合成生物学 ›› 2022, Vol. 3 ›› Issue (2): 302-319.DOI: 10.12211/2096-8280.2021-063
DNA纳米技术与合成生物学
施茜, 吴园园, 杨洋
- 上海交通大学医学院附属仁济医院,分子医学研究院,上海 200127
-
收稿日期:2021-06-04修回日期:2021-10-24出版日期:2022-04-30发布日期:2022-05-11 -
通讯作者:杨洋 -
作者简介:施茜 (1988—),女,博士后。研究方向为基于核酸纳米结构的药物递送。 E-mail:sshiqian@hotmail.com杨洋 (1983—),男,研究员,博士生导师。研究方向为核酸纳米自组装与磷脂膜工程,核酸信息存储与计算。 E-mail:yang.yang.nano@sjtu.edu.cn -
基金资助:国家重点研发计划(2018YFA0902600);国家自然科学基金(NSF21977069)
DNA nanotechnology and synthetic biology
SHI Qian, WU Yuanyuan, YANG yang
- Institute of Molecular Medicine,Renji Hospital,School of Medicine,Shanghai Jiao Tong University,Shanghai 200127,China
-
Received:2021-06-04Revised:2021-10-24Online:2022-04-30Published:2022-05-11 -
Contact:YANG yang
摘要:
合成生物学突破了经典生物学“格物致知”的研究范式,开启了“建物致知”“建物致用”的研究时代。合成生物学是以系统生物学为基础,结合工程学设计,运用现代生物学技术方法,通过构建新的生物体系以揭示生命规律和开发颠覆性技术的交叉学科。以DNA为主要建筑材料进行纳米尺度结构自组装的DNA纳米技术,具有高度可设计性、精确可寻址性、生物亲和性、模块化组装等独特优势,已经成为合成生物学重要的支持技术。本文介绍了利用DNA纳米结构实现核酸、蛋白质、磷脂等生物大分子的有序装配;构建仿生细胞元件(例如核孔、人工膜通道、网格蛋白),生物过程(例如膜融合、脂质转移、成管过程)和生化体系(例如RNA挤出纳米工厂、体外病毒衣壳蛋白合成和凝血系统);及其在药物递送、肿瘤治疗等领域的应用。此外,未来的研究有望通过DNA纳米结构来更好地合成、模拟和调节天然生物体系。例如,如何一定程度恢复和利用DNA纳米结构携载遗传信息的能力;如何提高结构设计复杂性的同时,兼顾人工体系的简单性和生产的高效性;如何扩大生产规模,降低成本;如何在细胞中生产结构并组装。同时,临床应用层面仍有许多亟待解决的问题,比如增加药物的搭载效率,增强结构的靶向性,维持机体中结构稳定性,以及通过修饰进行免疫治疗。DNA纳米技术在合成生物学具有广泛的应用前景,将有助于认识生命本质、模拟生命过程、建立人工体系、开发改变未来的技术。
中图分类号:
引用本文
施茜, 吴园园, 杨洋. DNA纳米技术与合成生物学[J]. 合成生物学, 2022, 3(2): 302-319.
SHI Qian, WU Yuanyuan, YANG yang. DNA nanotechnology and synthetic biology[J]. Synthetic Biology Journal, 2022, 3(2): 302-319.
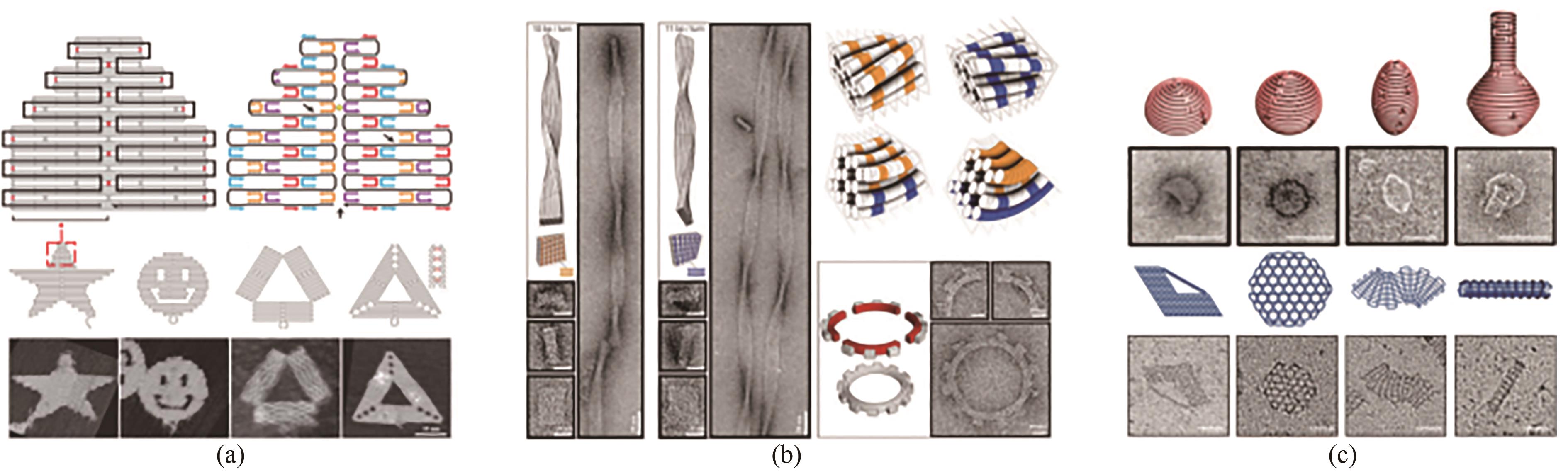
图2 DNA折纸术基本原理及其发展(标尺:50 nm)(a)DNA折纸术原理及二维组装[9];(b)三维组装与弯曲控制[11];(c)曲面设计与网状编织[12-13]
Fig. 2 Basic principle of DNA origami and its development (Scale bar: 50 nm)(a) Principle for the 2D assembly of DNA origami[9]; (b) 3D structure assembly with the bending control[11]; (c) Design for curved surface and DNA gridiron[12-13]
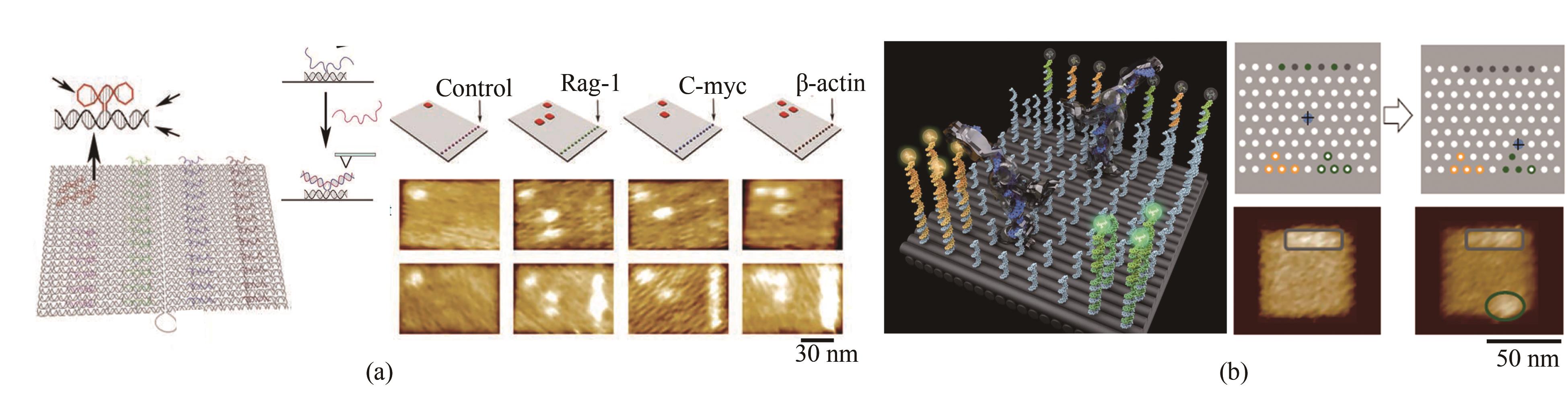
图3 核酸分子装配及应用(a)RNA识别阵列[34];(b)DNA机器与分子搬运[46]
Fig. 3 Assembly and applications of nucleic acid molecules(a) RNA recognition array[34];(b) DNA machines and molecular handling[46]
| 根据结合方式分类 | DNA修饰 | 蛋白修饰 | 优点 | 缺点 | |
|---|---|---|---|---|---|
| 非共价偶联 | 生物素-链亲和素[ | 生物素 | 链亲和素 | ①具有可逆性; | 结合不稳定 |
| Ni-NTA-Histag[ | NTA | Histag | ②可进行定点定位修饰 | ||
| 抗体-抗原[ | 抗原 | 抗体 | |||
| 核酸适配体-蛋白[ | 核酸适配体 | — | |||
| DNA结合蛋白[ | 特异的dsDNA | 锌指蛋白 | |||
| RNA-病毒蛋白[ | RNA | 病毒蛋白 | |||
| DNA-衣壳蛋白[ | — | 衣壳蛋白 | |||
| 共价偶联 | √非特异性 | ①结合较稳定; | 结合位点不易控制 | ||
| SPDP[ | 氨基 | 半胱氨酸残基 | ②反应温和,步骤简单 | ||
| Sulfo-SMCC[ | 氨基 | 半胱氨酸残基 | |||
| √特异性 | |||||
| SNAP-tag[ | O6-烷基鸟嘌呤 | SNAP-tag | ①结合较稳定; | 需要进行蛋白质工程 | |
| Halo-tag[ | 5-氯已烷 | Halo-tag | ②可控制结合位点 | 操作较复杂 |
表1 蛋白装配的分类
Tab. 1 Classification of protein assembly
| 根据结合方式分类 | DNA修饰 | 蛋白修饰 | 优点 | 缺点 | |
|---|---|---|---|---|---|
| 非共价偶联 | 生物素-链亲和素[ | 生物素 | 链亲和素 | ①具有可逆性; | 结合不稳定 |
| Ni-NTA-Histag[ | NTA | Histag | ②可进行定点定位修饰 | ||
| 抗体-抗原[ | 抗原 | 抗体 | |||
| 核酸适配体-蛋白[ | 核酸适配体 | — | |||
| DNA结合蛋白[ | 特异的dsDNA | 锌指蛋白 | |||
| RNA-病毒蛋白[ | RNA | 病毒蛋白 | |||
| DNA-衣壳蛋白[ | — | 衣壳蛋白 | |||
| 共价偶联 | √非特异性 | ①结合较稳定; | 结合位点不易控制 | ||
| SPDP[ | 氨基 | 半胱氨酸残基 | ②反应温和,步骤简单 | ||
| Sulfo-SMCC[ | 氨基 | 半胱氨酸残基 | |||
| √特异性 | |||||
| SNAP-tag[ | O6-烷基鸟嘌呤 | SNAP-tag | ①结合较稳定; | 需要进行蛋白质工程 | |
| Halo-tag[ | 5-氯已烷 | Halo-tag | ②可控制结合位点 | 操作较复杂 |
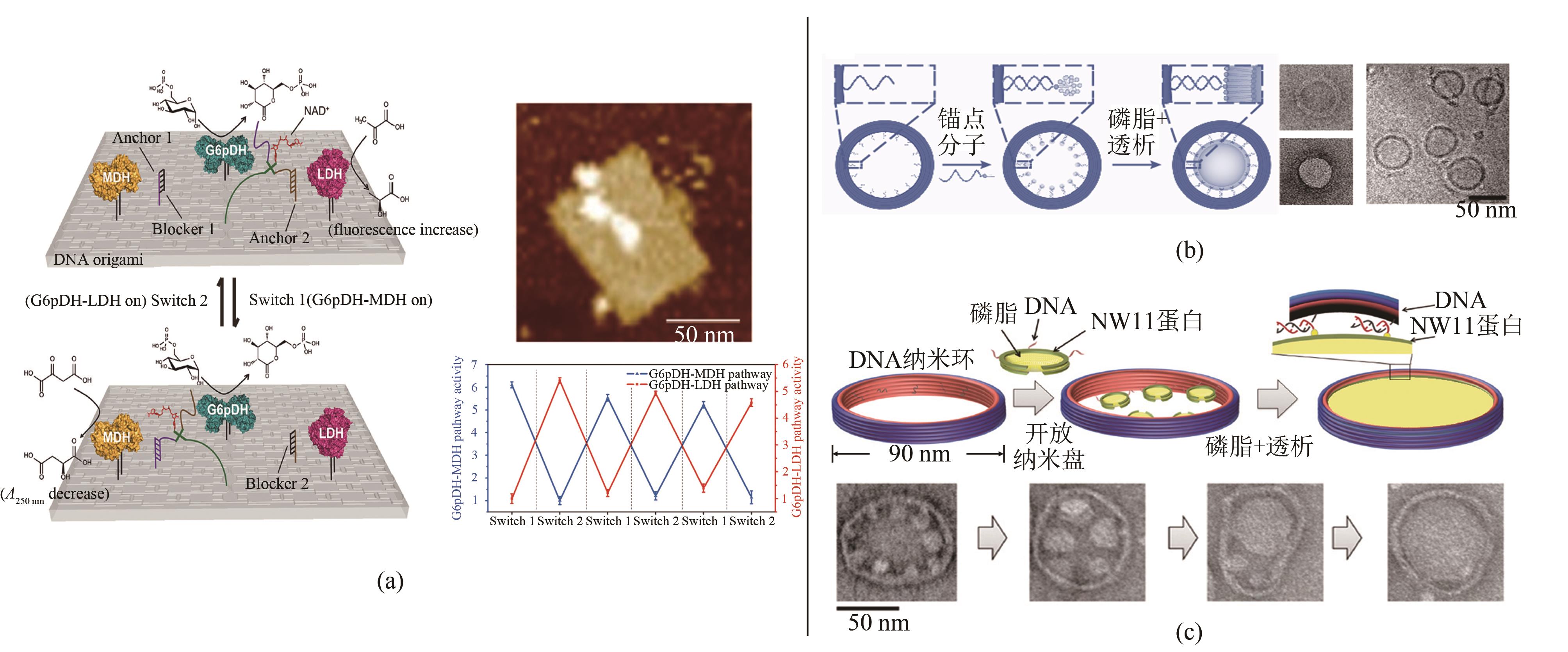
图4 蛋白组装与磷脂组装(a)酶促反应介导与调控[61];(b)可控尺寸囊泡组装[62];(c)二维磷脂膜组装[64]
Fig. 4 DNA nanoframe-directed assembly of proteins and phospholipids(a) DNA swing arm and regulation on cascade enzymatic reactions[61]; (b) Vesicles with controllable sizes [62]; (c) Two-dimensional lipid bilayer assembly[64]

图5 构建仿生细胞元件(a)模拟核孔复合体[55];(b)模拟细胞骨架蛋白[70]
Fig. 5 Mimic and construction of cell elements(a) Simulation of the nuclear pore complex [55]; (b) Simulation of cytoskeleton protein and network [70]

图6 模拟生物过程和生化体系(a)模拟蛋白锚定和膜融合[71];(b)囊泡成管[74];(c)RNA挤出纳米工厂[75];(d)自组装合成衣壳蛋白[52];(e)自主调控凝血系统[77];(f)肌动蛋白运动[78]
Fig. 6 Mimic king biological reactions and biochemical systems(a) Simulation of SNAREs protein-induced membrane fusion[71]; (b) Mimicking BAR protein-induced membrane tubulation [74]; (c) Producing RNA in a nanofactory[75]; (d) Assembly TMV capsid protein on the DNA template [52]; (e) Autonomous regulation of the thrombin dependent coagulation [77]; (f) Simulation of the G-actin movement [78]

图7 在生物医学领域应用(a)逻辑门控DNA纳米机器人[81];(b)病毒衣壳蛋白修饰DNA纳米结构[104];(c)仿病毒磷脂膜包裹DNA纳米结构[105]
Fig. 7 Applications in biomedicines(a) Logic-gate controlled DNA nanorobot[81]; (b) Viral capsid protein modified DNA nanostructure[104]; (c) Virus-inspired membrane-coated DNA nanostructure[105]
| 1 | JACOB F, MONOD J. On the regulation of gene activity[J]. Cold Spring Harbor Symposia on Quantitative Biology, 1961, 26: 193-211. |
| 2 | ELOWITZ M B, LEIBLER S. A synthetic oscillatory network of transcriptional regulators[J]. Nature, 2000, 403(6767): 335-338. |
| 3 | GARDNER T S, CANTOR C R, COLLINS J J. Construction of a genetic toggle switch in Escherichia coli [J]. Nature, 2000, 403(6767): 339-342. |
| 4 | FREDENS J, WANG K, DE LA TORRE D, et al. Total synthesis of Escherichia coli with a recoded genome[J]. Nature, 2019, 569(7757): 514-518. |
| 5 | 合成生物学全球初创公司图谱,万亿美金市场现状梳理[R/OL]. CB Insights, 2020. . |
| 6 | GROUP B F, BAKER D, CHURCH G, et al. Engineering life: Building a FAB for biology[J]. Scientific American, 2006, 294(6): 44-51. |
| 7 | SEEMAN N C. Nucleic acid junctions and lattices[J]. Journal of Theoretical Biology, 1982, 99(2): 237-247. |
| 8 | SEEMAN N C, SLEIMAN H F. DNA nanotechnology[J]. Nature Reviews Materials, 2018, 3: 17068. |
| 9 | ROTHEMUND P W K. Folding DNA to create nanoscale shapes and patterns[J]. Nature, 2006, 440(7082): 297-302. |
| 10 | SANDERSON K. Bioengineering: what to make with DNA origami[J]. Nature, 2010, 464(7286): 158-159. |
| 11 | DIETZ H, DOUGLAS S M, SHIH W M. Folding DNA into twisted and curved nanoscale shapes[J]. Science, 2009, 325(5941): 725-730. |
| 12 | HAN D R, PAL S, NANGREAVE J, et al. DNA origami with complex curvatures in three-dimensional space[J]. Science, 2011, 332(6027): 342-346. |
| 13 | HAN D R, PAL S, YANG Y, et al. DNA gridiron nanostructures based on four-arm junctions[J]. Science, 2013, 339(6126): 1412-1415. |
| 14 | Tikhomirov G, Petersen P, Qian L. Fractal assembly of micrometre-scale DNA origami arrays with arbitrary patterns[J]. Nature, 2017, 552(7683): 67. |
| 15 | WAGENBAUER K F, SIGL C, DIETZ H. Gigadalton-scale shape-programmable DNA assemblies[J]. Nature, 2017, 552(7683): 78-83. |
| 16 | YAN H, PARK S H, FINKELSTEIN G, et al. DNA-templated self-assembly of protein arrays and highly conductive nanowires[J]. Science, 2003, 301(5641): 1882-1884. |
| 17 | LIU W Y, ZHONG H, WANG R S, et al. Crystalline two-dimensional DNA-origami arrays[J]. Angewandte Chemie International Edition, 2011, 123(1): 278-281. |
| 18 | DOUGLAS S M, DIETZ H, LIEDL T, et al. Self-assembly of DNA into nanoscale three-dimensional shapes[J]. Nature, 2009, 459(7245): 414-418. |
| 19 | PRAETORIUS F, DIETZ H. Self-assembly of genetically encoded DNA-protein hybrid nanoscale shapes[J]. Science, 2017, 355(6331): eaam5488. |
| 20 | ZHANG J P, LIU Y, KE Y G, et al. Periodic square-like gold nanoparticle arrays templated by self-assembled 2D DNA Nanogrids on a surface[J]. Nano Letters, 2006, 6(2): 248-251. |
| 21 | LUND K, LIU Y, LINDSAY S, et al. Self-assembling a molecular pegboard[J]. Journal of the American Chemical Society, 2005, 127(50): 17606-17607. |
| 22 | DONG Y C, YANG Y R, ZHANG Y Y, et al. Cuboid vesicles formed by frame-guided assembly on DNA origami scaffolds[J]. Angewandte Chemie International Edition, 2017, 56(6): 1586-1589. |
| 23 | KE Y G, ONG L L, SHIH W M, et al. Three-dimensional structures self-assembled from DNA bricks[J]. Science, 2012, 338(6111): 1177-1183. |
| 24 | ONG L L, HANIKEL N, YAGHI O K, et al. Programmable self-assembly of three-dimensional nanostructures from 10, 000 unique components[J]. Nature, 2017, 552(7683): 72-77. |
| 25 | YAO G, ZHANG F, WANG F, et al. Meta-DNA structures[J]. Nature Chemistry, 2020, 12(11): 1067-1075. |
| 26 | CHANDRAN H, GOPALKRISHNAN N, YURKE B, et al. Meta-DNA: synthetic biology via DNA nanostructures and hybridization reactions[J]. Journal of the Royal Society, Interface, 2012, 9(72): 1637-1653. |
| 27 | YANG Y, ZHANG R, FAN C H. Shaping functional materials with DNA frameworks[J]. Trends in Chemistry, 2020, 2(2): 137-147. |
| 28 | ZHANG Y Y, MAO X H, LI F, et al. Nanoparticle-assisted alignment of carbon nanotubes on DNA origami[J]. Angewandte Chemie International Edition, 2020, 59(12): 4892-4896. |
| 29 | PEI H, SHA R J, WANG X W, et al. Organizing end-site-specific SWCNTs in specific loci using DNA[J]. Journal of the American Chemical Society, 2019, 141(30): 11923-11928. |
| 30 | SUN W, SHEN J, ZHAO Z, et al. Precise pitch-scaling of carbon nanotube arrays within three-dimensional DNA nanotrenches[J]. Science, 2020, 368(6493): 874-877. |
| 31 | ATSUMI H, BELCHER A M. DNA origami and G-quadruplex hybrid complexes induce size control of single-walled carbon nanotubes via biological activation[J]. ACS Nano, 2018, 12(8): 7986-7995. |
| 32 | KNUDSEN J B, LIU L, BANK KODAL A L, et al. Routing of individual polymers in designed patterns[J]. Nature Nanotechnology, 2015, 10(10): 892-898. |
| 33 | TOKURA Y, HARVEY S, CHEN C J, et al. Fabrication of defined polydopamine nanostructures by DNA origami-templated polymerization[J]. Angewandte Chemie International Edition, 2018, 57(6): 1587-1591. |
| 34 | KE Y G, LINDSAY S, CHANG Y, et al. Self-assembled water-soluble nucleic acid probe tiles for label-free RNA hybridization assays[J]. Science, 2008, 319(5860): 180-183. |
| 35 | LIN C, JUNGMANN R, LEIFER A M, et al. Submicrometre geometrically encoded fluorescent barcodes self-assembled from DNA[J]. Nature Chemistry, 2012, 4(10): 832-839. |
| 36 | SEELIG G, SOLOVEICHIK D, ZHANG D Y, et al. Enzyme-free nucleic acid logic circuits[J]. Science, 2006, 314(5805): 1585-1588. |
| 37 | QIAN L L, WINFREE E. Scaling up digital circuit computation with DNA strand displacement cascades[J]. Science, 2011, 332(6034): 1196-1201. |
| 38 | TIKHOMIROV G, PETERSEN P, QIAN L. Programmable disorder in random DNA tilings[J]. Nature Nanotechnology, 2017, 12(3): 251-259. |
| 39 | PETERSEN P, TIKHOMIROV G, QIAN L. Information-based autonomous reconfiguration in systems of interacting DNA nanostructures[J]. Nature Communications, 2018, 9: 5362. |
| 40 | CHERRY K M, QIAN L. Scaling up molecular pattern recognition with DNA-based winner-take-all neural networks[J]. Nature, 2018, 559(7714): 370-376. |
| 41 | WOODS D, DOTY D, MYHRVOLD C, et al. Diverse and robust molecular algorithms using reprogrammable DNA self-assembly[J]. Nature, 2019, 567(7748): 366-372. |
| 42 | SHERMAN W B, SEEMAN N C. A precisely controlled DNA biped walking device[J]. Nano Letters, 2004, 4(7): 1203-1207. |
| 43 | GU H, CHAO J, XIAO S J, et al. A proximity-based programmable DNA nanoscale assembly line[J]. Nature, 2010, 465(7295): 202-205. |
| 44 | TIAN Y, HE Y, CHEN Y, et al. A DNAzyme that walks processively and autonomously along a one-dimensional track[J]. Angewandte Chemie International Edition, 2005, 44(28): 4355-4358. |
| 45 | LUND K, MANZO A J, DABBY N, et al. Molecular robots guided by prescriptive landscapes[J]. Nature, 2010, 465(7295): 206-210. |
| 46 | THUBAGERE A J, LI W, JOHNSON R F, et al. A cargo-sorting DNA robot[J]. Science, 2017, 357(6356): 1112. |
| 47 | MALLIK L, DHAKAL S, NICHOLS J, et al. Electron microscopic visualization of protein assemblies on flattened DNA origami[J]. ACS Nano, 2015, 9(7): 7133-7141. |
| 48 | SHEN W Q, ZHONG H, NEFF D, et al. NTA directed protein nanopatterning on DNA origami nanoconstructs[J]. Journal of the American Chemical Society, 2009, 131(19): 6660-6661. |
| 49 | YAMAZAKI T, HEDDLE J G, KUZUYA A, et al. Orthogonal enzyme arrays on a DNA origami scaffold bearing size-tunable wells[J]. Nanoscale, 2014, 6(15): 9122-9126. |
| 50 | CHHABRA R, SHARMA J, KE Y G, et al. Spatially addressable multiprotein nanoarrays templated by aptamer-tagged DNA nanoarchitectures[J]. Journal of the American Chemical Society, 2007, 129(34): 10304-10305. |
| 51 | NGO T A, NAKATA E, SAIMURA M, et al. Spatially organized enzymes drive cofactor-coupled cascade reactions[J]. Journal of the American Chemical Society, 2016, 138(9): 3012-3021. |
| 52 | ZHOU K, KE Y G, WANG Q B. Selective in situ assembly of viral protein onto DNA origami[J]. Journal of the American Chemical Society, 2018, 140(26): 8074-8077. |
| 53 | KOPATZ I, ZALK R, LEVI-KALISMAN Y, et al. Packaging of DNA origami in viral capsids[J]. Nanoscale, 2019, 11(21): 10160-10166. |
| 54 | FU J L, LIU M H, LIU Y, et al. Interenzyme substrate diffusion for an enzyme cascade organized on spatially addressable DNA nanostructures[J]. Journal of the American Chemical Society, 2012, 134(12): 5516-5519. |
| 55 | FISHER P, SHEN Q, AKPINAR B, et al. A programmable DNA origami platform for organizing intrinsically disordered nucleoporins within nanopore confinement[J]. ACS Nano, 2018, 12(2): 1508-1518. |
| 56 | NAKATA E, DINH H, NGO T A, et al. A modular zinc finger adaptor accelerates the covalent linkage of proteins at specific locations on DNA nanoscaffolds[J]. Chemical Communications, 2015, 51(6): 1016-1019. |
| 57 | KOßMANN K J, ZIEGLER C, ANGELIN A, et al. A rationally designed connector for assembly of protein-functionalized DNA nanostructures[J]. ChemBioChem, 2016, 17(12): 1102-1106. |
| 58 | VOIGT N V, TØRRING T, ROTARU A, et al. Single-molecule chemical reactions on DNA origami[J]. Nature Nanotechnology, 2010, 5(3): 200-203. |
| 59 | RINKER S, KE Y G, LIU Y, et al. Self-assembled DNA nanostructures for distance-dependent multivalent ligand-protein binding[J]. Nature Nanotechnology, 2008, 3(7): 418-422. |
| 60 | FU J, YANG Y R, JOHNSON-BUCK A, et al. Multi-enzyme complexes on DNA scaffolds capable of substrate channelling with an artificial swinging arm[J]. Nature Nanotechnology, 2014, 9(7): 531-536. |
| 61 | KE G L, LIU M H, JIANG S X, et al. Directional regulation of enzyme pathways through the control of substrate channeling on a DNA origami scaffold[J]. Angewandte Chemie International Edition, 2016, 128(26): 7609-7612. |
| 62 | YANG Y, WANG J, SHIGEMATSU H, et al. Self-assembly of size-controlled liposomes on DNA nanotemplates[J]. Nature Chemistry, 2016, 8(5): 476-483. |
| 63 | ZHANG Z, YANG Y, PINCET F, et al. Placing and shaping liposomes with reconfigurable DNA nanocages[J]. Nature Chemistry, 2017, 9(7): 653-659. |
| 64 | ZHAO Z, ZHANG M, HOGLE J M, et al. DNA-corralled nanodiscs for the structural and functional characterization of membrane proteins and viral entry[J]. Journal of the American Chemical Society, 2018, 140(34): 10639-10643. |
| 65 | KETTERER P, ANANTH A N, LAMAN TRIP D S, et al. DNA origami scaffold for studying intrinsically disordered proteins of the nuclear pore complex[J]. Nature Communications, 2018, 9: 902. |
| 66 | LANGECKER M, ARNAUT V, MARTIN T G, et al. Synthetic lipid membrane channels formed by designed DNA nanostructures[J]. Science, 2012, 338(6109): 932-936. |
| 67 | LÜ C, GU X Y, LI H W, et al. Molecular transport through a biomimetic DNA channel on live cell membranes[J]. ACS Nano, 2020, 14(11): 14616-14626. |
| 68 | LANPHERE C, OFFENBARTL-STIEGERT D, DOREY A, et al. Design, assembly, and characterization of membrane-spanning DNA nanopores[J]. Nature Protocols, 2021, 16(1): 86-130. |
| 69 | DOHERTY G J, MCMAHON H T. Mechanisms of endocytosis[J]. Annual Review of Biochemistry, 2009, 78: 857-902. |
| 70 | JOURNOT C M A, RAMAKRISHNA V, WALLACE M I, et al. Modifying membrane morphology and interactions with DNA origami clathrin-mimic networks[J]. ACS Nano, 2019, 13(9): 9973-9979. |
| 71 | XU W M, NATHWANI B, LIN C X, et al. A programmable DNA origami platform to organize SNAREs for membrane fusion[J]. Journal of the American Chemical Society, 2016, 138(13): 4439-4447. |
| 72 | BIAN X, ZHANG Z, XIONG Q C, et al. A programmable DNA-origami platform for studying lipid transfer between bilayers[J]. Nature Chemical Biology, 2019, 15(8): 830-837 |
| 73 | FROST A, UNGER V M, DE CAMILLI P. The BAR domain superfamily: membrane-molding macromolecules[J]. Cell, 2009, 137(2): 191-196. |
| 74 | GROME M W, ZHANG Z, PINCET F, et al. Vesicle tubulation with self-assembling DNA nanosprings[J]. Angewandte Chemie International Edition, 2018, 57(19): 5330-5334. |
| 75 | HAHN J, CHOU L Y T, SØRENSEN R S, et al. Extrusion of RNA from a DNA-origami-based nanofactory[J]. ACS Nano, 2020, 14(2): 1550-1559. |
| 76 | ZHOU K, ZHOU Y H, PAN V, et al. Programming dynamic assembly of viral proteins with DNA origami[J]. Journal of the American Chemical Society, 2020, 142(13): 5929-5932. |
| 77 | YANG L L, ZHAO Y M, XU X M, et al. An intelligent DNA nanorobot for autonomous anticoagulation[J]. Angewandte Chemie International Edition, 2020, 59(40): 17697-17704. |
| 78 | FUJITA K, OHMACHI M, IKEZAKI K, et al. Direct visualization of human myosin II force generation using DNA origami-based thick filaments[J]. Communications Biology, 2019, 2: 437. |
| 79 | ANGELL C, XIE S B, ZHANG L F, et al. DNA nanotechnology for precise control over drug delivery and gene therapy[J]. Small, 2016, 12(9): 1117-1132. |
| 80 | HU Q Q, LI H, WANG L H, et al. DNA nanotechnology-enabled drug delivery systems[J]. Chemical Reviews, 2019, 119(10): 6459-6506. |
| 81 | DOUGLAS S M, BACHELET I, CHURCH G M. A logic-gated nanorobot for targeted transport of molecular payloads[J]. Science, 2012, 335(6070): 831-834. |
| 82 | AGUDELO D, BOURASSA P, BÉRUBÉ G, et al. Intercalation of antitumor drug doxorubicin and its analogue by DNA duplex: Structural features and biological implications[J]. International Journal of Biological Macromolecules, 2014, 66: 144-150. |
| 83 | KIM K R, KIM D R, LEE T, et al. Drug delivery by a self-assembled DNA tetrahedron for overcoming drug resistance in breast cancer cells[J]. Chemical Communications, 2013, 49(20): 2010. |
| 84 | NAITO Y, UI-TEI K. siRNA design software for a target gene-specific RNA interference[J]. Frontiers in Genetics, 2012, 3: 102. |
| 85 | GUO P X, COBAN O, SNEAD N M, et al. Engineering RNA for targeted siRNA delivery and medical application[J]. Advanced Drug Delivery Reviews, 2010, 62(6): 650-666. |
| 86 | LEE H, LYTTON-JEAN A K R, CHEN Y, et al. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery[J]. Nature Nanotechnology, 2012, 7(6): 389-393. |
| 87 | RAHMAN M A, WANG P F, ZHAO Z X, et al. Systemic delivery of Bc12-targeting siRNA by DNA nanoparticles suppresses cancer cell growth[J]. Angewandte Chemie International Edition, 2017, 56(50): 16023-16027. |
| 88 | LIU J B, SONG L L, LIU S L, et al. A DNA-based nanocarrier for efficient gene delivery and combined cancer therapy[J]. Nano Letters, 2018, 18(6): 3328-3334. |
| 89 | JIANG Q, SHI Y F, ZHANG Q, et al. A self-assembled DNA origami-gold nanorod complex for cancer theranostics[J]. Small, 2015, 11(38): 5134-5141. |
| 90 | WANG P F, RAHMAN M A, ZHAO Z X, et al. Visualization of the cellular uptake and trafficking of DNA origami nanostructures in cancer cells[J]. Journal of the American Chemical Society, 2018, 140(7): 2478-2484. |
| 91 | GE Z L, GUO L J, WU G Q, et al. DNA origami-enabled engineering of ligand-drug conjugates for targeted drug delivery[J]. Small, 2020, 16(16): 1904857. |
| 92 | RANIOLO S, VINDIGNI G, OTTAVIANI A, et al. Selective targeting and degradation of doxorubicin-loaded folate-functionalized DNA nanocages[J]. Nanomedicine: Nanotechnology, Biology and Medicine, 2018, 14(4): 1181-1190. |
| 93 | VINDIGNI G, RANIOLO S, OTTAVIANI A, et al. Receptor-mediated entry of pristine octahedral DNA nanocages in mammalian cells[J]. ACS Nano, 2016, 10(6): 5971-5979. |
| 94 | LIANG L, LI J, LI Q, et al. Single-particle tracking and modulation of cell entry pathways of a tetrahedral DNA nanostructure in live cells[J]. Angewandte Chemie International Edition, 2014, 53(30): 7745-7750. |
| 95 | KEUM J W, AHN J H, Design BERMUDEZ H., assembly, and activity of antisense DNA nanostructures[J]. Small, 2011, 7(24): 3529-3535. |
| 96 | BURNS J R, LAMARRE B, PYNE A L B, et al. DNA origami inside-out viruses[J]. ACS Synthetic Biology, 2018, 7(3): 767-773. |
| 97 | LIU K, XU C, LIU J Y. Regulation of cell binding and entry by DNA origami mediated spatial distribution of aptamers[J]. Journal of Materials Chemistry B, 2020, 8(31): 6802-6809. |
| 98 | BHATIA D, ARUMUGAM S, NASILOWSKI M, et al. Quantum dot-loaded monofunctionalized DNA icosahedra for single-particle tracking of endocytic pathways[J]. Nature Nanotechnology, 2016, 11(12): 1112-1119. |
| 99 | XIA K, KONG H T, CUI Y Z, et al. Systematic study in mammalian cells showing no adverse response to tetrahedral DNA nanostructure[J]. ACS Applied Materials & Interfaces, 2018, 10(18): 15442-15448. |
| 100 | BASTINGS M M C, ANASTASSACOS F M, PONNUSWAMY N, et al. Modulation of the cellular uptake of DNA origami through control over mass and shape[J]. Nano Letters, 2018, 18(6): 3557-3564. |
| 101 | ZENG Y, LIU J J, YANG S, et al. Time-lapse live cell imaging to monitor doxorubicin release from DNA origami nanostructures[J]. Journal of Materials Chemistry B, 2018, 6(11): 1605-1612. |
| 102 | KIM K R, LEE T, KIM B S, et al. Correction: Utilizing the bioorthogonal base-pairing system of l-DNA to design ideal DNA nanocarriers for enhanced delivery of nucleic acid cargos[J]. Chemical Science, 2015, 6(3): 2122. |
| 103 | LI Q S, ZHAO D, SHAO X R, et al. Aptamer-modified tetrahedral DNA nanostructure for tumor-targeted drug delivery[J]. ACS Applied Materials & Interfaces, 2017, 9(42): 36695-36701. |
| 104 | MIKKILÄ J, ESKELINEN A P, NIEMELÄ E H, et al. Virus-encapsulated DNA origami nanostructures for cellular delivery[J]. Nano Letters, 2014, 14(4): 2196-2200. |
| 105 | PERRAULT S D, SHIH W M. Virus-inspired membrane encapsulation of DNA nanostructures to achieve in vivo stability[J]. ACS Nano, 2014, 8(5): 5132-5140. |
| 106 | MA W J, ZHAN Y X, ZHANG Y X, et al. An intelligent DNA nanorobot with in vitro enhanced protein lysosomal degradation of HER2[J]. Nano Letters, 2019, 19(7): 4505-4517. |
| 107 | LI S P, JIANG Q, LIU S L, et al. A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo [J]. Nature Biotechnology, 2018, 36(3): 258-264. |
| 108 | LIU S L, JIANG Q, ZHAO X, et al. A DNA nanodevice-based vaccine for cancer immunotherapy[J]. Nature Materials, 2021, 20(3): 421-430. |
| [1] | 高歌, 边旗, 王宝俊. 合成基因线路的工程化设计研究进展与展望[J]. 合成生物学, 2025, 6(1): 45-64. |
| [2] | 李冀渊, 吴国盛. 合成生物学视域下有机体的两种隐喻[J]. 合成生物学, 2025, 6(1): 190-202. |
| [3] | 焦洪涛, 齐蒙, 邵滨, 蒋劲松. DNA数据存储技术的法律治理议题[J]. 合成生物学, 2025, 6(1): 177-189. |
| [4] | 唐兴华, 陆钱能, 胡翌霖. 人类世中对合成生物学的哲学反思[J]. 合成生物学, 2025, 6(1): 203-212. |
| [5] | 徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176. |
| [6] | 石婷, 宋展, 宋世怡, 张以恒. 体外生物转化(ivBT):生物制造的新前沿[J]. 合成生物学, 2024, 5(6): 1437-1460. |
| [7] | 柴猛, 王风清, 魏东芝. 综合利用木质纤维素生物转化合成有机酸[J]. 合成生物学, 2024, 5(6): 1242-1263. |
| [8] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [9] | 陈雨, 张康, 邱以婧, 程彩云, 殷晶晶, 宋天顺, 谢婧婧. 微生物电合成技术转化二氧化碳研究进展[J]. 合成生物学, 2024, 5(5): 1142-1168. |
| [10] | 郑皓天, 李朝风, 刘良叙, 王嘉伟, 李恒润, 倪俊. 负碳人工光合群落的设计、优化与应用[J]. 合成生物学, 2024, 5(5): 1189-1210. |
| [11] | 夏孔晨, 徐维华, 吴起. 光酶催化混乱性反应的研究进展[J]. 合成生物学, 2024, 5(5): 997-1020. |
| [12] | 张宣梁, 李青婷, 王飞. DNA存储系统中的数据写入[J]. 合成生物学, 2024, 5(5): 1125-1141. |
| [13] | 陈子苓, 向阳飞. 类器官技术与合成生物学协同研究进展[J]. 合成生物学, 2024, 5(4): 795-812. |
| [14] | 蔡冰玉, 谭象天, 李伟. 合成生物学在干细胞工程化改造中的研究进展[J]. 合成生物学, 2024, 5(4): 782-794. |
| [15] | 谢皇, 郑义蕾, 苏依婷, 阮静怡, 李永泉. 放线菌聚酮类化合物生物合成体系重构研究进展[J]. 合成生物学, 2024, 5(3): 612-630. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||