas reinhardtii via CRISPR-Cas9 ribonucleoproteins[J]. Scientific Reports, 2016, 6: 30620. |
| 148 | SCHRODA M, BLÖCKER D, BECK C F. The HSP70A promoter as a tool for the improved expression of transgenes in Chlamydomonas [J]. The Plant Journal, 2000, 21(2): 121-131. |
| 149 | FRANKLIN S, NGO B, EFUET E, et al. Development of a GFP reporter gene for Chlamydomonas reinhardtii chloroplast[J]. The Plant Journal, 2002, 30(6): 733-744. |
| 150 | JIA Y L, LI S K, ALLEN G, et al. A novel glyceraldehyde-3-phosphate dehydrogenase (GAPDH) promoter for expressing transgenes in the halotolerant alga Dunaliella salina [J]. Current Microbiology, 2012, 64(5): 506-513. |
| 151 | HOU Q T, QIU S, LIU Q, et al. Selenoprotein-transgenic Chlamydomonas reinhardtii [J]. Nutrients, 2013, 5(3): 624-636. |
| 152 | CROZET P, NAVARRO F J, WILLMUND F, et al. Birth of a photosynthetic chassis: a MoClo toolkit enabling synthetic biology in the microalga Chlamydomonas reinhardtii [J]. ACS Synthetic Biology, 2018, 7(9): 2074-2086. |
| 153 | SUN H, REN Y Y, FAN Y W, et al. Systematic metabolic tools reveal underlying mechanism of product biosynthesis in Chromochloris zofingiensis [J]. Bioresource Technology, 2021, 337: 125406. |
| 154 | SUN H, WANG Y X, HE Y J, et al. Microalgae-derived pigments for the food industry[J]. Marine Drugs, 2023, 21(2): 82. |
| 155 | SCHUHMANN H, LIM D K, SCHENK P M. Perspectives on metabolic engineering for increased lipid contents in microalgae[J]. Biofuels, 2012, 3(1): 71-86. |
| 156 | YAMORI W, SHIKANAI T. Physiological functions of cyclic electron transport around photosystemⅠin sustaining photosynthesis and plant growth[J]. Annual Review of Plant Biology, 2016, 67: 81-106. |
| 157 | XUE J, BALAMURUGAN S, LI D W, et al. Glucose-6-phosphate dehydrogenase as a target for highly efficient fatty acid biosynthesis in microalgae by enhancing NADPH supply[J]. Metabolic Engineering, 2017, 41: 212-221. |
| 158 | SCHIEVANO A, SCIARRIA T P, VANBROEKHOVEN K, et al. Electro-fermentation-merging electrochemistry with fermentation in industrial applications[J]. Trends in Biotechnology, 2016, 34(11): 866-878. |
| 159 | CHOI O, KIM T, WOO H M, et al. Electricity-driven metabolic shift through direct electron uptake by electroactive heterotroph Clostridium pasteurianum [J]. Scientific Reports, 2014, 4: 6961. |
| 160 | MICHELET L, ZAFFAGNINI M, MORISSE S, et al. Redox regulation of the Calvin-Benson cycle: something old, something new[J]. Frontiers in Plant Science, 2013, 4: 470. |
| 161 | KUREK I, CHANG T K, BERTAIN S M, et al. Enhanced thermostability of Arabidopsis Rubisco activase improves photosynthesis and growth rates under moderate heat stress[J]. The Plant Cell, 2007, 19(10): 3230-3241. |
| 162 | YAMORI W, MASUMOTO C, FUKAYAMA H, et al. Rubisco activase is a key regulator of non-steady-state photosynthesis at any leaf temperature and, to a lesser extent, of steady-state photosynthesis at high temperature[J]. The Plant Journal, 2012, 71(6): 871-880. |
| 163 | VUORISTO K S, MARS A E, SANDERS J P M, et al. Metabolic engineering of TCA cycle for production of chemicals[J]. Trends in Biotechnology, 2016, 34(3): 191-197. |
| 164 | SAINI R, KAPOOR R, KUMAR R, et al. CO2 utilizing microbes—a comprehensive review[J]. Biotechnology Advances, 2011, 29(6): 949-960. |
| 165 | XIONG W, MORGAN J A, UNGERER J, et al. The plasticity of cyanobacterial metabolism supports direct CO2 conversion to ethylene[J]. Nature Plants, 2015, 1: 15053. |
| [1] | 高歌, 边旗, 王宝俊. 合成基因线路的工程化设计研究进展与展望[J]. 合成生物学, 2025, 6(1): 45-64. |
| [2] | 李冀渊, 吴国盛. 合成生物学视域下有机体的两种隐喻[J]. 合成生物学, 2025, 6(1): 190-202. |
| [3] | 焦洪涛, 齐蒙, 邵滨, 蒋劲松. DNA数据存储技术的法律治理议题[J]. 合成生物学, 2025, 6(1): 177-189. |
| [4] | 唐兴华, 陆钱能, 胡翌霖. 人类世中对合成生物学的哲学反思[J]. 合成生物学, 2025, 6(1): 203-212. |
| [5] | 徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176. |
| [6] | 石婷, 宋展, 宋世怡, 张以恒. 体外生物转化(ivBT):生物制造的新前沿[J]. 合成生物学, 2024, 5(6): 1437-1460. |
| [7] | 柴猛, 王风清, 魏东芝. 综合利用木质纤维素生物转化合成有机酸[J]. 合成生物学, 2024, 5(6): 1242-1263. |
| [8] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [9] | 陈雨, 张康, 邱以婧, 程彩云, 殷晶晶, 宋天顺, 谢婧婧. 微生物电合成技术转化二氧化碳研究进展[J]. 合成生物学, 2024, 5(5): 1142-1168. |
| [10] | 郑皓天, 李朝风, 刘良叙, 王嘉伟, 李恒润, 倪俊. 负碳人工光合群落的设计、优化与应用[J]. 合成生物学, 2024, 5(5): 1189-1210. |
| [11] | 夏孔晨, 徐维华, 吴起. 光酶催化混乱性反应的研究进展[J]. 合成生物学, 2024, 5(5): 997-1020. |
| [12] | 陈子苓, 向阳飞. 类器官技术与合成生物学协同研究进展[J]. 合成生物学, 2024, 5(4): 795-812. |
| [13] | 蔡冰玉, 谭象天, 李伟. 合成生物学在干细胞工程化改造中的研究进展[J]. 合成生物学, 2024, 5(4): 782-794. |
| [14] | 谢皇, 郑义蕾, 苏依婷, 阮静怡, 李永泉. 放线菌聚酮类化合物生物合成体系重构研究进展[J]. 合成生物学, 2024, 5(3): 612-630. |
| [15] | 查文龙, 卜兰, 訾佳辰. 中药药效成分群的合成生物学研究进展[J]. 合成生物学, 2024, 5(3): 631-657. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||