合成生物学 ›› 2021, Vol. 2 ›› Issue (4): 528-542.DOI: 10.12211/2096-8280.2020-091
人造蛋白功能材料的生物合成及应用
曾丹1, 储建林2, 陈燕茹1, 范代娣1
- 1.西北大学化工学院,陕西省可降解生物医用材料重点实验室,陕西省生物材料与发酵工程技术研究中心,陕西 西安 710069
2.南京工业大学药学院,江苏 南京 210000
-
收稿日期:2020-12-28修回日期:2021-05-29出版日期:2021-08-31发布日期:2021-09-10 -
通讯作者:范代娣 -
作者简介:曾丹 (1989—),女,博士,副教授。主要研究方向为生物医用材料。E-mail:zengdan0301@nwu.edu.cn范代娣 (1965—),女,博士,教授。主要研究方向为可降解生物材料,预防医学和营养医学。E-mail:fandaidi@nwu.edu.cn -
基金资助:国家重点研发计划“合成生物学”重点专项;人造蛋白质合成的细胞设计构建及应用(2019YFA0905200)
Biological synthesis and applications of artificial protein functional materials
ZENG Dan1, CHU Jianlin2, CHEN Yanru1, FAN Daidi1
- 1.School of Chemical Engineering,Shaanxi Key Laboratory of Degradable Biomedical Materials,Shaanxi R&D Center of Biomaterials and Fermentation Engineering,Northwest University,Xi’an 710069,Shaanxi,China
2.School of Pharmacy,Nanjing University of Technology,Nanjing 210000,Jiangsu,China
-
Received:2020-12-28Revised:2021-05-29Online:2021-08-31Published:2021-09-10 -
Contact:FAN Daidi
摘要:
人造蛋白功能材料具有良好的生物相容性和生物可降解性,来源广泛且功能多样,是一类理想的生物材料,在生物工程、医药、军事和纺织等领域具有广泛的应用前景。然而,现阶段蛋白功能材料的微生物合成仍存在表达量低、性能不稳定等问题,严重限制了这些蛋白材料的高效生产与应用。合成生物学在工程化理念的指导下,为人造蛋白功能材料提供了“精准设计—系统构建—调控表达—工程应用”的研究策略。本文介绍了蛛丝蛋白、蚕丝蛋白、类人胶原蛋白和贻贝蛋白等主要蛋白功能材料的生物合成研究进展,并阐述了人造蛋白细胞工厂的构建、蛋白的调控表达和其在组织工程材料等领域的应用现状。具有动态响应特性的人造蛋白功能材料是未来的研究方向。
中图分类号:
引用本文
曾丹, 储建林, 陈燕茹, 范代娣. 人造蛋白功能材料的生物合成及应用[J]. 合成生物学, 2021, 2(4): 528-542.
ZENG Dan, CHU Jianlin, CHEN Yanru, FAN Daidi. Biological synthesis and applications of artificial protein functional materials[J]. Synthetic Biology Journal, 2021, 2(4): 528-542.
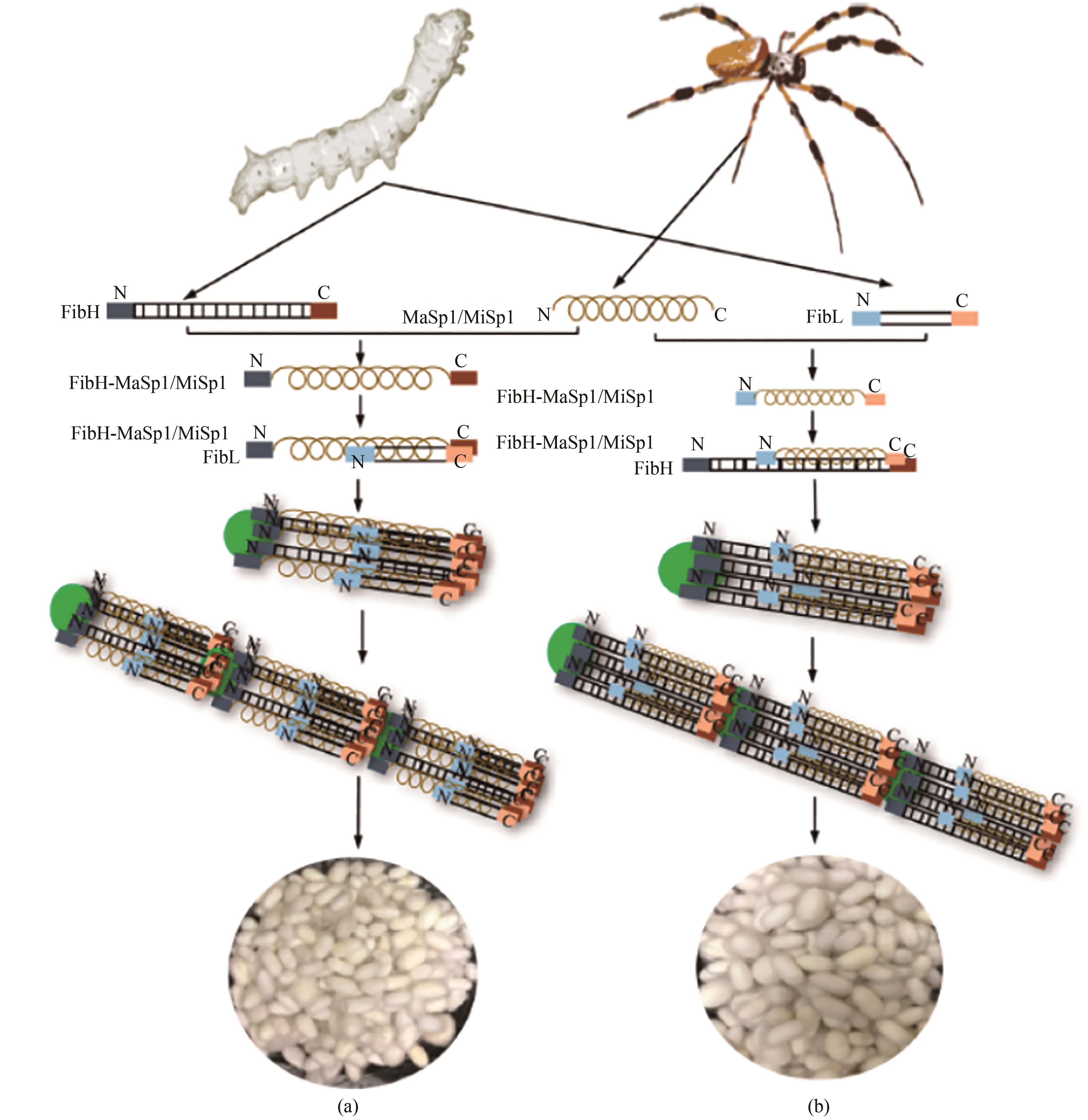
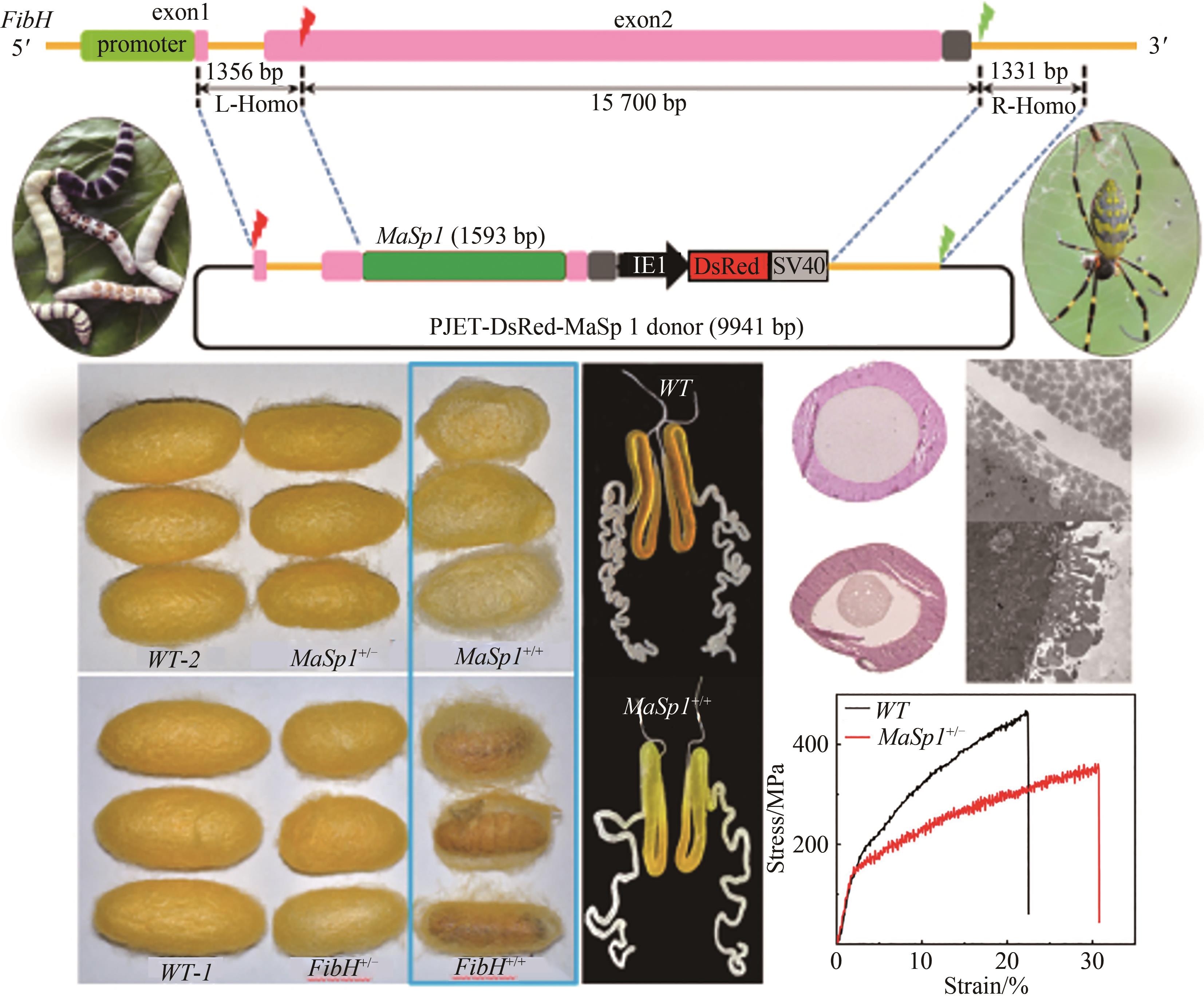
图2 转基因家蚕制成的蜘蛛丝状纤维[46](a)替换天然大小蜘蛛丝蛋白(MaSp1/MiSp1)对丝素重链的替换及其吐丝结茧;(b)蜘蛛丝蛋白MaSp1/MiSp1掺入丝素重链融合表达丝蛋白及其吐丝结茧
Fig. 2 Spider silk fiber made from transgenic silkworm [46](a) Replacement of the natural spider silk protein (MaSp1/MiSp1) through engineering silk fibroin heavy chain and cocooning; (b) Incorporation of the spider silk protein MaSp1/MiSp1 fibroin heavy chain for the fusion expression of silk protein and its cocooning
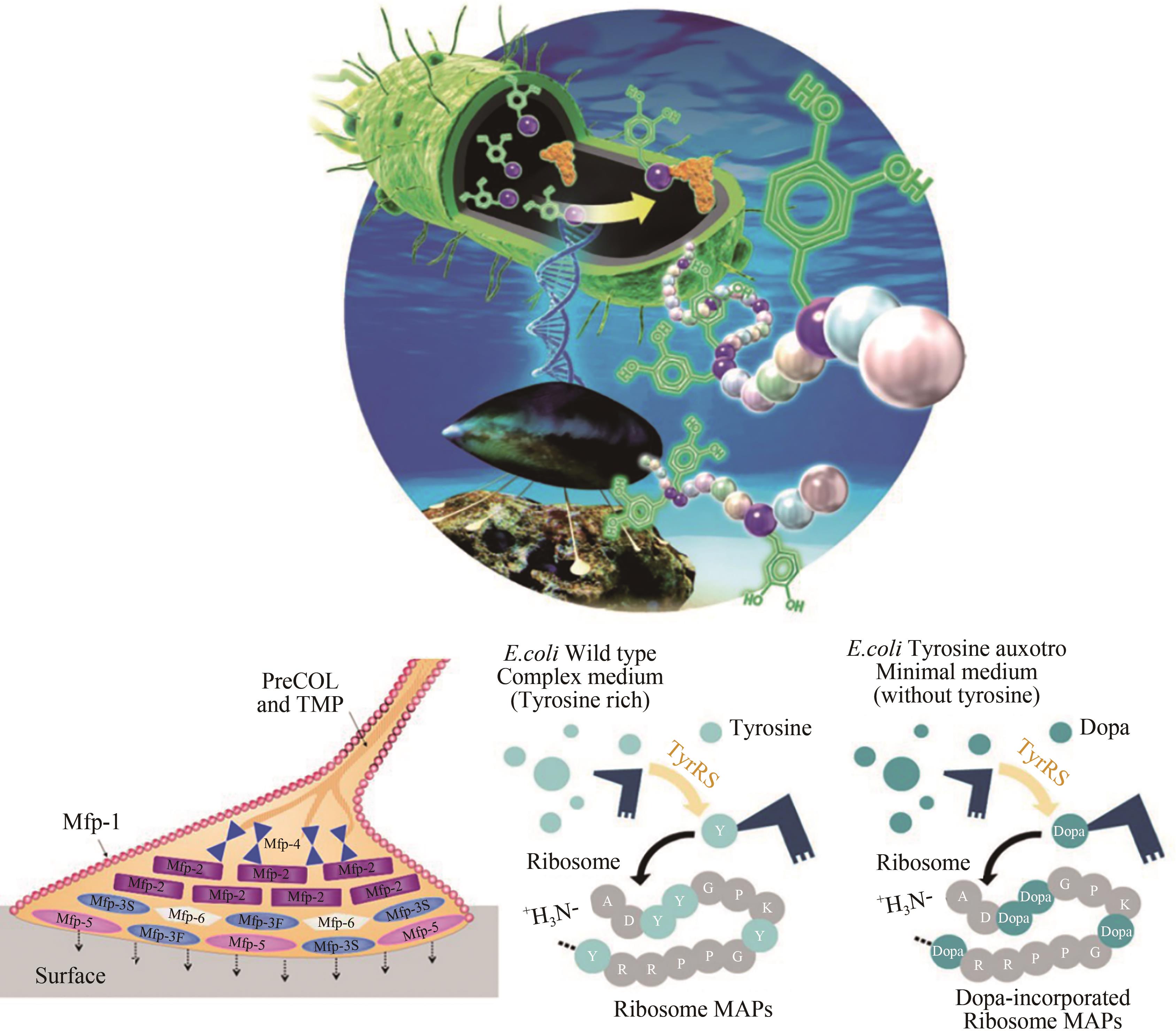
图4 Tyr营养缺陷型大肠杆菌宿主构建及其表达贻贝黏附蛋白Mfp-3和Mfp-5[53]
Fig. 4 Construction of Tyr auxotrophic E. coli host for the expression of mussel adhesion proteins Mfp-3 and Mfp-5[53]
| 应用领域 | 体系 | 材料特性 | 参考文献 |
|---|---|---|---|
| 人工肌腱 | 丝素蛋白 蛛丝蛋白 | 机械强度高,组织相容性好; 无明显排斥反应,衔接部牢固 | [ |
| 人工皮肤 | 丝素蛋白 | 高柔性、良好的非侵入性、环保; 集成度和灵敏度高 | [ |
| 可降解止血材料及人工骨 | HLC | 优良的凝胶性、吸水性和生物相容性;材料可降解 | [ |
| 黏附抗污涂层 | 贻贝黏附蛋白 | 高强度和韧性; 防水抗污性质良好; 生物相容性高 | [ |
表1 人造蛋白功能材料的应用
Tab. 1 Applications of artificial protein functional materials
| 应用领域 | 体系 | 材料特性 | 参考文献 |
|---|---|---|---|
| 人工肌腱 | 丝素蛋白 蛛丝蛋白 | 机械强度高,组织相容性好; 无明显排斥反应,衔接部牢固 | [ |
| 人工皮肤 | 丝素蛋白 | 高柔性、良好的非侵入性、环保; 集成度和灵敏度高 | [ |
| 可降解止血材料及人工骨 | HLC | 优良的凝胶性、吸水性和生物相容性;材料可降解 | [ |
| 黏附抗污涂层 | 贻贝黏附蛋白 | 高强度和韧性; 防水抗污性质良好; 生物相容性高 | [ |
| 1 | 李春. 合成生物学 [M]. 北京:化学工业出版社, 2019: 9. |
| LI C. Synthetic biology [M]. Beijing: Chemical Industry Press, 2019: 9. | |
| 2 | ENDY D. Foundations for engineering biology [J]. Nature, 2005, 438(7067): 449-453. |
| 3 | KHALIL A S, COLLINS J J. Synthetic biology: applications come of age [J]. Nature Reviews Genetics, 2010, 11(5): 367-379. |
| 4 | GIROTTI A, FERNáNDEZ-COLINO A, LÓPEZ I M, et al. Elastin-like recombinamers: biosynthetic strategies and biotechnological applications [J]. Biotechnology Journal, 2011, 6(10): 1174-1186. |
| 5 | YIGIT S, DINJASKI N, KAPLAN D L. Fibrous proteins: at the crossroads of genetic engineering and biotechnological applications [J]. Biotechnology and Bioengineering, 2016, 113(5): 913-929. |
| 6 | DESAI M S, LEE S-W. Protein-based functional nanomaterial design for bioengineering applications [J]. WIREs Nanomedicine and Nanotechnology, 2015, 7(1): 69-97. |
| 7 | CHANG Y, JIAO Y, SYMONS H E, et al. Molecular engineering of polymeric supra-amphiphiles [J]. Chemical Society Reviews, 2019, 48(4): 989-1003. |
| 8 | SUN J, SU J, MA C, et al. Fabrication and mechanical properties of engineered protein-based adhesives and fibers [J]. Advanced Materials, 2020, 32(6): e1906360. |
| 9 | HAGN F, EISOLDT L, HARDY J G, et al. A conserved spider silk domain acts as a molecular switch that controls fibre assembly [J]. Nature, 2010, 465: 239-242. |
| 10 | ADACHI T, WANG X, MURATA T, et al. Production of a non-triple helical collagen alpha chain in transgenic silkworms and its evaluation as a gelatin substitute for cell culture [J]. Biotechnology and Bioengineering, 2010, 106(6): 860-870. |
| 11 | HE J, MA X, ZHANG F, et al. New strategy for expression of recombinant hydroxylated human collagen alpha1(Ⅲ) chains in Pichia pastoris GS115 [J]. Biotechnology and Applied Biochemistry, 2015, 62(3): 293-299. |
| 12 | CHRISTOPH R, STEPHAN B, JÜRG C, et al. Recombinant expression of hydroxylated human collagen in Escherichia coli [J]. Applied Microbiology and Biotechnology, 2014, 98(10): 4445-4455. |
| 13 | RISING A, JOHANSSON J. Toward spinning artificial spider silk [J]. Nature Chemical Biology, 2015, 11(5): 309-315. |
| 14 | XU J, WANG L N, ZHU C H, et al. Co-expression of recombinant human prolyl with human collagen alpha1 (Ⅲ) chains in two yeast systems [J]. Letters in Applied Microbiology, 2015, 61(3): 259-266. |
| 15 | TANG Y, YANG X, HANG B, et al. Efficient production of hydroxylated human-like collagen via the co-expression of three key genes in Escherichia coli origami (DE3) [J]. Applied Biochemistry and Biotechnology, 2016, 178(7): 1458-1470. |
| 16 | WANG Y, KATYAL P, MONTCLARE J K. Protein-engineered functional materials [J]. Advanced Healthcare Materials, 2019, 8(11): 1801374. |
| 17 | HUANG P S, BOYKEN S E, BAKER D. The coming of age of de novo protein design [J]. Nature, 2016, 537(7620): 320-327. |
| 18 | LANGAN R A, BOYKEN S E, NG A H, et al. De novo design of bioactive protein switches [J]. Nature, 2019, 572(7768): 205-210. |
| 19 | ALM E, BAKER D. Prediction of protein-folding mechanisms from free-energy landscapes derived from native structures [J]. Proceedings of the National Academy of Sciences of the United States of America, 1999, 96(20): 11305-11310. |
| 20 | LAI Y T, READING E, HURA G L, et al. Structure of a designed protein cage that self-assembles into a highly porous cube [J]. Nature Chemistry, 2014, 6(12): 1065-1071. |
| 21 | LIN S, RYU S, TOKAREVA O, et al. Predictive modelling-based design and experiments for synthesis and spinning of bioinspired silk fibres [J]. Nature Communications, 2015, 6(1): 1-12. |
| 22 | LI Y, LI J, SUN J, et al. Bio-inspired and mechanically strong fibers based on engineered non-spider chimeric proteins [J]. Angewandte Chemie International Edition, 2020, 59(21): 8148-8152. |
| 23 | EISOLDT L, SMITH A, SCHEIBEL T. Decoding the secrets of spider silk [J]. Materials Today, 2011, 14(3): 80-86. |
| 24 | YARGER J L, CHERRY B R, VAART A VAN DER. Uncovering the structure-function relationship in spider silk [J]. Nature Reviews Materials, 2018, 3(3): 18008. |
| 25 | ALLEY E C, KHIMULYA G, BISWAS S, et al. Unified rational protein engineering with sequence-based deep representation learning [J]. Nature Methods, 2019, 16(12): 1315-1322. |
| 26 | YANG K K, WU Z, ARNOLD F H. Machine-learning-guided directed evolution for protein engineering [J]. Nature Methods, 2019, 16(8): 687-694. |
| 27 | NG A H, NGUYEN T H, GOMEZ-SCHIAVON M, et al. Modular and tunable biological feedback control using a de novo protein switch [J]. Nature, 2019, 572(7768): 265-269. |
| 28 | 刘凯, 李敬敬, 马俊, 等. 力学功能蛋白生物合成及材料应用 [J]. 高分子学报, 2020, 51: 698-709. |
| LIU K, LI J J, MA J, et al. Biological synthesis of structural proteins and applications [J]. Acta Polymerica Sinica, 2020, 51:698-709. | |
| 29 | ZHENG K, LING S. De novo design of recombinant spider silk proteins for material applications [J]. Biotechnology Journal, 2019, 14(1): 1700753. |
| 30 | FRANCO A R, FERNANDES E M, MARCIA T R, et al. Antimicrobial coating of spider silk to prevent bacterial attachment on silk surgical sutures[J]. Acta Biomaterialia, 2019, 99:236-246. |
| 31 | SALEHI S, KOECK K, SCHEIBEL T. Spider silk for tissue engineering applications [J]. Molecules, 2020, 25(3): 737. |
| 32 | SCHELLER J, K-H GÜHRS, GROSSE F, et al. Production of spider silk proteins in tobacco and potato [J]. Nature Biotechnology, 2001, 19(6): 573-577. |
| 33 | LAZARIS A, ARCIDIACONO S, HUANG Y, et al. Spider silk fibers spun from soluble recombinant silk produced in mammalian cells [J]. Science, 2002, 295(5554): 472-476. |
| 34 | FOO C W P, BINI E, HENSMAN J, et al. Role of pH and charge on silk protein assembly in insects and spiders [J]. Applied Physics A, 2006, 82(2): 223-233. |
| 35 | LIU B, WANG T, XIAO L, et al. A directed self-assembly quasi-spider silk protein expressed in Pichia pastoris [J]. Biotechnology & Biotechnological Equipment, 2018, 32(2): 451-461. |
| 36 | ARCIDIACONO S, MELLO C, KAPLAN D, et al. Purification and characterization of recombinant spider silk expressed in Escherichia coli [J]. Applied Microbiology and Biotechnology, 1998, 49(1): 31-38. |
| 37 | XIA X X, QIAN Z G, KI C S, et al. Native-sized recombinant spider silk protein produced in metabolically engineered Escherichia coli results in a strong fiber [J]. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(32): 14059-14063. |
| 38 | ANDERSSON M, JIA Q, ABELLA A, et al. Biomimetic spinning of artificial spider silk from a chimeric minispidroin [J]. Nature Chemical Biology, 2017, 13(3): 262-264. |
| 39 | BOWEN C H, DAI B, SARGENT C J, et al. Recombinant spidroins fully replicate primary mechanical properties of natural spider silk [J]. Biomacromolecules, 2018, 19(9): 3853-3860. |
| 40 | KOH L D, YEO J, LEE Y Y, et al. Advancing the frontiers of silk fibroin protein-based materials for futuristic electronics and clinical wound-healing (invited review) [J]. Materials Science and Engineering: C, 2018, 86: 151-172. |
| 41 | VEPARI C, KAPLAN D L. Silk as a biomaterial [J]. Progress in Polymer Science, 2007, 32(8/9): 991-1007. |
| 42 | PANDEY V, HAIDER T, JAIN P, et al. Silk as a leading-edge biological macromolecule for improved drug delivery [J]. Journal of Drug Delivery Science Technology, 2019, 55: 101294. |
| 43 | SINGH M, BOLLELLA P, GORTON L, et al. Conductive and enzyme-like silk fibers for soft sensing application [J]. Biosensors & Bioelectronics, 2019, 150: 111859. |
| 44 | ANDERSSON M, JOHANSSON J, RISING A. Silk spinning in silkworms and spiders [J]. International Journal of Molecular Sciences, 2016, 17(8): 1290. |
| 45 | XU J, DONG Q, YU Y, et al. Mass spider silk production through targeted gene replacement in Bombyx mori [J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(35): 8757-8762. |
| 46 | ZHANG X, XIA L, DAY B A, et al. CRISPR/Cas9 initiated transgenic silkworms as a natural spinner of spider silk [J]. Biomacromolecules, 2019, 20(6): 2252-2264. |
| 47 | LU T K, KHALIL A S, COLLINS J J. Next-generation synthetic gene networks [J]. Nature Biotechnology, 2009, 27(12): 1139. |
| 48 | WAN X, PINTO F, YU L, et al. Synthetic protein-binding DNA sponge as a tool to tune gene expression and mitigate protein toxicity [J]. Nature Communications, 2020, 11:5961. |
| 49 | SHI J, MA X, GAO Y, et al. Hydroxylation of human type Ⅲ collagen alpha chain by recombinant coexpression with a viral prolyl 4-hydroxylase in Escherichia coli [J]. The Protein Journal, 2017, 36:322-331. |
| 50 | XU R, LUO Y-E, FAN D-D, et al. Improving the production of human-like collagen by pulse-feeding glucose during the fed-batch culture of recombinant Escherichia coli [J]. Biotechnology Applied Biochemistry, 2012, 59(5): 330-337. |
| 51 | FAN D D, LUO Y, MI Y, et al. Characteristics of fed-batch cultures of recombinant Escherichia coli containing human-like collagen cDNA at different specific growth rates [J]. Biotechnology Letters, 2005, 27(12): 865-870. |
| 52 | CHEN Z Y, FAN D D, SHANG L J. Exploring the potential of the recombinant human collagens for biomedical and clinical applications: a short review [J]. Biomedical Materials, 2020, 16(1): 012001. |
| 53 | YANG B, AYYADURAI N, YUN H, et al. In vivo residue-specific dopa-incorporated engineered mussel bioglue with enhanced adhesion and water resistance [J]. Angewandte Chemie International Edition, 2014, 53:13360 –13364. |
| 54 | SPOHNER S C, MÜLLER H, QUITMANN H, et al. Expression of enzymes for the usage in food and feed industry with Pichia pastoris [J]. Journal of Biotechnology, 2015, 202:118-134. |
| 55 | ZHU J, GUO M, ZHUANG Y, et al. Efficient generation of multi-copy strains for optimizing secretory expression of porcine insulin precursor in yeast Pichia pastoris [J]. Journal of Applied Microbiology, 2009, 107: 954-963. |
| 56 | YANG J, LU Z, CHEN J, et al. Efficient of cooperation of chaperones and gene dosage on the expression of porcine PGLYRP-1 in Pichia pastoris [J]. Applied Microbiology & Biotechnology, 2016, 100: 5453-5465. |
| 57 | PAN R, ZHANG J, SHEN W-L, et al. Sequential deletion of Pichia pastoris genes by a self-excisable cassette [J].Federation of European Microbiological Societies Yeast Research, 2011, 11(3): 292-298. |
| 58 | VERMA M, CHOI J, COTTRELL K A, et al. A short translational ramp determines the efficiency of protein synthesis [J]. Nature Communications, 2019, 10:5774. |
| 59 | 李佳, 王软林, 梁爱华. 真核翻译起始因子5A在蛋白质合成中的功能与机制 [J]. 中国生物化学与分子生物学报, 2021,37(2): 161-168. |
| LI J, WANG R L, LIANG A H. Function and mechanism of eukaryotic translation initiation factor 5A in protein synthesis [J]. Chinese Journal of Biochemistry and Molecular Biology, 2021,37(2):161-168. | |
| 60 | JIA Q L, LUO Y E, FAN D D. Application of molecular chaperone to increase the expression of soluble human-like collagen in Escherichia coli [J]. Biotechnology an Indian Journal, 2013, 7(12): 531-536. |
| 61 | JIA Q L, LUO Y E, FAN D D, et al. The different roles of chaperone teams on over-expression of human-like collagen in recombinant Escherichia coli [J]. Journal of the Taiwan Institute of Chemical Engineers, 2014, 45(6): 2843-2850. |
| 62 | WILDT S, GERNGROSS T U. The humanization of N-glycosylation pathways in yeast [J]. Nature Reviews Microbiology, 2005, 3(2): 119-128. |
| 63 | ZHU B, SHEN J, ZHAO T, et al. Intact glycopeptide analysis of influenza A/H1N1/09 neuraminidase revealing the effects of host and glycosite location on site-specific glycan structures [J]. Proteomics, 2019, 19(3): 1800202. |
| 64 | BERN M, KIL Y J, BECKER C. Byonic: advanced peptide and protein identification software [J]. Current Protocols in Bioinformatics, 2012,40(1): 13.20.1-13.20.14. |
| 65 | ESHGHI S T, SHAH P, YANG W M, et al. GPQuest: a spectral library matching algorithm for site-specific assignment of tandem mass spectra to intact N-glycopeptides [J]. Analytical Chemistry, 2015, 87(10): 5181-5188. |
| 66 | LIU M Q, ZENG W F, FANG P, et al. pGlyco 2.0 enables precision N-glycoproteomics with comprehensive quality control and one-step mass spectrometry for intact glycopeptide identification [J]. Nature Communications, 2017, 8(1): 438. |
| 67 | XIAO K, TIAN Z. GPSeeker enables quantitative structural N-glycoproteomics for site-and structure-specific characterization of differentially expressed N-glycosylation in hepatocellular carcinoma [J]. Journal of Proteome Research, 2019, 18(7): 2885-2895. |
| 68 | POLASKY D A, YU F, TEO G C, et al. Fast and comprehensive N- and O-glycoproteomics analysis with MSFragger-Glyco [J]. Nature Methods, 2020, 17(11): 1125-1132. |
| 69 | BELDJILALI-LABRO M, GARCIA A G, FARHAT F, et al. Biomaterials in tendon and skeletal muscle tissue engineering: current trends and challenges [J]. Materials, 2018, 11(7): 1116. |
| 70 | JIANG J, ZHANG S, QIAN Z, et al. Protein bricks: 2D and 3D Bio-nanostructures with shape and function on demand [J]. Advanced Materials, 2018, 30(20): 1705919. |
| 71 | ZHI Y, LIU W, ZHANG P, et al. Electrospun silk fibroin mat enhances tendon-bone healing in a rabbit extra-articular model [J]. Biotechnology Letters, 2016, 38(10): 1827-1835. |
| 72 | NAGHASHZARGAR E, FARÈ S, CATTO V, et al. Nano/Micro hybrid scaffold of PCL or P3HB nanofibers combined with silk fibroin for tendon and ligament tissue engineering [J]. Journal of Applied Biomaterials & Functional Materials, 2015, 13(2): 156-168. |
| 73 | TELLADO S T, BONANI W, BALMAYOR E R, et al. Fabrication and characterization of biphasic silk fibroin scaffolds for tendon/ligament-to-bone tissue engineering [J]. Tissue Engineering Part A, 2017, 23(15-16): 859-872. |
| 74 | WANG C, XIA K, ZHANG Y, et al. Silk-based advanced materials for soft electronics [J]. Accounts of Chemical Research, 2019, 52(10): 2916-2927. |
| 75 | GAO B, GUO M, LYU K, et al. Intelligent silk fibroin based microneedle dressing (i-SMD) [J]. Advanced Functional Materials, 2021, 31(3): 2006839. |
| 76 | GHOLIPOURMALEKABADI M, SAMADIKUCHAKSARAEI A, SEIFALIAN A M, et al. Silk fibroin/amniotic membrane 3D bi-layered artificial skin [J]. Biomedical Materials, 2018, 13(3): 035003. |
| 77 | KEIROUZ A, ZAKHAROVA M, KWON J, et al. High-throughput production of silk fibroin-based electrospun fibers as biomaterial for skin tissue engineering applications [J]. Materials Science and Engineering: C, 2020, 112: 110939. |
| 78 | LI X, FAN D, ZHU C, et al. Effects of self-assembled fibers on the synthesis, characteristics and biomedical applications of CCAG hydrogels [J]. Journal of Materials Chemistry B, 2014, 2(9): 1234-1249. |
| 79 | LI X, XUE W, LIU Y, et al. Novel multifunctional PB and PBH hydrogels as soft filler for tissue engineering [J]. Journal of Materials Chemistry B, 2015, 3(23): 4742-4755. |
| 80 | JIANG X, WANG Y, FAN D, et al. A novel human-like collagen hemostatic sponge with uniform morphology, good biodegradability and biocompatibility [J]. Journal of Biomaterials Applications, 2017, 31(8): 1099-1107. |
| 81 | 范代娣, 马晓轩, 米钰, 等. 一种可生物降解止血海绵材料及其制备方法: CN200610041913.3[P]. 2006-08-23. |
| FAN D D, MA X X, MI Y, et al. A biodegradable hemostatic sponge material and its preparation method: CN200610041913.3[P]. 2006-08-23. | |
| 82 | PAN H, FAN D D, CAO W, et al. Preparation and characterization of breathable hemostatic hydrogel dressings and determination of their effects on full-thickness defects [J]. Polymers, 2017, 9(12): 727. |
| 83 | FAN H, MI Y, HUI J, et al. Cytocompatibility of human-like collagen/nano-hydroxyapatite porous scaffolds using cartilages [J]. Biotechnology, 2013, 12(2): 99-103. |
| 84 | JIA L P, DUAN Z G, FAN D D, et al. Human-like collagen/nano-hydroxyapatite scaffolds for the culture of chondrocytes [J]. Materials Science and Engineering C, 2013, 33(2): 727-734. |
| 85 | LIU K Q, LIU Y N, DUAN Z G, et al. A biomimetic bi-layered tissue engineering scaffolds for osteochondral defects repair [J]. Science China Technological Sciences, 2020,64(4): 793-805. |
| 86 | SONG X, ZHU C H, FAN D D, et al. A novel human-like collagen hydrogel scaffold with porous structure and sponge-like properties [J]. Polymers, 2017, 9(12): 638. |
| 87 | LIU Y N, GU J, FAN D D. Fabrication of high-strength and porous hybrid scaffolds based on nano-hydroxyapatite and human-like collagen for bone tissue regeneration [J]. Polymers, 2020, 12(1): 61. |
| 88 | XIE J H, FAN D D. A high-toughness and high cell adhesion polyvinyl alcohol (PVA-hyaluronic acid (HA)-human-like collagen (HLC) composite hydrogel for cartilage repair [J]. International Journal of Polymeric Materials and Polymeric Biomaterials, 2020, 69(14): 928-937. |
| 89 | ZHOU J, GUO X, ZHENG Q, et al. Improving osteogenesis of three-dimensional porous scaffold based on mineralized recombinant human-like collagen via mussel-inspired polydopamine and effective immobilization of BMP-2-derived peptide [J]. Colloids and Surfaces B: Biointerfaces, 2017, 152: 124-132. |
| 90 | DAMODARAN V B, MURTHY N S. Bio-inspired strategies for designing antifouling biomaterials [J]. Biomaterials Research, 2016, 20: 18. |
| 91 | 吕亚维, 王睿劼, 张雨靖 等. 重组贻贝黏蛋白 Mgfp-5 的表达及功能评价 [J]. 基因组学与应用生物学, 2017, 36(10): 4108-4115. |
| LYU Y W, WANG R J, ZHANG Y J, et al. The expression and function evaluation of the recombinated mussel adhesive protein Mgfp-5 [J]. Genomics and Applied Biology, 2017, 36(10): 4108-4115. | |
| 92 | 薛瑞, 姚林, 王瑞, 等. 重组贻贝足蛋白的研究进展与应用 [J]. 中国生物工程杂志, 2020, 40(11): 82-89. |
| XUE R, YAO L, WANG R, et al. Advances and applications of recombinant mussel foot proteins [J]. China Biotechnology, 2020, 40(11): 82-89. | |
| 93 | QI H S, ZHENG W W, ZHOU X, et al. A mussel-inspired chimeric protein as a novel facile antifouling coating [J]. Chemical Communications, 2018, 54: 11328-11331. |
| 94 | QI H S, ZHENG W W, ZHANG C, et al. Novel mussel-inspired universal surface functionalization strategy: protein-based coating with residue-specific post-translational modification in vivo [J]. American Chemical Society Applied Materials & Interfaces, 2019, 11(13): 12846-12853. |
| 95 | QI H S, ZHANG C, GUO H S, et al. Bioinspired multifunctional protein coating for antifogging, self-cleaning, and antimicrobial properties [J]. American Chemical Society Applied Materials & Interfaces, 2019, 11(27): 24504-24511. |
| 96 | WANG J X, CAO H L, ZHANG J Z H, et al. Computational protein design with deep learning neural networks [J]. Scientific Reports, 2018, 8(1): 6349-6358. |
| 97 | 胡如云, 张嵩亚, 蒙海林, 等. 面向合成生物学的机器学习方法及应用 [J]. 科学通报,2021, 66(3): 284-299. |
| HU R Y, ZHANG S Y, MENG H L, et al. Machine learning for synthetic biology: methods and applications [J]. Chinese Science Bulletin, 2021, 66(3): 284-299. | |
| 98 | STAPLES M, CHAN L, SI D, et al. Artificial intelligence for bioinformatics: applications in protein folding prediction [C]// Proceedings of the 2019 IEEE Technology & Engineering Management Conference (TEMSCON), Atlanta, GA, USA: 2019. |
| 99 | DRIENOVSKá I, ROELFES G. Expanding the enzyme universe with genetically encoded unnatural amino acids [J]. Nature Catalysis, 2020, 3: 193-202. |
| [1] | 应汉杰, 柳东, 王振宇, 沈涛, 庄伟, 朱晨杰. 工业生物制造与“碳中和”目标探讨[J]. 合成生物学, 2025, 6(1): 1-7. |
| [2] | 高歌, 边旗, 王宝俊. 合成基因线路的工程化设计研究进展与展望[J]. 合成生物学, 2025, 6(1): 45-64. |
| [3] | 李冀渊, 吴国盛. 合成生物学视域下有机体的两种隐喻[J]. 合成生物学, 2025, 6(1): 190-202. |
| [4] | 焦洪涛, 齐蒙, 邵滨, 蒋劲松. DNA数据存储技术的法律治理议题[J]. 合成生物学, 2025, 6(1): 177-189. |
| [5] | 任家卫, 张金鹏, 徐国强, 张晓梅, 许正宏, 张晓娟. 大肠杆菌中终止子对下游转录单元基因表达的影响[J]. 合成生物学, 2025, 6(1): 213-227. |
| [6] | 唐兴华, 陆钱能, 胡翌霖. 人类世中对合成生物学的哲学反思[J]. 合成生物学, 2025, 6(1): 203-212. |
| [7] | 徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176. |
| [8] | 石婷, 宋展, 宋世怡, 张以恒. 体外生物转化(ivBT):生物制造的新前沿[J]. 合成生物学, 2024, 5(6): 1437-1460. |
| [9] | 柴猛, 王风清, 魏东芝. 综合利用木质纤维素生物转化合成有机酸[J]. 合成生物学, 2024, 5(6): 1242-1263. |
| [10] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [11] | 陈雨, 张康, 邱以婧, 程彩云, 殷晶晶, 宋天顺, 谢婧婧. 微生物电合成技术转化二氧化碳研究进展[J]. 合成生物学, 2024, 5(5): 1142-1168. |
| [12] | 郑皓天, 李朝风, 刘良叙, 王嘉伟, 李恒润, 倪俊. 负碳人工光合群落的设计、优化与应用[J]. 合成生物学, 2024, 5(5): 1189-1210. |
| [13] | 夏孔晨, 徐维华, 吴起. 光酶催化混乱性反应的研究进展[J]. 合成生物学, 2024, 5(5): 997-1020. |
| [14] | 陈子苓, 向阳飞. 类器官技术与合成生物学协同研究进展[J]. 合成生物学, 2024, 5(4): 795-812. |
| [15] | 蔡冰玉, 谭象天, 李伟. 合成生物学在干细胞工程化改造中的研究进展[J]. 合成生物学, 2024, 5(4): 782-794. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||