合成生物学 ›› 2020, Vol. 1 ›› Issue (5): 570-582.DOI: 10.12211/2096-8280.2020-038
烟酰胺类辅酶依赖型氧化还原酶的辅酶偏好性改造及其在合成生物学中的应用
刘美霞1,2, 李强子1,2, 孟冬冬2, 魏欣蕾2, 游淳1,2
- 1.中国科学院大学,北京 100049
2.中国科学院天津工业生物技术研究所,天津 300308
-
收稿日期:2020-04-03修回日期:2020-09-24出版日期:2020-10-31发布日期:2020-12-03 -
作者简介:作者简介:刘美霞(1990—),女,博士研究生。研究方向为生物化工。
作者简介:李强子(1993—),男,博士研究生。研究方向为生物化工。 -
基金资助:国家自然科学基金面上项目(21778073)
Protein engineering of nicotinamide coenzyme-dependent oxidoreductases for coenzyme preference and its application in synthetic biology
LIU Meixia1,2, LI Qiangzi1,2, MENG Dongdong2, WEI Xinlei2, YOU Chun1,2
- 1.University of Chinese Academy of Sciences,Beijing 100049,China
2.Tianjin Institute of Industrial Biotechnology,Chinese Academy of Sciences,Tianjin 300308,China
-
Received:2020-04-03Revised:2020-09-24Online:2020-10-31Published:2020-12-03
摘要:
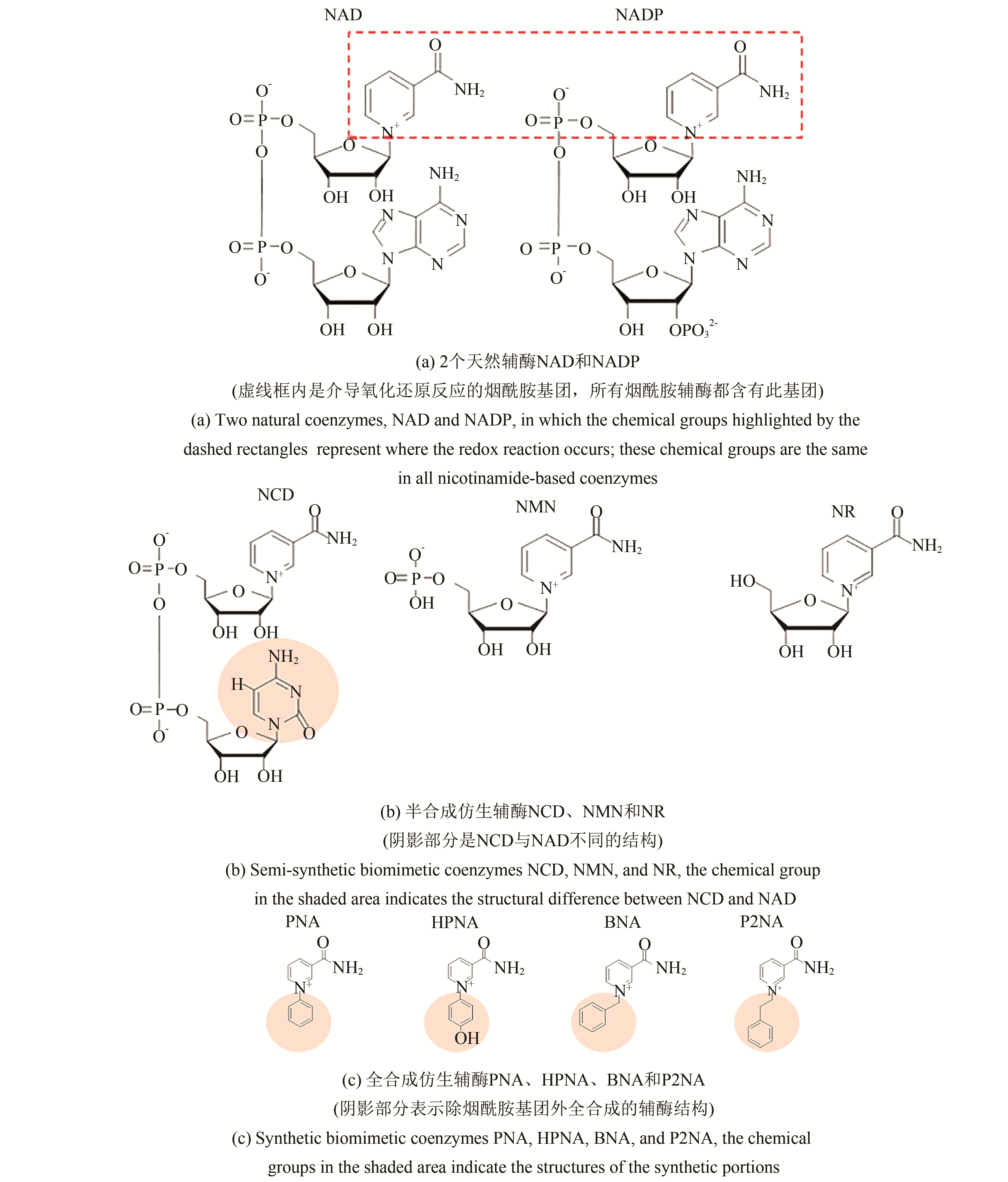
烟酰胺类辅酶NAD(P)是生命体氧化还原过程中最常见的电子中介体。绝大多数氧化还原酶都需要NAD或NADP进行分解代谢和合成代谢,但根据其在代谢过程中的作用,不同氧化还原酶表现出不同的辅酶偏好性。为了解决辅酶在代谢过程中的供需平衡、生产成本和稳定性等问题,对氧化还原酶的辅酶偏好性改造是蛋白质工程的研究热点。本文综述了氧化还原酶的天然烟酰胺辅酶偏好性的改造方法和研究进展,以及天然辅酶偏好性改造工程在提高产品收率和降低生产成本方面的应用。仿生烟酰胺辅酶因成本低、稳定性高,常被用来代替天然烟酰胺辅酶参与氧化还原反应,因此本文还详细介绍了改造酶元件使其偏好仿生烟酰胺辅酶以及其在合成生物学领域构建体内生物正交氧化还原途径和体外合成途径等方面的研究进展。随着更多天然酶元件以及新型优良仿生辅酶的挖掘和设计,从天然辅酶到仿生辅酶的辅酶偏好性改造已成为辅酶工程的一个重要方向。
中图分类号:
引用本文
刘美霞, 李强子, 孟冬冬, 魏欣蕾, 游淳. 烟酰胺类辅酶依赖型氧化还原酶的辅酶偏好性改造及其在合成生物学中的应用[J]. 合成生物学, 2020, 1(5): 570-582.
LIU Meixia, LI Qiangzi, MENG Dongdong, WEI Xinlei, YOU Chun. Protein engineering of nicotinamide coenzyme-dependent oxidoreductases for coenzyme preference and its application in synthetic biology[J]. Synthetic Biology Journal, 2020, 1(5): 570-582.

图2 不同来源的6PGDH辅酶结合域的多序列比对,框内是辅酶识别的loop区
Fig. 2 Amino acid sequence alignment of coenzyme-binding motifs of various 6PGDH enzymes, residues composing loop region and responsible for coenzyme recognition are shown in red box
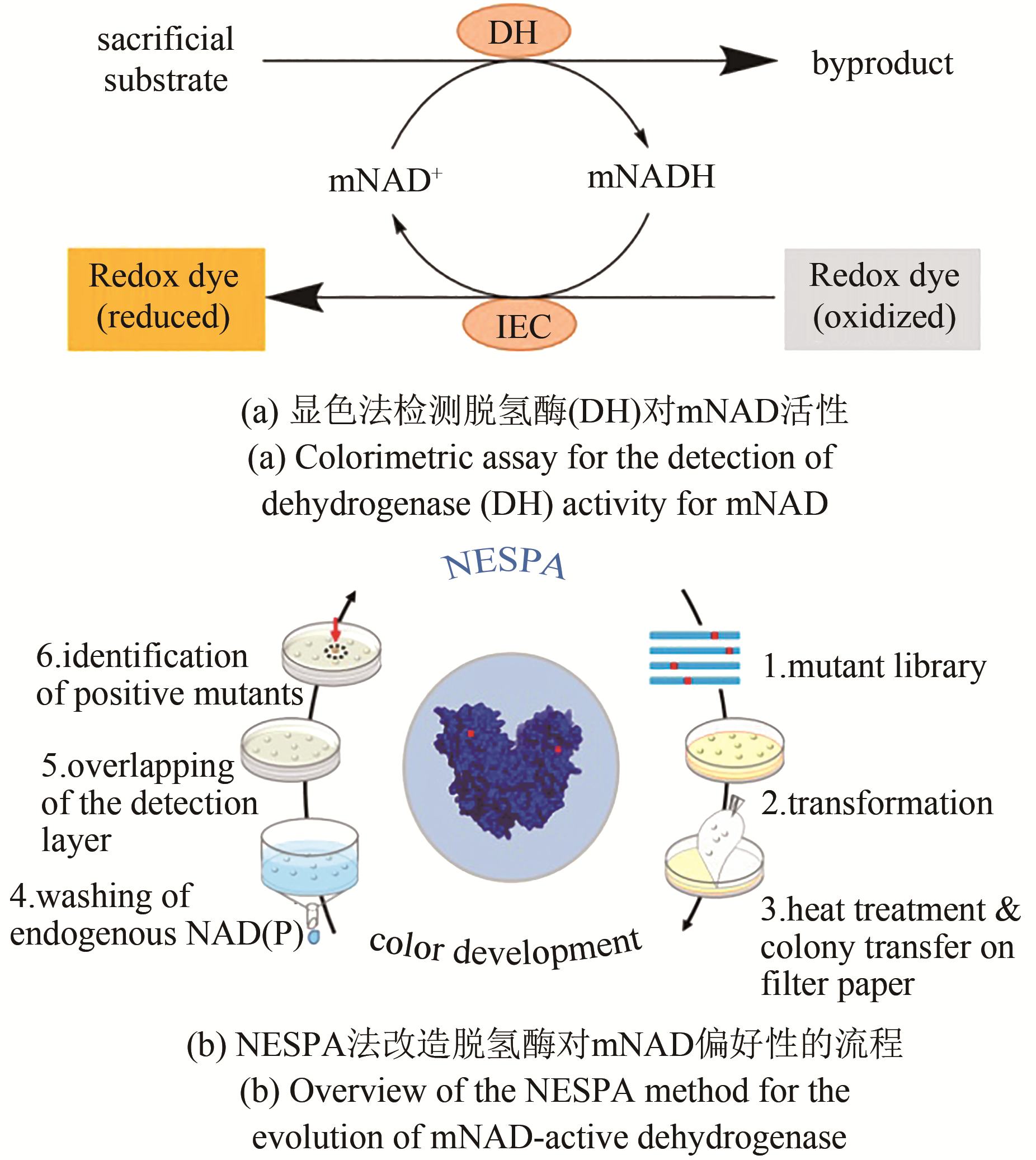
图3 使用NESPA法改造NAD(P)依赖型脱氢酶偏好仿生辅酶(mNAD)
Fig. 3 Schematic presentation of NESPA method for evolution of NAD(P)-dependent dehydrogenases to utilize biomimetic coenzyme mNAD
| Enzyme | Source | Specificity | Mutations | Cofactor specificity shift toward mNADs① | Reference |
|---|---|---|---|---|---|
| alcohol dehydrogenase | Pyrococcus furiosus | NAD→NMN | K249G/H255R | 180-fold | [ |
| phosphite dehydrogenase | Ralstonia sp. 4506 | NAD→NCD | I151R/P176R/M207A | 770-fold | [ |
| malic enzyme | Escherichia coli | NAD→NCD | L310R/Q401C | 33000-fold | [ |
| malic enzyme | Escherichia coli | NAD→NFCD | L310R/Q401C | 30000-fold | [ |
| D-lactate dehydrogenase | Lactobacillus helveticus | NAD→NFCD | V152R | NA | [ |
| glucose dehydrogenase | Bacillus subtilis | NAD→NMN | I195R/A93K/Y39Q/S17E | 2×107-fold | [ |
| glucose dehydrogenase | Bacillus subtilis | NADP→NMN | I195R/A93K/Y39Q/S17E | 1×107-fold | [ |
| glucose dehydrogenase | Sulfolobus solfataricus | NAD→BNA | I192T/V306I | 29-fold | [ |
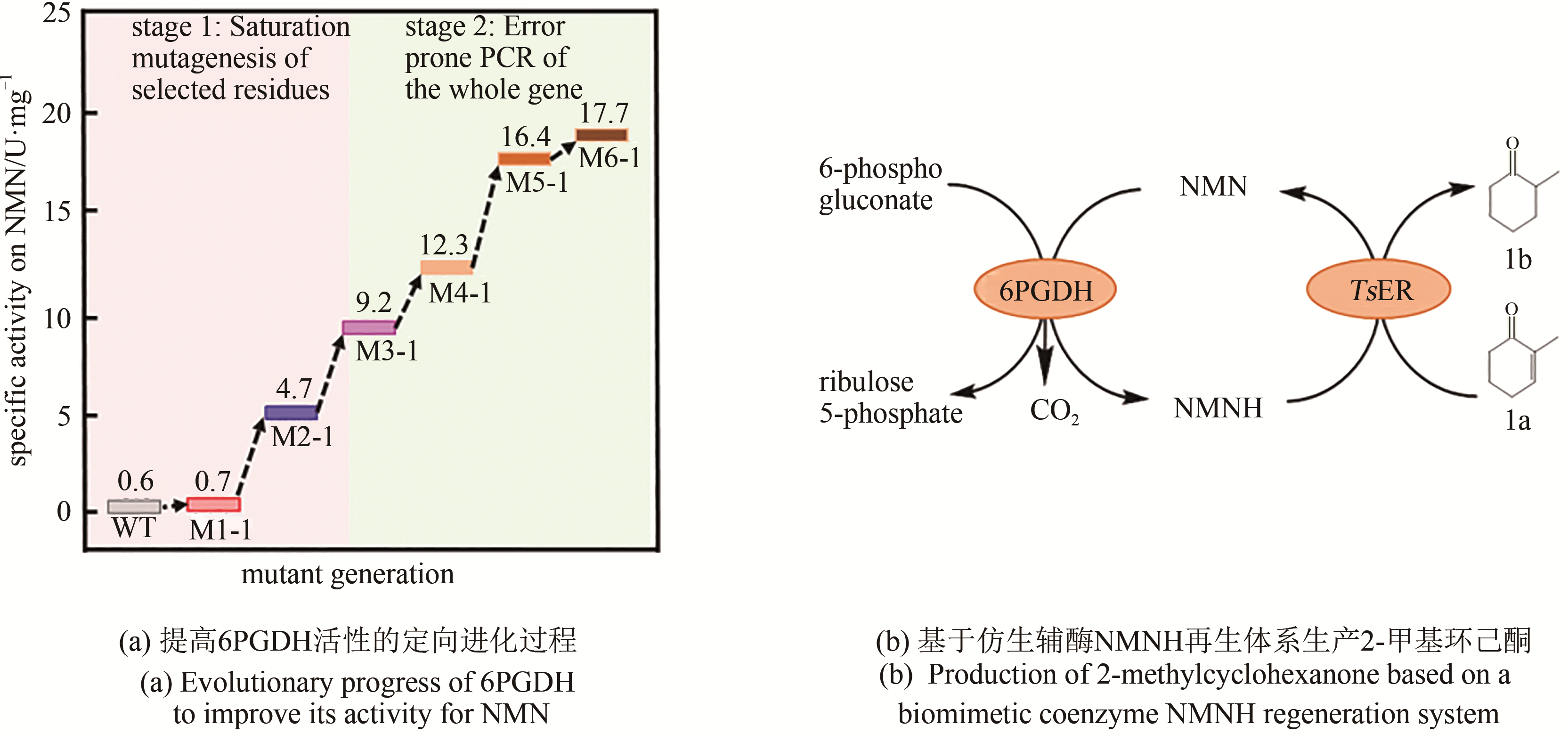
| 6-phosphogluconate dehydrogenase | Thermotoga maritima | NADP→NMN | A11G/K27R/R33I/T34I/F60Y/D82L/T83L/Q86L/K118N/I120F/D251E/D294V/F326S/F329Y/Y383C/N387S/V390G/A447V | 4300-fold | [ |
| formate dehydrogenase | Pseudomonas sp. 101 | NAD→NCD | V198I/C256I/P260S/E261P/S381N/S383F | 3500-fold | [ |
表1 氧化还原酶对仿生烟酰胺辅酶mNADs偏好性的改造
Tab. 1 List of coenzyme engineering of oxidoreductases on biomimetic nicotinamide coenzymes mNADs
| Enzyme | Source | Specificity | Mutations | Cofactor specificity shift toward mNADs① | Reference |
|---|---|---|---|---|---|
| alcohol dehydrogenase | Pyrococcus furiosus | NAD→NMN | K249G/H255R | 180-fold | [ |
| phosphite dehydrogenase | Ralstonia sp. 4506 | NAD→NCD | I151R/P176R/M207A | 770-fold | [ |
| malic enzyme | Escherichia coli | NAD→NCD | L310R/Q401C | 33000-fold | [ |
| malic enzyme | Escherichia coli | NAD→NFCD | L310R/Q401C | 30000-fold | [ |
| D-lactate dehydrogenase | Lactobacillus helveticus | NAD→NFCD | V152R | NA | [ |
| glucose dehydrogenase | Bacillus subtilis | NAD→NMN | I195R/A93K/Y39Q/S17E | 2×107-fold | [ |
| glucose dehydrogenase | Bacillus subtilis | NADP→NMN | I195R/A93K/Y39Q/S17E | 1×107-fold | [ |
| glucose dehydrogenase | Sulfolobus solfataricus | NAD→BNA | I192T/V306I | 29-fold | [ |
| 6-phosphogluconate dehydrogenase | Thermotoga maritima | NADP→NMN | A11G/K27R/R33I/T34I/F60Y/D82L/T83L/Q86L/K118N/I120F/D251E/D294V/F326S/F329Y/Y383C/N387S/V390G/A447V | 4300-fold | [ |
| formate dehydrogenase | Pseudomonas sp. 101 | NAD→NCD | V198I/C256I/P260S/E261P/S381N/S383F | 3500-fold | [ |
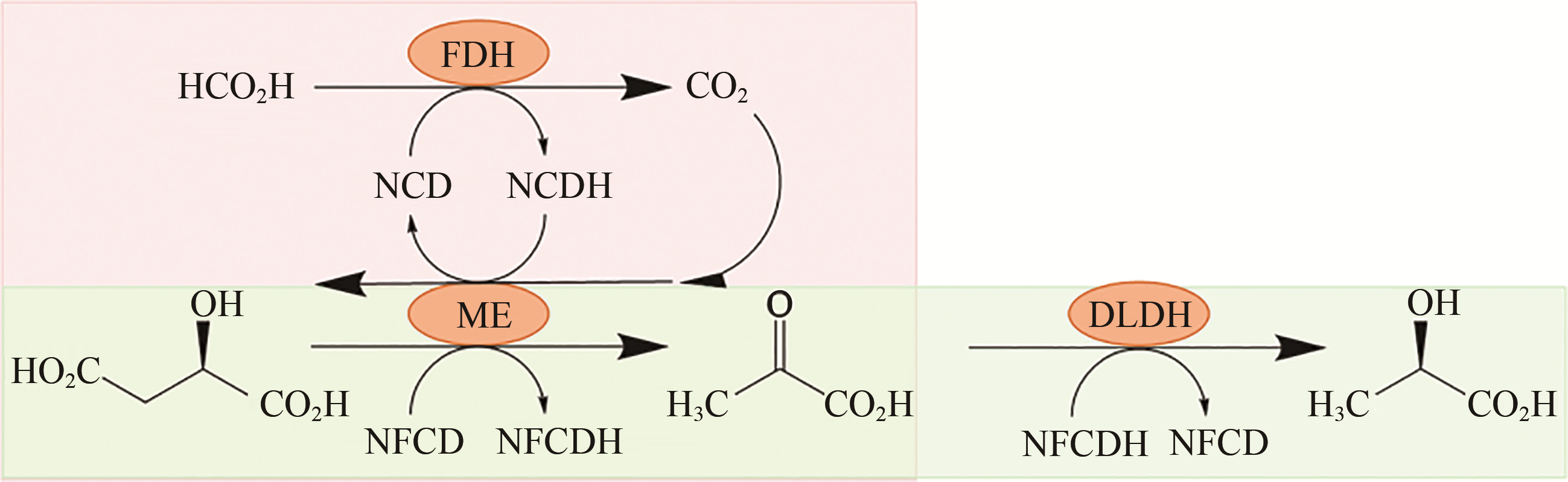
图5 由仿生辅酶NCD和NFCD与突变体酶苹果酸酶(ME)、D-乳酸脱氢酶(DLDH)和甲酸脱氢酶(FDH)组成的仿生辅酶正交体系
Fig. 5 Cofactor orthogonal system of NCD and NFCD with mutated malic enzyme (ME), D-lactate dehydrogenase (DLDH) and formate dehydrogenase (FDH)
| 1 | GUARNERI A, BERKEL W J VAN, PAUL C E. Alternative coenzymes for biocatalysis [J]. Current Opinion in Biotechnology, 2019, 60: 63-71. |
| 2 | WU Hong, TIAN Chunyong, SONG Xiaokai, et al. Methods for the regeneration of nicotinamide coenzymes [J]. Green Chemistry, 2013, 15(7): 1773-1789. |
| 3 | YOU Chun, ZHANG Yi-Heng P. Biomanufacturing by in vitro biosystems containing complex enzyme mixtures [J]. Process Biochemistry, 2017, 52: 106-114. |
| 4 | HUFFMAN M A, FRYSZKOWSKA A, ALVIZO O, et al. Design of an in vitro biocatalytic cascade for the manufacture of islatravir [J]. Science, 2019, 366(6470): 1255-1259. |
| 5 | WANG Yu, TAPPERTZHOFEN N, MENDEZ-SANCHEZ D, et al. Design and use of de novo cascades for the biosynthesis of new benzylisoquinoline alkaloids [J]. Angewandte Chemie-International Edition, 2019, 58(30): 10120-10125. |
| 6 | 应向贤, 毛王伟, 张丽, 等. 氧化还原酶在多酶级联反应中的应用进展[J]. 发酵科技通讯, 2018, 47(3): 6-10. |
| YING Xiangxian, MAO Wangwei, ZHANG Li, et al. Multi-enzymatic cascade reation involving oxidoreductases [J]. Bulletin of Fermentation Science and Technology. 2018, 47(3): 6-10. | |
| 7 | WICHMANN R, VASIC-RACKI D. Cofactor regeneration at the lab scale [J]. Advances in Biochemical Engineering /Biotechnology, 2005, 92: 225-260. |
| 8 | WASYLENKO T M, STEPHANOPOULOS G. Metabolomic and 13C-metabolic flux analysis of a xylose-consuming Saccharomyces cerevisiae strain expressing xylose isomerase [J]. Biotechnology Bioengineering, 2015, 112(3): 470-483. |
| 9 | 李慧敏, 李宁宁, 翁建清, 等. 酶促还原过程中辅因子的再生研究[J]. 杭州师范大学学报(自然科学版), 2019, 18(6): 605-609. |
| LI Huimin, LI Ningning, WENG Jianqing, et al. Regenaration of cofactor in enzymatic reduction [J]. Journal of Hangzhou Normal University (Natural Science Edition), 2019, 18(6): 605-609. | |
| 10 | 吉爱国, 高培基. 烟酰胺类辅因子的保留和再生研究进展[J]. 药物生物技术, 1997(2): 59-65. |
| JI Aiguo, GAO Peiji. Advances in retention and regeneration of nicotinamide cofactor [J]. Chinese Journal of Pharmaceutical Biotechnology, 1997(2): 59-65. | |
| 11 | 张红缨, 孔祥铎, 张今. 蛋白质工程的新策略——酶的体外定向进化[J]. 科学通报, 1999(11): 3-9. |
| ZHANG Hongying, KONG Xiangduo, ZHANG Jin. A new strategy of protein engineering — directed evolution of enzymes in vitro [J]. Chinese Science Bulletin, 1999(11): 3-9. | |
| 12 | YOU Chun, HUANG Rui, WEI Xinlei, et al. Protein engineering of oxidoreductases utilizing nicotinamide-based coenzymes, with applications in synthetic biology [J]. Synthetic and Systems Biotechnology, 2017, 2(3): 208-218. |
| 13 | BASTIAN S, LIU Xiang, MEYEROWITZ J T, et al. Engineered ketol-acid reductoisomerase and alcohol dehydrogenase enable anaerobic 2-methylpropan-1-ol production at theoretical yield in Escherichia coli [J]. Metablic Engineering, 2011, 13(3): 345-352. |
| 14 | HUANG Rui, CHEN Hui, ZHONG Chao, et al. High-throughput screening of coenzyme preference change of thermophilic 6-phosphogluconate dehydrogenase from NADP+ to NAD+ [J]. Scientific Reports, 2016, 6: 32644. |
| 15 | CAMPBELL E, MEREDITH M, MINTEER S D, et al. Enzymatic biofuel cells utilizing a biomimetic cofactor [J]. Chemical Communications, 2012, 48(13): 1898-1900. |
| 16 | LEIGH J A, ALBERS S V, ATOMI H, et al. Model organisms for genetics in the domain Archaea: methanogens, halophiles, Thermococcales and Sulfolobales [J]. FEMS Microbiology Reviews, 2011, 35(4): 577-608. |
| 17 | PAUL C E, ARENDS I W C E, HOLLMANN F. Is simpler better? Synthetic nicotinamide cofactor analogues for redox chemistry [J]. ACS Catalysis, 2014, 4(3): 788-797. |
| 18 | PAUL C E, HOLLMANN F. A survey of synthetic nicotinamide cofactors in enzymatic processes [J]. Applied Microbiology and Biotechnology, 2016, 100(11): 4773-4778. |
| 19 | LIU Yuxue, FENG Yanbin, WANG Lei, et al. Structural insights into phosphite dehydrogenase variants favoring a non-natural redox cofactor [J]. ACS Catalysis, 2019, 9(3): 1883-1887. |
| 20 | JI Debin, WANG Lei, HOU Shuhua, et al. Creation of bioorthogonal redox systems depending on nicotinamide flucytosine dinucleotide [J]. Journal of the American Chemical Society, 2011, 133(51): 20857-20862. |
| 21 | WANG Lei, LIU Bin, LIU Yuxue, et al. Escherichia coli strain designed for characterizing in vivo functions of nicotinamide adenine dinucleotide analogues [J]. Organic Letters, 2019, 21(9): 3218-3222. |
| 22 | CHICA R A, NICOLAS D, JOELLE N P. Semi-rational approaches to engineering enzyme activity: combining the benefits of directed evolution and rational design [J]. Current Oponion in Biotechnology, 2005, 16(4): 378-384. |
| 23 | LÜ Jing, SUN Honglei, HE Hao, et al. Synthetic biology: its applications in biotechnology [J]. Progress in Biochemistry & Biophysics, 2012, 39(2): 95-118. |
| 24 | 曲戈, 朱彤, 蒋迎迎, 等. 蛋白质工程: 从定向进化到计算设计[J]. 生物工程学报, 2019, 35(10): 1843-1856. |
| QU Ge, ZHU Tong, ZHENG Yingying, ta el. Protein engineering: from directed evolution to computational design [J]. Chinese Journal of Biotechnology, 2019, 35(10): 1843-1856. | |
| 25 | AN Yingfeng, WU Wenfang, Anguo LÜ. A convenient and robust method for construction of combinatorial and random mutant libraries [J]. Biochimie, 2010, 92(8): 1081-1084. |
| 26 | KIM Jae-Eung, HUANG Rui, CHEN Hui, et al. Facile construction of random gene mutagenesis library for directed evolution without the use of restriction enzyme in Escherichia coli [J]. Biotechnology Journal, 2016, 11(9): 1142-1150. |
| 27 | CUI Dongbing, ZHANG Lujia, JIANG Shuiqin, et al. A computational strategy for altering an enzyme in its cofactor preference to NAD(H) and/or NADP(H) [J]. FEBS Journal, 2015, 282(12): 2339-2351. |
| 28 | BLACK W B, ZHANG Linyue, Wai Shun MAK, et al. Engineering a nicotinamide mononucleotide redox cofactor system for biocatalysis [J]. Nature Chemical Biology, 2020, 16(1): 87-94. |
| 29 | LESK A M. NAD-binding domains of dehydrogenases [J]. Current Opinion in Structural Biology, 1996, 5(6): 775-783. |
| 30 | RAO S T, ROSSMANN M G. Comparison of super-secondary structures in proteins [J]. Journal of Molecular Biology, 1973, 76(2): 241-256. |
| 31 | NOWAK C, PICK A, LOMMES P, et al. Enzymatic reduction of nicotinamide biomimetic cofactors using an engineered glucose dehydrogenase: providing a regeneration system for artificial cofactors [J]. ACS Catalysis, 2017, 7(8): 5202-5208. |
| 32 | WULF H, MALLIN H, BORNSCHEUER U T. Protein engineering of a thermostable polyol dehydrogenase [J]. Enzyme and Microbial Technology, 2012, 51(4): 217-224. |
| 33 | BUBNER P, KLIMACEK M, NIDETZKY B. Structure-guided engineering of the coenzyme specificity of Pseudomonas fluorescens mannitol 2-dehydrogenase to enable efficient utilization of NAD(H) and NADP(H) [J]. FEBS Letters, 2008, 582(2): 233-237. |
| 34 | ROSELL A, VALENCIA E, OCHOA W F, et al. Complete reversal of coenzyme specificity by concerted mutation of three consecutive residues in alcohol dehydrogenase [J]. Journal of Biological Chemistry, 2003, 278(42): 40573-40580. |
| 35 | MORIKAWA S, NAKAI T, YASOHARA Y, et al. Highly active mutants of carbonyl reductase S1 with inverted coenzyme specificity and production of optically active alcohols [J]. Bioscience Biotechnology & Biochemistry, 2005, 69(3): 544-552. |
| 36 | CHEN Hui, ZHU Zhiguang, HUANG Rui, et al. Coenzyme engineering of a hyperthermophilic 6-phosphogluconate dehydrogenase from NADP+ to NAD+ with its application to biobatteries [J]. Scientific Reports, 2016, 6: 36311. |
| 37 | ROSADO L A, CACERES R A, DE AZEVEDO W F JR, et al. Role of Serine140 in the mode of action of Mycobacterium tuberculosis β-ketoacyl-ACP reductase (MabA) [J]. BMC Research Notes, 2012, 5: 526. |
| 38 | BRINKMANN-CHEN S, FLOCK T, CAHN J K B, et al. General approach to reversing ketol-acid reductoisomerase cofactor dependence from NADPH to NADH [J]. Proeedings of the National Academy of Sciences of United States of America, 2013, 110(27): 10946-10951. |
| 39 | GEERTZ-HANSEN H M, BLOM N, FEIST A M, et al. Cofactory: Sequence-based prediction of cofactor specificity of Rossmann folds [J]. Proteins Structure Function & Bioinformatics, 2014, 82(9): 1819-1828. |
| 40 | LEVY H R, VALARIE E V, YIN Xiaohong, et al. Identification of an arginine residue in the dual coenzyme-specific glucose-6-phosphate dehydrogenase from Leuconostoc mesenteroides that plays a key role in binding NADP+ but not NAD+ [J]. Archives of Biochemistry and Biophysics, 1996, 326(1): 145-151. |
| 41 | LI Lei, COOK P F. The 2ʹ-phosphate of NADP is responsible for proper orientation of the nicotinamide ring in the oxidative decarboxylation reaction catalyzed by sheep liver 6-phosphogluconate dehydrogenase [J]. Journal of Biological Chemistry, 2006, 281: 36803-36810. |
| 42 | TETAUD E, HANAU S, WELLS J M, et al. 6-Phosphogluconate dehydrogenase from Lactococcus lactis: a role for arginine residues in binding substrate and coenzyme [J]. Biochemical Journal, 1999, 338(1): 55-60. |
| 43 | HUANG Rui, CHEN Hui, UPP D M, et al. A high-throughput method for directed evolution of NAD(P)+-dependent dehydrogenases for the reduction of biomimetic nicotinamide analogues [J]. ACS Catalysis, 2019, 9(12): 11709-11719. |
| 44 | WANG Xueying, ZHOU Yongjin, WANG Lei, et al. Engineering Escherichia coli nicotinic acid mononucleotide adenylyltransferase for fully active amidated NAD biosynthesis [J]. Applied and Environmental Microbiology, 2017, 83(13): e00692-17. |
| 45 | EHSANI M, FERNANDEZ M R, BIOSCA J A, et al. Reversal of coenzyme specificity of 2,3-butanediol dehydrogenase from Saccharomyces cerevisae and in vivo functional analysis [J]. Biotechnology and Bioengineering, 2009, 104(2): 381-389. |
| 46 | 刘云, 徐琪寿. 氨基酸发酵生产的研究进展[J]. 氨基酸和生物资源, 1999(4): 21-25. |
| LIU Yun, XU Qishou. Progress in amino acid production by ferment method [J]. Amino Acids and Biotic Resources, 1999(4): 21-25. | |
| 47 | BOMMAREDDY, REDDY R, ZHEN Chen, et al. A de novo NADPH generation pathway for improving lysine production of Corynebacterium glutamicum by rational design of the coenzyme specificity of glyceraldehyde 3-phosphate dehydrogenase [J]. Metabolic Engineering, 2014, 25: 30-37. |
| 48 | MATSUSHIKA A, WATANABE S, KODAKI T, et al. Bioethanol production from xylose by recombinant Saccharomyces cerevisiae expressing xylose reductase, NADP+-dependent xylitol dehydrogenase, and xylulokinase [J]. Journal Bioscience and Bioengineering, 2008, 105(3): 296-299. |
| 49 | MATSUSHIKA A, WATANABE S, KODAKI T, et al. Expression of protein engineered NADP+-dependent xylitol dehydrogenase increases ethanol production from xylose in recombinant Saccharomyces cerevisiae [J]. Applied Microbioligy and Biotechnology, 2008, 81(2): 243-255. |
| 50 | CAHN J K B, WERLANG C A, BAUMSCHLAGER A, et al. A general tool for engineering the NAD/NADP cofactor preference of oxidoreductases [J]. ACS Synthetic Biology, 2017, 6(2): 326-333. |
| 51 | LIN P P, RABE K S, TAKASUMI J L, et al. Isobutanol production at elevated temperatures in thermophilic Geobacillus thermoglucosidasius [J]. Metabolic Engineering, 2014, 24: 1-8. |
| 52 | 郭媛, 林丽华, 郭玲, 等. 基因敲除提高大肠杆菌工程菌生产异丁醇产量的研究[J]. 生物技术通报, 2012(7): 170-175. |
| GUO Yuan, LIN Lihua, GUO Ling, et al. Influence of isobutanol production in gene defective mutant of Escherichia coli [J]. Biotechnology Bulletin, 2012(7): 170-175. | |
| 53 | PETSCHACHER B, NIDETZKY B. Altering the coenzyme preference of xylose reductase to favor utilization of NADH enhances ethanol yield from xylose in a metabolically engineered strain of Saccharomyces cerevisiae [J]. Microbial Cell Factories, 2008, 7(1): 9. |
| 54 | WATANABE S, SALEH A A, PACK Seung Pil, et al. Ethanol production from xylose by recombinant Saccharomyces cerevisiae expressing protein-engineered NADH-preferring xylose reductase from Pichia stipitis [J]. Microbiology, 2007, 153(9): 3044-3054. |
| 55 | HOFMANN D, WIRTZ A, SANTIAGO-SCHUBEL B, et al. Structure elucidation of the thermal degradation products of the nucleotide cofactors NADH and NADPH by nano-ESI-FTICR-MS and HPLC-MS [J]. Analytical and Bioanalytical Chemistry, 2010, 398(7/8): 2803-2811. |
| 56 | 茹祥莉, 郝二军, 贺燕燕, 等. 烟酰胺辅酶NADH新型模型物的合成与表征[J]. 河南师范大学学报(自然版), 2013(3): 109-110. |
| RU Xiangli, HAO Erjun, HE Yanyan, et al. Synthesis and characterization of the new coenzyme NADH model compounds [J]. Journal of Henan Normal University (Natural Science Dition), 2013(3): 109-110. | |
| 57 | ZACHOS I, NOWAK C, SIEBER V. Biomimetic cofactors and methods for their recycling [J]. Current Opinion in Chemical Biology, 2019, 49: 59-66. |
| 58 | GUO Xiaojia, LIU Yuxue, WANG Qian, et al. Non-natural cofactor and formate driven reductive carboxylation of pyruvate [J]. Angewandte Chemie-International Edition, 2019, 59(8): 3143-3146. |
| 59 | ZHANG Y-H P. Simpler is better: high-yield and potential low-cost biofuels production through cell-free synthetic pathway biotransformation (SyPaB) [J]. ACS Catalysis, 2011, 1(9): 998-1009. |
| 60 | 刘阳, 郭小翠, 耿金慧, 等. 体外合成生物学: 无细胞蛋白合成系统研究进展[J]. 科学通报, 2017(33): 61-70. |
| LIU Yang, GUO Xiaocui, GENG Jinhui, et al. In vitro synthetic biology: cell-free protein synthesis [J]. Chinese Science Bulletin, 2017(33): 61-70. | |
| 61 | 王文方, 钟建江. 合成生物学驱动的智能生物制造研究进展[J]. 生命科学, 2019, 31(4): 95-104. |
| WANG Wenfang, ZHONG Jianjiang. Recent advances in smart biomanufacturing driven by synthetic biology [J]. Chinese Bulletin of Life Sciences, 2019, 31(4): 95-104. | |
| 62 | ZHU Zhiguang, Tsz Kin TAM, SUN Fangfang, et al. A high-energy-density sugar biobattery based on a synthetic enzymatic pathway [J]. Nature Communications, 2014, 5: 3026. |
| 63 | ROLLIN J A, CAMPO J M DEL, MYUNG Suwan, et al. High-yield hydrogen production from biomass by in vitro metabolic engineering: mixed sugars coutilization and kinetic modeling [J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(16): 4964-4969. |
| 64 | YOU Chun, SHI Ting, LI Yunjie, et al. An in vitro synthetic biology platform for the industrial biomanufacturing of myo-inositol from starch [J]. Biotechnology and Bioengineering, 2017, 114(8): 1855-1864. |
| 65 | WANG Wei, LIU Meixia, YOU Chun, et al. ATP-free biosynthesis of a high-energy phosphate metabolite fructose 1,6-diphosphate by in vitro metabolic engineering [J]. Metabolic Engineering, 2017, 42: 168-174. |
| 66 | LI Zhong, YAN Jinxin, SUN Jinkai, et al. Production of value-added chemicals from glycerol using in vitro enzymatic cascades [J]. Communications Chemistry, 2018, 1(71). |
| 67 | BAI Xue, MENG Dongdong, WEI Xinlei, et al. Facile synthesis of (-)-vibo-quercitol from maltodextrin via an in vitro synthetic enzymatic biosystem [J]. Biotechnology Bioengineering, 2019, 116(10): 2710-2719. |
| 68 | BEER B, PICK A, SIEBER V. In vitro metabolic engineering for the production of alpha-ketoglutarate [J]. Metablic Engineering, 2017, 40: 5-13. |
| 69 | LI Yuan, LIU Shan, YOU Chun. Permeabilized Escherichia coli whole cells containing co-expressed two thermophilic enzymes facilitate the synthesis of scyllo-Inositol from myo-Inositol [J]. Biotechnology Journal, 2020, 15(2): e1900191. |
| 70 | ZHU Zhiguang, MA Chunling, ZHANG Yi Heng Percival. Co-utilization of mixed sugars in an enzymatic fuel cell based on an in vitro enzymatic pathway [J]. Electrochimica Acta, 2018, 263: 184-191. |
| 71 | ZHU Zhiguang, ZHANG Yi Heng Percival. In vitro metabolic engineering of bioelectricity generation by the complete oxidation of glucose [J]. Metabolic Engineering, 2017, 39: 110-116. |
| 72 | KIM Eui-Jin, KIM Jae-Eung, ZHANG Yi-Heng P Job. Ultra-rapid rates of water splitting for biohydrogen gas production through in vitro artificial enzymatic pathways [J]. Energy & Environmental Science, 2018, 11(8): 2064-2072. |
| 73 | KIM Jae-Eung, KIM Eui-Jin, CHEN Hui, et al. Advanced water splitting for green hydrogen gas production through complete oxidation of starch by in vitro metabolic engineering [J]. Metabolic Engineering, 2017, 44: 246-252. |
| 74 | RUPP S. Next-generation bioproduction systems: cell-free conversion concepts for industrial biotechnology [J]. Engineering in Life Sciences, 2013, 13(1): 19-25. |
| 75 | 谢希贤, 马倩, 陈宁. 氨基酸生产菌代谢工程研究发展趋势[J]. 生物产业技术, 2016(4): 32-36. |
| XIE Xixian, MA Qian, CHEN Ning. Development trend of metabolic engineering of amino acid producing bacteria [J]. Biotechnology & Business, 2016(4): 32-36. | |
| 76 | 许晟, 郝宁, 许琳, 等. 谷氨酸棒杆菌串联生产氨基酸和有机酸的发酵工艺[J]. 南京工业大学学报(自然科学版), 2014(6): 53. |
| XU Sheng, HAO Ning, XU Lin, et al. Production of amino acids and organic acids in series fermentation process by Corynebacterium glutamicum [J]. Journal of Nanjing Tech University (Natural Science Edition), 2014(6): 53. | |
| 77 | CHEN Xianzhong, ZHOU Junbo, ZHANG Lihua, et al. Development of an Escherichia coli-based biocatalytic system for the efficient synthesis of N-acetyl-D-neuraminic acid [J]. Metabolic Engineering, 2018, 47: 374-382. |
| 78 | LI Zhengjun, HONG Penghui, Yangyang DA, et al. Metabolic engineering of Escherichia coli for the production of L-malate from xylose [J]. Metabolic Engineering, 2018, 48: 25-32. |
| 79 | NEMR K, MULLER J E N, Jeong Chan JOO, et al. Engineering a short, aldolase-based pathway for (R)-1,3-butanediol production in Escherichia coli [J]. Metabolic Engineering, 2018, 48: 13-24. |
| 80 | QU Yanan, YAN Haojie, GUO Qiang, et al. Biosynthesis of D-glucaric acid from sucrose with routed carbon distribution in metabolically engineered Escherichia coli [J]. Metabolic Engineering, 2018, 47: 393-400. |
| 81 | 何煦昌. 手性药物的发展[J]. 中国医药工业杂志, 1997(11): 519-524. |
| HE Xuchang. Development of chiral drugs [J]. Chinese Journal of Pharmaceuticals, 1997(11): 519-524. | |
| 82 | 王丹, 李亚, 何浪. 手性药物及其开发与应用[J]. 现代医药卫生, 2007(6): 51-52. |
| WANG Dan, LI Ya, HE Lang. Chiral drugs and their development and Application [J]. Journal of Modern Medicine & Health, 2007(6): 51-52. | |
| 83 | WANG Lei, JI Debin, LIU Yuxue, et al. Synthetic cofactor-linked metabolic circuits for selective energy transfer [J]. ACS Catalysis, 2017, 7(3): 1977-1983. |
| 84 | 黄玉梅, 刘雅, 张海丽, 等. 氨基硅烷磁性纳米粒子修饰碳糊电极固定血红蛋白的过氧化氢传感器[J]. 分析化学, 2009, 37(a03): 67-67. |
| HUANG Yumei, LIU Ya, ZHANG Haili, et al. Hydrogen peroxide sensor based on hemoglobin immobilized on carbon paste electrode modified by aminosilane magnetic nanoparticles [J]. Chinese Journal of Analytical Chemistry, 2009, 37(a03): 67-67. | |
| 85 | 宋娟, 杨亲正. 固定化酶阳极修饰在酶生物燃料电池研究中的进展[J]. 齐鲁工业大学学报(自然科学版), 2010, 24(3): 71-73. |
| SONG Juan, YANG Qinzheng. Progress in enzyme immobilization anode of enzymatic fuel cell [J]. Journal of Qilu University of Technology (Natural Science Edition), 2010, 24(3): 71-73. | |
| 86 | 宋荣斌, 朱俊杰. 酶生物燃料电池自供能传感器的研究现状及应用[J]. 分析科学学报, 2017(5): 101-105. |
| SONG Rongbin, ZHU Junjie. Recent progress in self-powered sensor based on enzymatic biofuel cells [J]. Journal of Analytical Science, 2017(5): 101-105. | |
| 87 | 邹琼, 刘娟, 朱刚兵, 等. 一种新型酶生物燃料电池阳极构建及性能研究[J]. 化学学报, 2013, 71(8): 1154-1160. |
| ZHOU Qiong, LIU Juan, ZHU Gangbing, et al. Construction and performance of a new bioanode for biofuel cells [J]. Acta Chimica Sinica, 2013, 71(8): 1154-1160. | |
| 88 | MOORE C M, AKERS N L, HILL A D, et al. Improving the environment for immobilized dehydrogenase enzymes by modifying Nafion with tetraalkylammonium bromides [J]. Biomacromolecules, 2004, 5(4): 1241-1247. |
| 89 | AKERS N L, MOORE C M, MINTEER S D. Development of alcohol/O2 biofuel cells using salt-extracted tetrabutylammonium bromide/Nafion membranes to immobilize dehydrogenase enzymes [J]. Electrochimica Acta, 2005, 50(12): 2521-2525. |
| 90 | MOORE C M, MINTEER S D, MARTIN R S. Microchip-based ethanol/oxygen biofuel cell [J]. Lab on a Chip, 2005, 5(2): 218-225. |
| 91 | PAUL C E, GARGIULO S, OPPERMAN D J, et al. Mimicking nature: synthetic nicotinamide cofactors for C=C bioreduction using enoate reductases [J]. Organic Letters, 2013, 15(1): 180-183. |
| 92 | LÖW S A, LÖW I M, WEISSENBORN M J, et al. Enhanced ene-reductase activity through alteration of artificial nicotinamide cofactor substituents [J]. ChemCatChem, 2016, 8(5): 911-915. |
| 93 | LUTZ J, HOLLMANN F, HO T V, et al. Bioorganometallic chemistry: biocatalytic oxidation reactions with biomimetic NAD+/NADH co-factors and [Cp*Rh(bpy)H]+ for selective organic synthesis [J]. Journal Organometallic Chemistry, 2004, 689(25): 4783-4790. |
| 94 | ZHANG Lei, Jun JUN, XU Yufang, et al. New artificial fluoro-cofactor of hydride transfer with novel fluorescence assay for redox biocatalysis [J]. Chemical Communications, 2016, 52: 6471-6474. |
| 95 | KNOX R J, FRIEDLOS F, JARMAN M, et al. Virtual cofactors for an Escherichia coli nitroreductase enzyme: Relevance to reductively activated prodrugs in antibody directed enzyme prodrug therapy (ADEPT) [J]. Biochemical Pharmacology, 1995, 49(11): 1641-1647. |
| 通讯作者:游淳(1983—),男,博士,研究员,博士生导师。主要研究方向为构建和优化体外合成生物体系高效生产高值化学品,例如单糖、寡糖、肌醇及其衍生物、人造淀粉等。 |
| [1] | 高歌, 边旗, 王宝俊. 合成基因线路的工程化设计研究进展与展望[J]. 合成生物学, 2025, 6(1): 45-64. |
| [2] | 李冀渊, 吴国盛. 合成生物学视域下有机体的两种隐喻[J]. 合成生物学, 2025, 6(1): 190-202. |
| [3] | 焦洪涛, 齐蒙, 邵滨, 蒋劲松. DNA数据存储技术的法律治理议题[J]. 合成生物学, 2025, 6(1): 177-189. |
| [4] | 唐兴华, 陆钱能, 胡翌霖. 人类世中对合成生物学的哲学反思[J]. 合成生物学, 2025, 6(1): 203-212. |
| [5] | 徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176. |
| [6] | 温艳华, 刘合栋, 曹春来, 巫瑞波. 蛋白质工程在医药产业中的应用[J]. 合成生物学, 2025, 6(1): 65-86. |
| [7] | 石婷, 宋展, 宋世怡, 张以恒. 体外生物转化(ivBT):生物制造的新前沿[J]. 合成生物学, 2024, 5(6): 1437-1460. |
| [8] | 柴猛, 王风清, 魏东芝. 综合利用木质纤维素生物转化合成有机酸[J]. 合成生物学, 2024, 5(6): 1242-1263. |
| [9] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [10] | 王子渊, 杨立荣, 吴坚平, 郑文隆. 酶促合成手性氨基酸的研究进展[J]. 合成生物学, 2024, 5(6): 1319-1349. |
| [11] | 陈雨, 张康, 邱以婧, 程彩云, 殷晶晶, 宋天顺, 谢婧婧. 微生物电合成技术转化二氧化碳研究进展[J]. 合成生物学, 2024, 5(5): 1142-1168. |
| [12] | 郑皓天, 李朝风, 刘良叙, 王嘉伟, 李恒润, 倪俊. 负碳人工光合群落的设计、优化与应用[J]. 合成生物学, 2024, 5(5): 1189-1210. |
| [13] | 夏孔晨, 徐维华, 吴起. 光酶催化混乱性反应的研究进展[J]. 合成生物学, 2024, 5(5): 997-1020. |
| [14] | 陈子苓, 向阳飞. 类器官技术与合成生物学协同研究进展[J]. 合成生物学, 2024, 5(4): 795-812. |
| [15] | 蔡冰玉, 谭象天, 李伟. 合成生物学在干细胞工程化改造中的研究进展[J]. 合成生物学, 2024, 5(4): 782-794. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||