合成生物学 ›› 2024, Vol. 5 ›› Issue (3): 631-657.DOI: 10.12211/2096-8280.2023-082
中药药效成分群的合成生物学研究进展
查文龙, 卜兰, 訾佳辰
- 中国医学科学院&北京协和医学院药物研究所,天然药物活性物质与功能国家重点实验室,国家卫生健康委员会天然药物生物合成重点实验室,中国医学科学院酶与天然药物生物催化重点实验室,北京 100050
-
收稿日期:2023-11-21修回日期:2024-03-18出版日期:2024-06-30发布日期:2024-07-12 -
通讯作者:訾佳辰 -
作者简介:查文龙 (1994—),男,博士后。研究方向为合成生物学,酶工程,代谢工程。E-mail: zhawenlong@imm.ac.cn卜兰 (1997—),女,博士研究生。研究方向为合成生物学,酶工程,代谢工程。E-mail:bulan@imm.ac.cn訾佳辰 (1978—),男,研究员,博士生导师。研究方向为合成生物学,酶工程,代谢工程。E-mail:zijiachen@imm.ac.cn -
基金资助:国家自然科学基金(82293682);中国博士后科学基金(2023M730332);中国医学科学院中央级公益性科研院所基本科研业务费(2021-RC350-009)
Advances in synthetic biology for producing potent pharmaceutical ingredients of traditional Chinese medicine
ZHA Wenlong, BU Lan, ZI Jiachen
- State Key Laboratory of Bioactive Substance and Function of Natural Medicines,NHC Key Laboratory of Biosynthesis of Natural Products,CAMS Key Laboratory of Enzyme and Biocatalysis of Natural Drugs,Institute of Materia Medica,Chinese Academy of Medical Sciences & Peking Union Medical College,Beijing 100050,China
-
Received:2023-11-21Revised:2024-03-18Online:2024-06-30Published:2024-07-12 -
Contact:ZI Jiachen
摘要:
中药是中华民族的文化瑰宝,也是我国在新药创制领域的重要驱动力。许多中药材来源于稀缺物种,其药效物质的规模化获取困难,是制约中药新药创制研究的重要瓶颈。合成生物学的出现和快速发展为解决这一瓶颈问题提供了新的途径。目前,中药药效物质的合成生物学研究在单个药效分子的生物制备方法上取得了重要进展。中药的药效主要源于多成分作用的叠加和协同,所以药效成分群是中药药效物质的主要形式,然而针对药效成分群的合成生物学研究鲜有报道。建立中药药效成分群合成生物技术的关键是精确调控组成分子的比例,从而产出优质药效成分群。本文首先总结了挥发油、总皂苷、总黄酮、总木脂素、总生物碱等重要类型中药药效成分群形成机制的研究进展。然后,重点以檀香挥发油为例,介绍如何通过酶工程和代谢工程的联合运用实现药效成分群成分比例和产量的双重优化。最后,对中药药效成分群合成生物学领域的未来研究重点进行了展望,包括:(1)加强中药药效成分群生物合成途径解析方面的研究,重点深入阐明复杂药效成分群的形成机制;(2)加强代谢优化手段方面的创新研究,重点揭示未知代谢调控机制并基于此发展创新调控策略;(3)加强酶工程方法学的创新研究,重点发展新型理性设计和定向进化的联用技术以及人工智能辅助的酶工程技术。
中图分类号:
引用本文
查文龙, 卜兰, 訾佳辰. 中药药效成分群的合成生物学研究进展[J]. 合成生物学, 2024, 5(3): 631-657.
ZHA Wenlong, BU Lan, ZI Jiachen. Advances in synthetic biology for producing potent pharmaceutical ingredients of traditional Chinese medicine[J]. Synthetic Biology Journal, 2024, 5(3): 631-657.
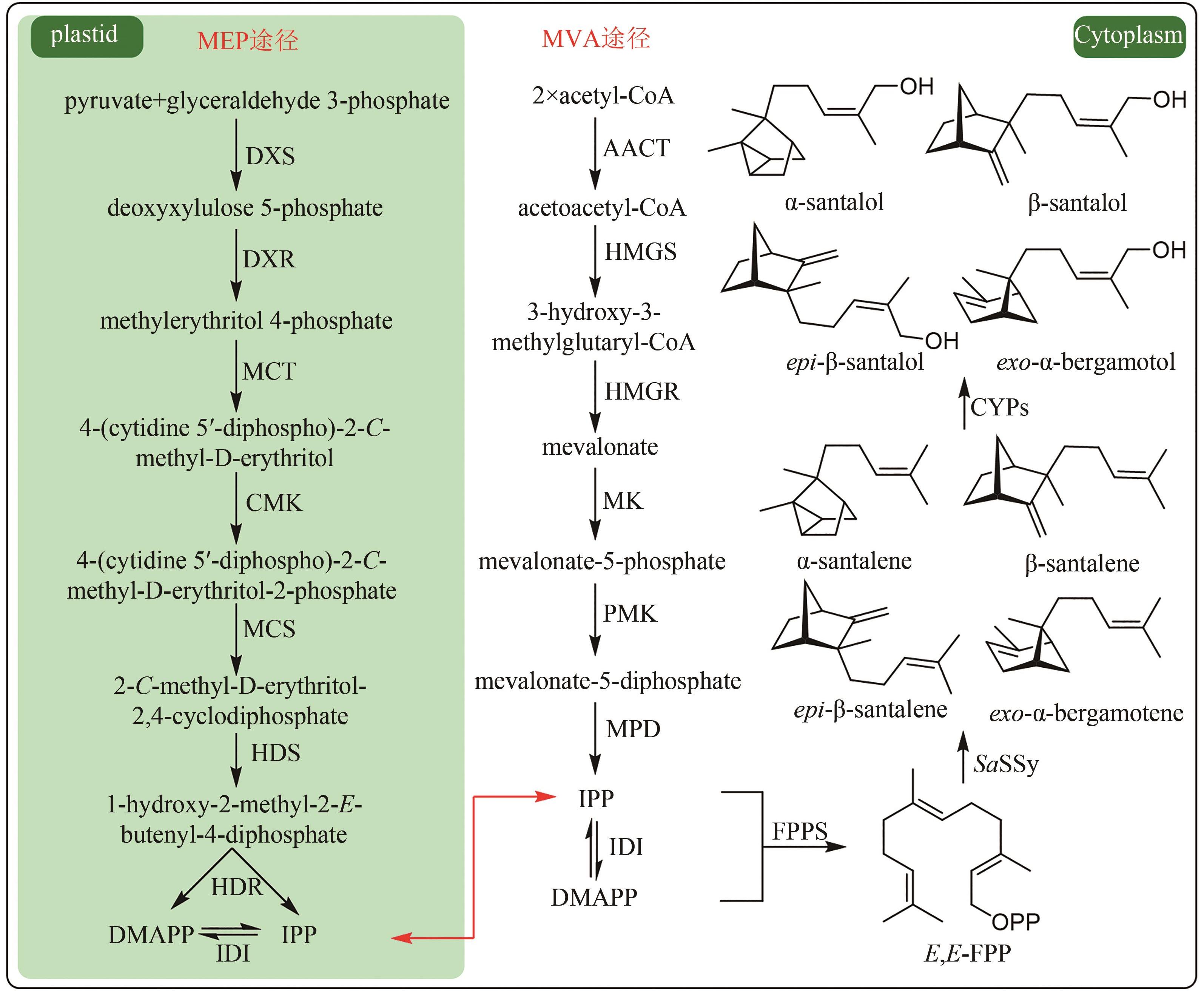
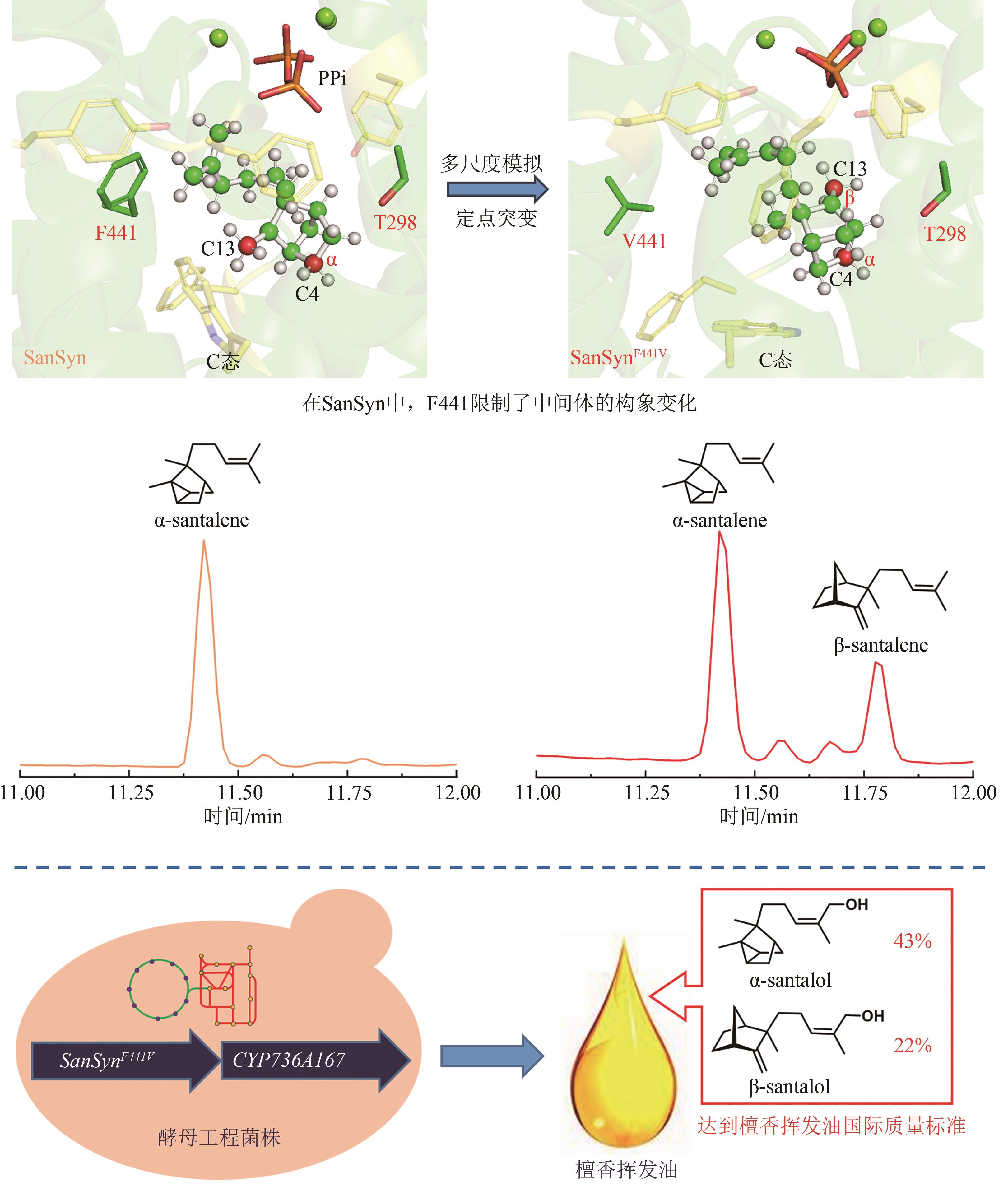
图1 檀香挥发油成分群的生物合成途径(DXS—1-脱氧-D-木酮糖-5-磷酸合酶;DXR—1-脱氧-D-木酮糖-5-磷酸还原异构酶;MCT—2-C-甲基-D-赤藻醇-4-磷酸胞苷酰转移酶;CMK—4-(5′-焦磷酸胞苷)-2-C-甲基-D-赤藓醇激酶;MCS—2-C-甲基-D-赤藓醇-2,4-环焦磷酸合成酶;HDS—1-羟基-2-甲基-2-丁烯-4-焦磷酸合成酶;HDR—1-羟基-2-甲基-2-丁烯-4-焦磷酸还原酶;IDI—异戊烯基焦磷酸异构酶;AACT—乙酰辅酶A酰基转移酶;HMGS—3-羟基-3-甲基戊二酰辅酶A合成酶;HMGR—3-羟基-3-甲基戊二酰辅酶A还原酶;MK—甲羟戊酸激酶;PMK—磷酸甲羟戊酸激酶;MPD—焦磷酸甲羟戊酸脱羧酶)
Fig. 1 Biosynthetic pathway for the production of sandalwood oil(DXS—1-deoxy-D-xylulose 5-phosphate synthase; DXR—1-deoxy-D-lxylulose 5-phosphate reductoisomerase; MCT—2-C-methyl-D-erythritol-4-phosphate cytidyltransferase; CMK—4-(cytidine 5′-diphospho)-2-C-methyl-D-erythritol kinase; MCS—2-C-methyl-D-erythritol 2,4-diphosphate synthase; HDS—1-hydroxy-2-methyl-2-E-butenyl-4-diphosphate synthase; HDR—1-hydroxy-2-methyl-2-butenyl-4-diphosphate reductase; IDI—isopentenyl pyrophosphate isomerase; AACT—acetyl-coenzyme A acetyltransferase; HMGS—3-hydroxy-3-methylglutaryl-CoA synthase; HMGR—3-hydroxy-3-methylglutaryl-CoA reductase; MK—mevalonate kinase; PMK—phosphate mevalonate kinase; MPD—mevalonate pyrophosphate decarboxylase)
| 基因 | 底物 | 产物 | 参考文献 |
|---|---|---|---|
| CYP76F41、CYP76F42、CYP76F39v1 | α-檀香烯、 β-檀香烯、 epi-β-檀香烯、 exo-α-香柠檬烯 | Z/E-α-檀香醇、 Z/E-β-檀香醇、 Z/E-epi-β-檀香醇、 Z/E-exo-α-香柠檬醇 | [ |
| CYP76F39v2 | Z/E-α-檀香醇、 Z/E-β-檀香醇、 E-epi-β-檀香醇、 Z/E-exo-α-香柠檬醇 | ||
| CYP76F40 | Z-α-檀香醇、 E-β-檀香醇、 E-exo-α-香柠檬醇 | ||
| CYP76F37v1、CYP76F37v2、CYP76F38v1、CYP76F38v2 | E-α-檀香醇、 E-β-檀香醇、 E-exo-α-香柠檬醇 | ||
| CYP736A167 | Z-α-檀香醇、 Z-β-檀香醇、 Z-epi-β-檀香醇、 Z-exo-α-香柠檬醇 | [ |
表1 檀香醇生物合成相关细胞色素P450酶
Table 1 Cytochrome P450 enzymes for santalols biosynthesis
| 基因 | 底物 | 产物 | 参考文献 |
|---|---|---|---|
| CYP76F41、CYP76F42、CYP76F39v1 | α-檀香烯、 β-檀香烯、 epi-β-檀香烯、 exo-α-香柠檬烯 | Z/E-α-檀香醇、 Z/E-β-檀香醇、 Z/E-epi-β-檀香醇、 Z/E-exo-α-香柠檬醇 | [ |
| CYP76F39v2 | Z/E-α-檀香醇、 Z/E-β-檀香醇、 E-epi-β-檀香醇、 Z/E-exo-α-香柠檬醇 | ||
| CYP76F40 | Z-α-檀香醇、 E-β-檀香醇、 E-exo-α-香柠檬醇 | ||
| CYP76F37v1、CYP76F37v2、CYP76F38v1、CYP76F38v2 | E-α-檀香醇、 E-β-檀香醇、 E-exo-α-香柠檬醇 | ||
| CYP736A167 | Z-α-檀香醇、 Z-β-檀香醇、 Z-epi-β-檀香醇、 Z-exo-α-香柠檬醇 | [ |
| 1 | QU L, LIANG X C, TIAN G Q, et al. Efficacy and safety of Mulberry twig alkaloids tablet for the treatment of type 2 diabetes: a multicenter, randomized, double-blind, double-dummy, and parallel controlled clinical trial[J]. Diabetes Care, 2021, 44(6): 1324-1333. |
| 2 | CHEN Y M, LIAN C F, SUN Q W, et al. Ramulus mori (Sangzhi) alkaloids alleviate high-fat diet-induced obesity and nonalcoholic fatty liver disease in mice[J]. Antioxidants, 2022, 11(5): 905. |
| 3 | VELLAS B, COLEY N, OUSSET P J, et al. Long-term use of standardised Ginkgo biloba extract for the prevention of Alzheimer’s disease (GuidAge): a randomised placebo-controlled trial[J]. The Lancet Neurology, 2012, 11(10): 851-859. |
| 4 | WANG L Y, TANG J Y, LIU J, et al. Dynamic changes in phenotypic groups in patients with stable angina pectoris after treatment with Xinxuekang capsule: a randomized controlled trial[J]. Current Vascular Pharmacology, 2015, 13(4): 492-503. |
| 5 | 魏玉蓉. 濒危民族药新疆阿魏分布的环境需求及适生区研究[D]. 石河子: 石河子大学, 2019. |
| WEI Y R. The distribution of Ferula sinkiangensis K.M.Shen and its response to climate change [D].Shihezi: Shihezi University, 2019. | |
| 6 | 朱军, 李晓瑾, 张际昭, 等. 一种药用阿魏人工种植栽培的方法: CN108338043B[P]. 2020-04-28. |
| ZHU J, LI X J, ZHANG J Z, et al. A method for artificial planting and cultivation of Ferula sinkiangensis : CN108338043B[P]. 2020-04-28. | |
| 7 | LUO H, ZHAO Y, HUA H, et al. Research progress on quality assurance of genuine Chinese medicinal in Sichuan[J]. Chinese Medicine, 2021, 16(1): 19. |
| 8 | TEIXEIRA DA SILVA J A, KHER M M, SONER D, et al. Sandalwood: basic biology, tissue culture, and genetic transformation[J]. Planta, 2016, 243(4): 847-887. |
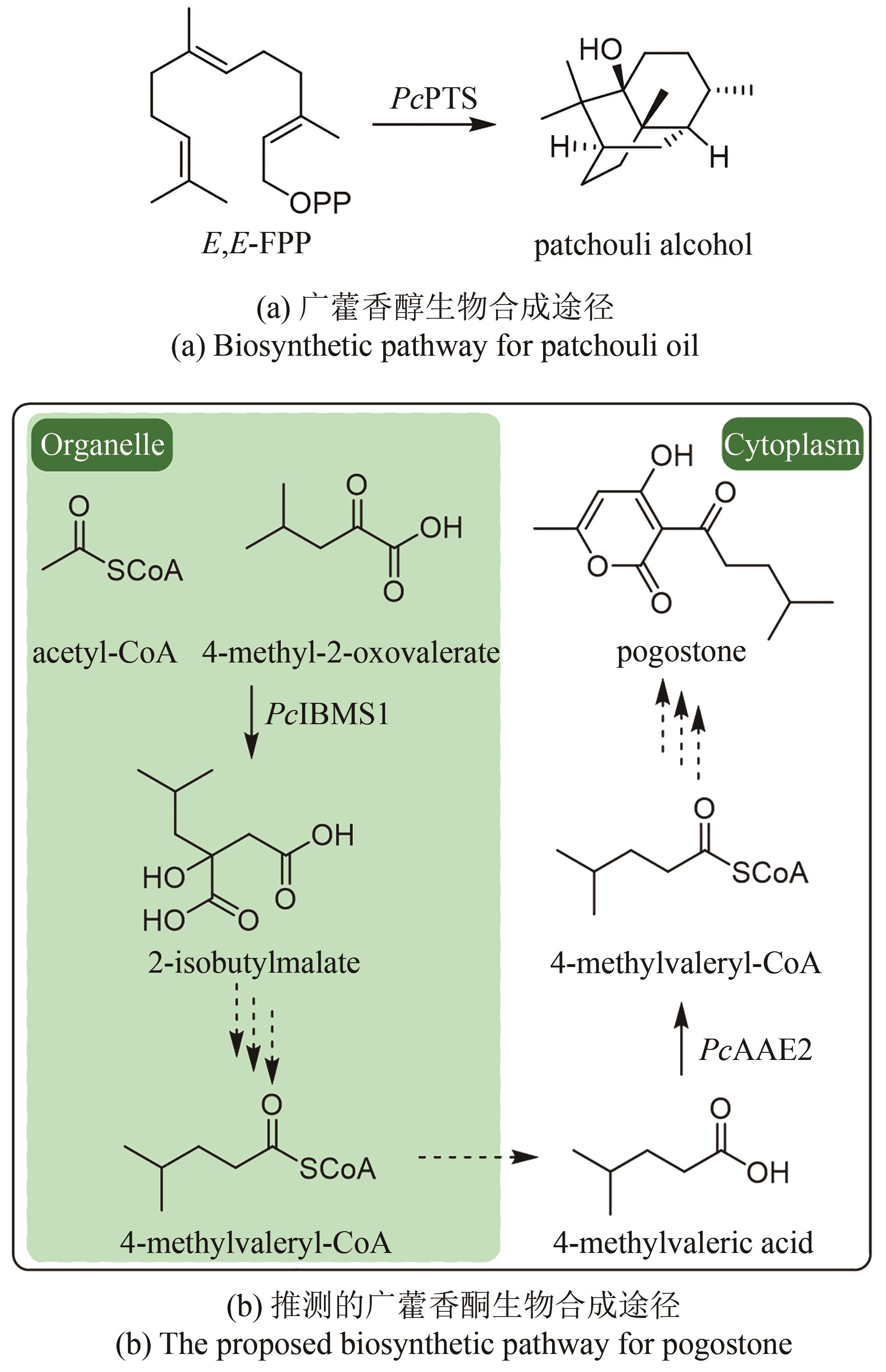
| 9 | ZHANG Y, MENG H, LYU F F, et al. Temporal characteristics of agarwood formation in Aquilaria sinensis after applying whole-tree agarwood-inducing technique[J]. Chinese Herbal Medicines, 2023, 15(1): 37-44. |
| 10 | ZHANG Y, ZHENG Y J, XIA P G, et al. Impact of continuous Panax notoginseng plantation on soil microbial and biochemical properties[J]. Scientific Reports, 2019, 9(1): 13205. |
| 11 | XIAO J J, XU X, WANG F, et al. Analysis of exposure to pesticide residues from traditional Chinese medicine[J]. Journal of Hazardous Materials, 2019, 365: 857-867. |
| 12 | WANG X, ZHANG Z, WU S C. Health benefits of Silybum marianum: phytochemistry, pharmacology, and applications[J]. Journal of Agricultural and Food Chemistry, 2020, 68(42): 11644-11664. |
| 13 | NETT R S, LAU W, SATTELY E S. Discovery and engineering of colchicine alkaloid biosynthesis[J]. Nature, 2020, 584(7819): 148-153. |
| 14 | HAGEL J M, FACCHINI P J. Dioxygenases catalyze the O-demethylation steps of morphine biosynthesis in opium poppy[J]. Nature Chemical Biology, 2010, 6(4): 273-275. |
| 15 | GAO W, HILLWIG M L, HUANG L Q, et al. A functional genomics approach to tanshinone biosynthesis provides stereochemical insights[J]. Organic Letters, 2009, 11(22): 5170-5173. |
| 16 | GUO J, ZHOU Y J, HILLWIG M L, et al. CYP76AH1 catalyzes turnover of miltiradiene in tanshinones biosynthesis and enables heterologous production of ferruginol in yeasts[J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(29): 12108-12113. |
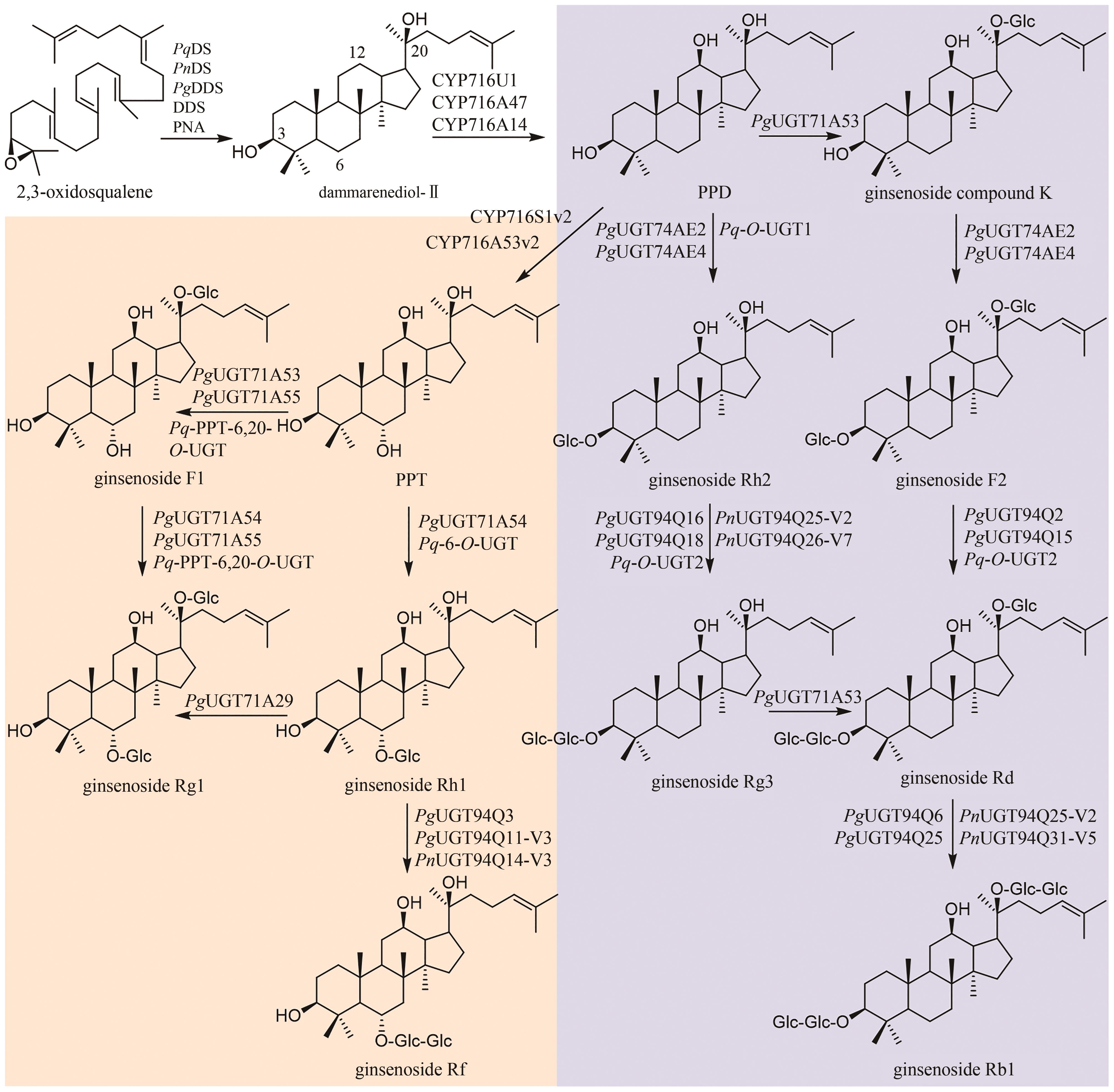
| 17 | GUO J, MA X H, CAI Y, et al. Cytochrome P450 promiscuity leads to a bifurcating biosynthetic pathway for tanshinones[J]. The New Phytologist, 2016, 210(2): 525-534. |
| 18 | MA Y, CUI G H, CHEN T, et al. Expansion within the CYP71D subfamily drives the heterocyclization of tanshinones synthesis in Salvia miltiorrhiza [J]. Nature Communications, 2021, 12(1): 685. |
| 19 | SONG J J, FANG X, LI C Y, et al. A 2-oxoglutarate-dependent dioxygenase converts dihydrofuran to furan in Salvia diterpenoids[J]. Plant Physiology, 2022, 188(3): 1496-1506. |
| 20 | REN L, LUO L L, HU Z M, et al. Functional characterization of CYP81C16 involved in the tanshinone biosynthetic pathway in Salvia miltiorrhiza [J]. Chinese Journal of Natural Medicines, 2023, 21(12): 938-949. |
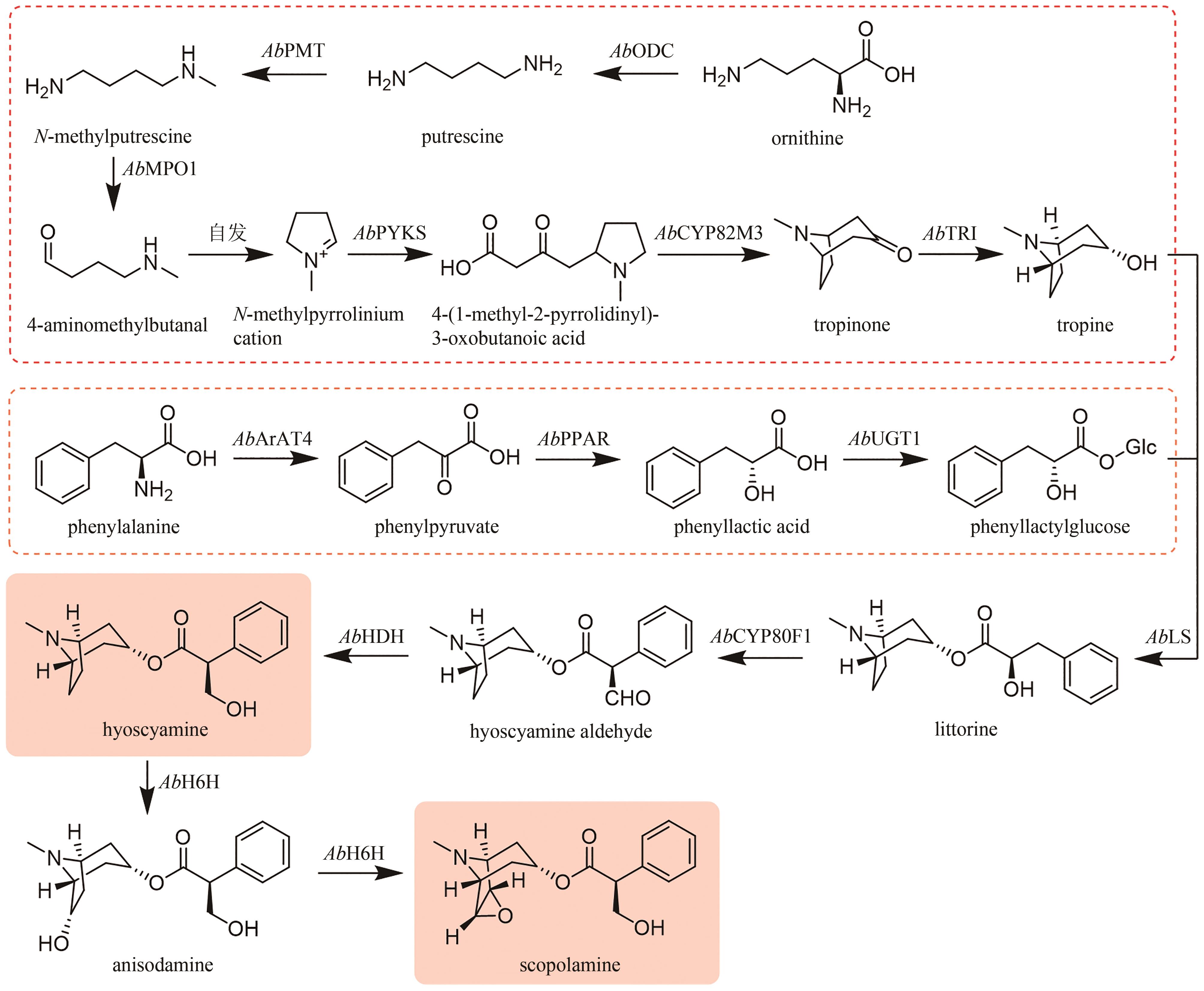
| 21 | ZHAO T F, LI S Q, WANG J, et al. Engineering tropane alkaloid production based on metabolic characterization of ornithine decarboxylase in Atropa belladonna [J]. ACS Synthetic Biology, 2020, 9(2): 437-448. |
| 22 | SUZUKI K, YAMADA Y, HASHIMOTO T. Expression of Atropa belladonna putrescine N-methyltransferase gene in root pericycle[J]. Plant & Cell Physiology, 1999, 40(3): 289-297. |
| 23 | ZHANG F Y, QIU F, ZENG J L, et al. Revealing evolution of tropane alkaloid biosynthesis by analyzing two genomes in the Solanaceae family[J]. Nature Communications, 2023, 14(1): 1446. |
| 24 | DE LUCA V, PIERRE B ST. The cell and developmental biology of alkaloid biosynthesis[J]. Trends in Plant Science, 2000, 5(4): 168-173. |
| 25 | BEDEWITZ M A, JONES A D, D’AURIA J C, et al. Tropinone synthesis via an atypical polyketide synthase and P450-mediated cyclization[J]. Nature Communications, 2018, 9(1): 5281. |
| 26 | HUANG J P, FANG C L, MA X Y, et al. Tropane alkaloids biosynthesis involves an unusual type Ⅲ polyketide synthase and non-enzymatic condensation[J]. Nature Communications, 2019, 10(1): 4036. |
| 27 | PADDON C J, WESTFALL P J, PITERA D J, et al. High-level semi-synthetic production of the potent antimalarial artemisinin[J]. Nature, 2013, 496(7446): 528-532. |
| 28 | ZHA W L, ZHANG F, SHAO J Q, et al. Rationally engineering santalene synthase to readjust the component ratio of sandalwood oil[J]. Nature Communications, 2022, 13(1): 2508. |
| 29 | NEWMAN J D, CHAPPELL J. Isoprenoid biosynthesis in plants: carbon partitioning within the cytoplasmic pathway[J]. Critical Reviews in Biochemistry and Molecular Biology, 1999, 34(2): 95-106. |
| 30 | ROHMER M, KNANI M, SIMONIN P, et al. Isoprenoid biosynthesis in bacteria: a novel pathway for the early steps leading to isopentenyl diphosphate[J]. The Biochemical Journal, 1993, 295(Pt 2): 517-524. |
| 31 | KIRBY J, KEASLING J D. Biosynthesis of plant isoprenoids: perspectives for microbial engineering[J]. Annual Review of Plant Biology, 2009, 60: 335-355. |
| 32 | MOHANKUMAR A, SHANMUGAM G, KALAISELVI D, et al. East Indian sandalwood (Santalum album L.) oil confers neuroprotection and geroprotection in Caenorhabditis elegans via activating SKN-1/Nrf2 signaling pathway[J]. RSC Advances, 2018, 8(59): 33753-33774. |
| 33 | GUO H M, ZHANG J Z, GAO W Y, et al. Anti-diarrhoeal activity of methanol extract of Santalum album L. in mice and gastrointestinal effect on the contraction of isolated jejunum in rats[J]. Journal of Ethnopharmacology, 2014, 154(3): 704-710. |
| 34 | BURDOCK G A, CARABIN I G. Safety assessment of sandalwood oil (Santalum album L.)[J]. Food and Chemical Toxicology, 2008, 46(2): 421-432. |
| 35 | KUCHARSKA M, FRYDRYCH B, WESOLOWSKI W, et al. A comparison of the composition of selected commercial sandalwood oils with the international standard[J]. Molecules, 2021, 26(8): 2249. |
| 36 | DARAMWAR P P, SRIVASTAVA P L, PRIYADARSHINI B, et al. Preparative separation of α- and β-santalenes and (Z)-α- and (Z)-β-santalols using silver nitrate-impregnated silica gel medium pressure liquid chromatography and analysis of sandalwood oil[J]. The Analyst, 2012, 137(19): 4564-4570. |
| 37 | JONES C G, MONIODIS J, ZULAK K G, et al. Sandalwood fragrance biosynthesis involves sesquiterpene synthases of both the terpene synthase (TPS)-a and TPS-b subfamilies, including santalene synthases[J]. The Journal of Biological Chemistry, 2011, 286(20): 17445-17454. |
| 38 | BEEKWILDER M J, VAN HOUWELINGEN A M M L, BOSCH H J, et al. Santalene synthase: US0010822A1 [P]. 2020-07-19. |
| 39 | SCHALK M. Method for producing alpha-santalene: US0008836A1[P]. 2013-03-27. |
| 40 | SALLAUD C, RONTEIN D, ONILLON S, et al. A novel pathway for sesquiterpene biosynthesis from Z,Z-farnesyl pyrophosphate in the wild tomato Solanum habrochaites [J]. The Plant Cell, 2009, 21(1): 301-317. |
| 41 | MATSUBA Y, NGUYEN T T, WIEGERT K, et al. Evolution of a complex locus for terpene biosynthesis in Solanum [J]. The Plant Cell, 2013, 25(6): 2022-2036. |
| 42 | DIAZ-CHAVEZ M L, MONIODIS J, MADILAO L L, et al. Biosynthesis of sandalwood oil: Santalum album CYP76F cytochromes P450 produce santalols and bergamotol[J]. PLoS One, 2013, 8(9): e75053. |
| 43 | CELEDON J M, CHIANG A, YUEN M M, et al. Heartwood-specific transcriptome and metabolite signatures of tropical sandalwood (Santalum album) reveal the final step of (Z)- santalol fragrance biosynthesis[J]. The Plant Journal, 2016, 86(4): 289-299. |
| 44 | GAN Y X, AI G X, WU J Z, et al. Patchouli oil ameliorates 5-fluorouracil-induced intestinal mucositis in rats via protecting intestinal barrier and regulating water transport[J]. Journal of Ethnopharmacology, 2020, 250: 112519. |
| 45 | FATIMA S, FARZEEN I, ASHRAF A, et al. A comprehensive review on pharmacological activities of pachypodol: a bioactive compound of an aromatic medicinal plant Pogostemon cablin Benth[J]. Molecules, 2023, 28(8): 3469. |
| 46 | DEGUERRY F, PASTORE L, WU S Q, et al. The diverse sesquiterpene profile of patchouli, Pogostemon cablin, is correlated with a limited number of sesquiterpene synthases[J]. Archives of Biochemistry and Biophysics, 2006, 454(2): 123-136. |
| 47 | CHEN X Z, WANG X B, WU D D, et al. PatDREB transcription factor activates patchoulol synthase gene promoter and positively regulates jasmonate-induced patchoulol biosynthesis[J]. Journal of Agricultural and Food Chemistry, 2022, 70(23): 7188-7201. |
| 48 | YU Z X, WANG L J, ZHAO B, et al. Progressive regulation of sesquiterpene biosynthesis in Arabidopsis and Patchouli (Pogostemon cablin) by the miR156-targeted SPL transcription factors[J]. Molecular Plant, 2015, 8(1): 98-110. |
| 49 | WANG C, WANG Y, CHEN J, et al. Synthesis of 4-methylvaleric acid, a precursor of pogostone, involves a 2-isobutylmalate synthase related to 2-isopropylmalate synthase of leucine biosynthesis[J]. The New Phytologist, 2022, 235(3): 1129-1145. |
| 50 | 刘浪. 广藿香酮生物合成关键基因的鉴定[D]. 重庆: 重庆大学, 2022. |
| LIU L. Identification of key genes that catalyze pogostone biosynthesis[D]. Chongqing: Chongqing University, 2022. | |
| 51 | GUO D M, WANG H Y, ZHANG S M, et al. The type Ⅲ polyketide synthase supergene family in plants: complex evolutionary history and functional divergence[J]. The Plant Journal, 2022, 112(2): 414-428. |
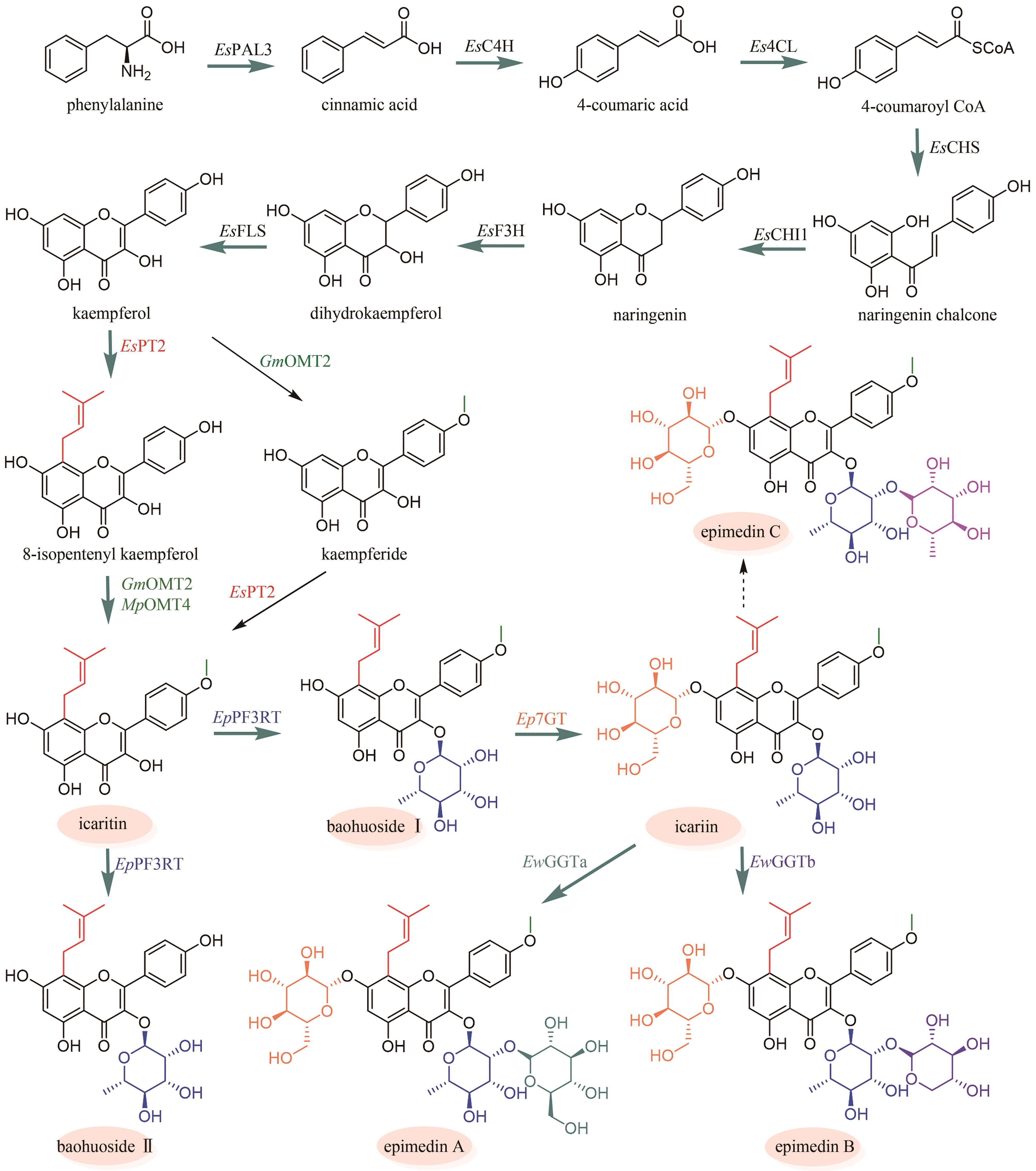
| 52 | GHOSH S. Triterpene structural diversification by plant cytochrome P450 enzymes[J]. Frontiers in Plant Science, 2017, 8: 1886. |
| 53 | CÁRDENAS P D, ALMEIDA A, BAK S. Evolution of structural diversity of triterpenoids[J]. Frontiers in Plant Science, 2019, 10: 1523. |
| 54 | CHEN W, BALAN P, POPOVICH D G. Review of ginseng anti-diabetic studies[J]. Molecules, 2019, 24(24): 4501. |
| 55 | BAIK I H, KIM K H, LEE K A. Antioxidant, anti-inflammatory and antithrombotic effects of ginsenoside compound K enriched extract derived from ginseng sprouts[J]. Molecules, 2021, 26(13): 4102. |
| 56 | ZHOU P, XIE W J, HE S B, et al. Ginsenoside Rb1 as an anti-diabetic agent and its underlying mechanism analysis[J]. Cells, 2019, 8(3): 204. |
| 57 | HOU M Q, WANG R F, ZHAO S J, et al. Ginsenosides in Panax genus and their biosynthesis[J]. Acta Pharmaceutica Sinica B, 2021, 11(7): 1813-1834. |
| 58 | TANSAKUL P, SHIBUYA M, KUSHIRO T, et al. Dammarenediol-Ⅱ synthase, the first dedicated enzyme for ginsenoside biosynthesis, in Panax ginseng [J]. FEBS Letters, 2006, 580(22): 5143-5149. |
| 59 | HAN J Y, KWON Y S, YANG D C, et al. Expression and RNA interference-induced silencing of the dammarenediol synthase gene in Panax ginseng [J]. Plant & Cell Physiology, 2006, 47(12): 1653-1662. |
| 60 | LEE M H, HAN J Y, KIM H J, et al. Dammarenediol-Ⅱ production confers TMV tolerance in transgenic tobacco expressing Panax ginseng dammarenediol-Ⅱ synthase[J]. Plant & Cell Physiology, 2012, 53(1): 173-182. |
| 61 | NIU Y Y, LUO H M, SUN C, et al. Expression profiling of the triterpene saponin biosynthesis genes FPS, SS, SE, and DS in the medicinal plant Panax notoginseng [J]. Gene, 2014, 533(1): 295-303. |
| 62 | WANG L, ZHAO S J, CAO H J, et al. The isolation and characterization of dammarenediol synthase gene from Panax quinquefolius and its heterologous co-expression with cytochrome P450 gene PqD12H in yeast[J]. Functional & Integrative Genomics, 2014, 14(3): 545-557. |
| 63 | HAN J Y, HWANG H S, CHOI S W, et al. Cytochrome P450 CYP716A53v2 catalyzes the formation of protopanaxatriol from protopanaxadiol during ginsenoside biosynthesis in Panax ginseng [J]. Plant & Cell Physiology, 2012, 53(9): 1535-1545. |
| 64 | HAN J Y, KIM H J, KWON Y S, et al. The Cyt P450 enzyme CYP716A47 catalyzes the formation of protopanaxadiol from dammarenediol-Ⅱ during ginsenoside biosynthesis in Panax ginseng [J]. Plant & Cell Physiology, 2011, 52(12): 2062-2073. |
| 65 | HAN J Y, KIM M J, BAN Y W, et al. The involvement of β-amyrin 28-oxidase (CYP716A52v2) in oleanane-type ginsenoside biosynthesis in Panax ginseng [J]. Plant & Cell Physiology, 2013, 54(12): 2034-2046. |
| 66 | JUNG S C, KIM W, PARK S C, et al. Two ginseng UDP-glycosyltransferases synthesize ginsenoside Rg3 and Rd[J]. Plant & Cell Physiology, 2014, 55(12): 2177-2188. |
| 67 | LU C, ZHAO S J, WEI G N, et al. Functional regulation of ginsenoside biosynthesis by RNA interferences of a UDP-glycosyltransferase gene in Panax ginseng and Panax quinquefolius [J]. Plant Physiology and Biochemistry, 2017, 111: 67-76. |
| 68 | YANG C S, LI C J, WEI W, et al. The unprecedented diversity of UGT94-family UDP-glycosyltransferases in Panax plants and their contribution to ginsenoside biosynthesis[J]. Scientific Reports, 2020, 10(1): 15394. |
| 69 | WANG P P, WEI Y J, FAN Y, et al. Production of bioactive ginsenosides Rh2 and Rg3 by metabolically engineered yeasts[J]. Metabolic Engineering, 2015, 29: 97-105. |
| 70 | LU C, ZHAO S J, WANG X S. Functional regulation of a UDP-glucosyltransferase gene (Pq3-O-UGT1) by RNA interference and overexpression in Panax quinquefolius [J]. Plant Cell, Tissue and Organ Culture (PCTOC), 2017, 129(3): 445-456. |
| 71 | YAN X, FAN Y, WEI W, et al. Production of bioactive ginsenoside compound K in metabolically engineered yeast[J]. Cell Research, 2014, 24(6): 770-773. |
| 72 | WEI W, WANG P P, WEI Y J, et al. Characterization of Panax ginseng UDP-glycosyltransferases catalyzing protopanaxatriol and biosyntheses of bioactive ginsenosides F1 and Rh1 in metabolically engineered yeasts[J]. Molecular Plant, 2015, 8(9): 1412-1424. |
| 73 | FENG P C, LI G X, WANG X S, et al. Identification and RNAi-based gene silencing of a novel UDP-glycosyltransferase from Panax quinquefolius [J]. Plant Cell, Tissue and Organ Culture (PCTOC), 2021, 144(3): 567-576. |
| 74 | LU J, YAO L, LI J X, et al. Characterization of UDP-glycosyltransferase involved in biosynthesis of ginsenosides Rg1 and Rb1 and identification of critical conserved amino acid residues for its function[J]. Journal of Agricultural and Food Chemistry, 2018, 66(36): 9446-9455. |
| 75 | FU J, WANG Z H, HUANG L F, et al. Review of the botanical characteristics, phytochemistry, and pharmacology of Astragalus membranaceus (Huangqi)[J]. Phytotherapy Research, 2014, 28(9): 1275-1283. |
| 76 | 北京大学药学院 天然药物及仿生药物国家重点实验室. 叶敏/乔雪团队在黄芪皂苷生物合成研究中取得系列进展[J]. 中国药学(英文版), 2022, 31(12): 963-966. |
| State Key Laboratory of Natural and Biomimetic Drugs, School of Pharmaceutical Sciences, Peking University Health Science Center. The research group of Prof. Min Ye and Prof. Xue Qiao made a series of progress in the biosynthesis of Astragalus saponins [J]. Journal of Chinese Pharmaceutical Sciences, 2022, 31(12): 963-966. | |
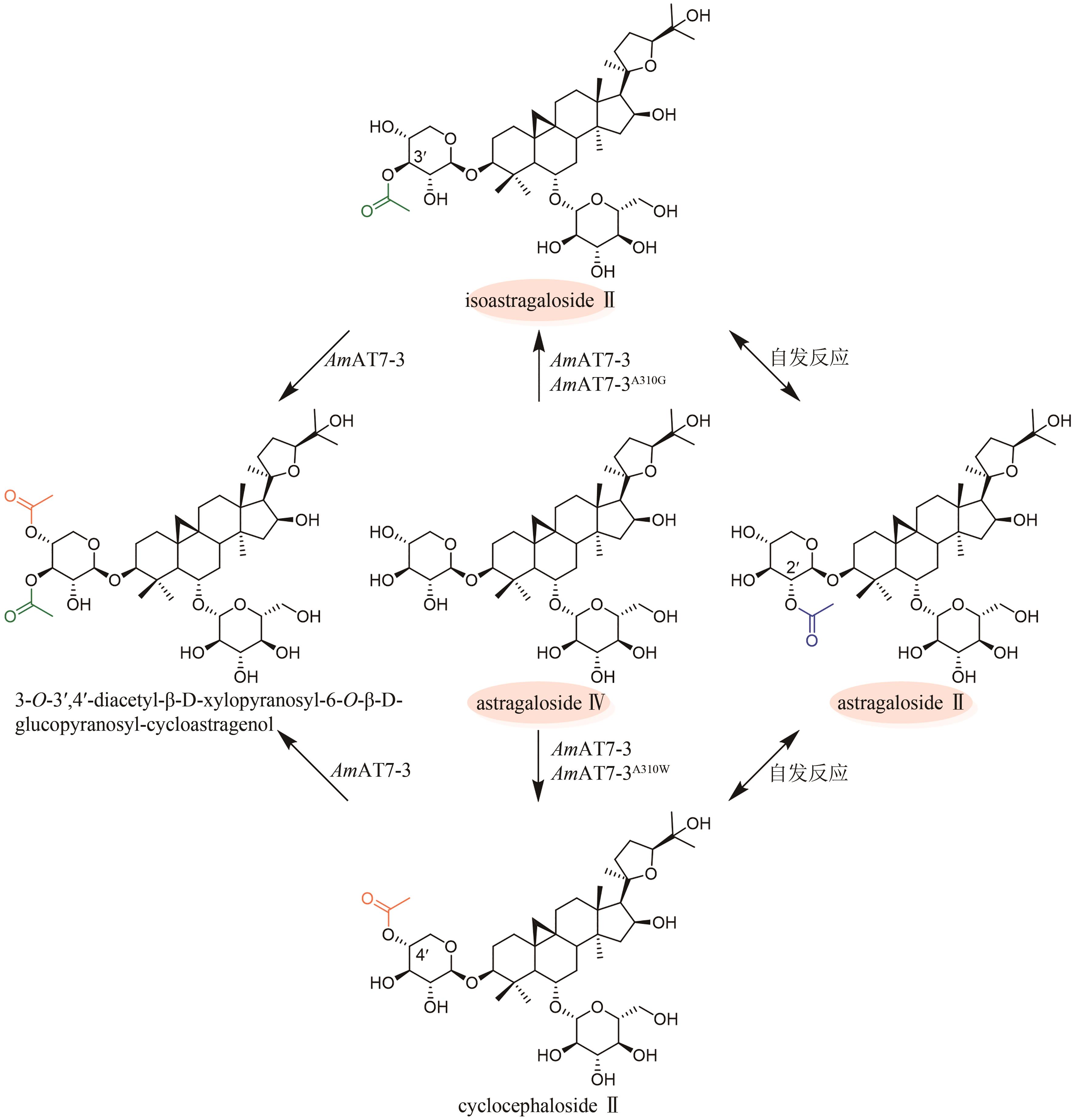
| 77 | CHEN K, ZHANG M, XU L L, et al. Identification of oxidosqualene cyclases associated with saponin biosynthesis from Astragalus membranaceus reveals a conserved motif important for catalytic function[J]. Journal of Advanced Research, 2023, 43: 247-257. |
| 78 | ZHANG M, YI Y, GAO B H, et al. Functional characterization and protein engineering of a triterpene 3-/6-/2'-O-glycosyltransferase reveal a conserved residue critical for the regiospecificity[J]. Angewandte Chemie International Edition, 2022, 61(8): e202113587. |
| 79 | CHEN K, ZHANG M, GAO B H, et al. Characterization and protein engineering of glycosyltransferases for the biosynthesis of diverse hepatoprotective cycloartane-type saponins in Astragalus membranaceus [J]. Plant Biotechnology Journal, 2023, 21(4): 698-710. |
| 80 | DUAN Y Y, DU W Y, SONG Z J, et al. Functional characterization of a cycloartenol synthase and four glycosyltransferases in the biosynthesis of cycloastragenol-type astragalosides from Astragalus membranaceus [J]. Acta Pharmaceutica Sinica B, 2023, 13(1): 271-283. |
| 81 | WANG L L, JIANG Z H, ZHANG J H, et al. Characterization and structure-based protein engineering of a regiospecific saponin acetyltransferase from Astragalus membranaceus [J]. Nature Communications, 2023, 14(1): 5969. |
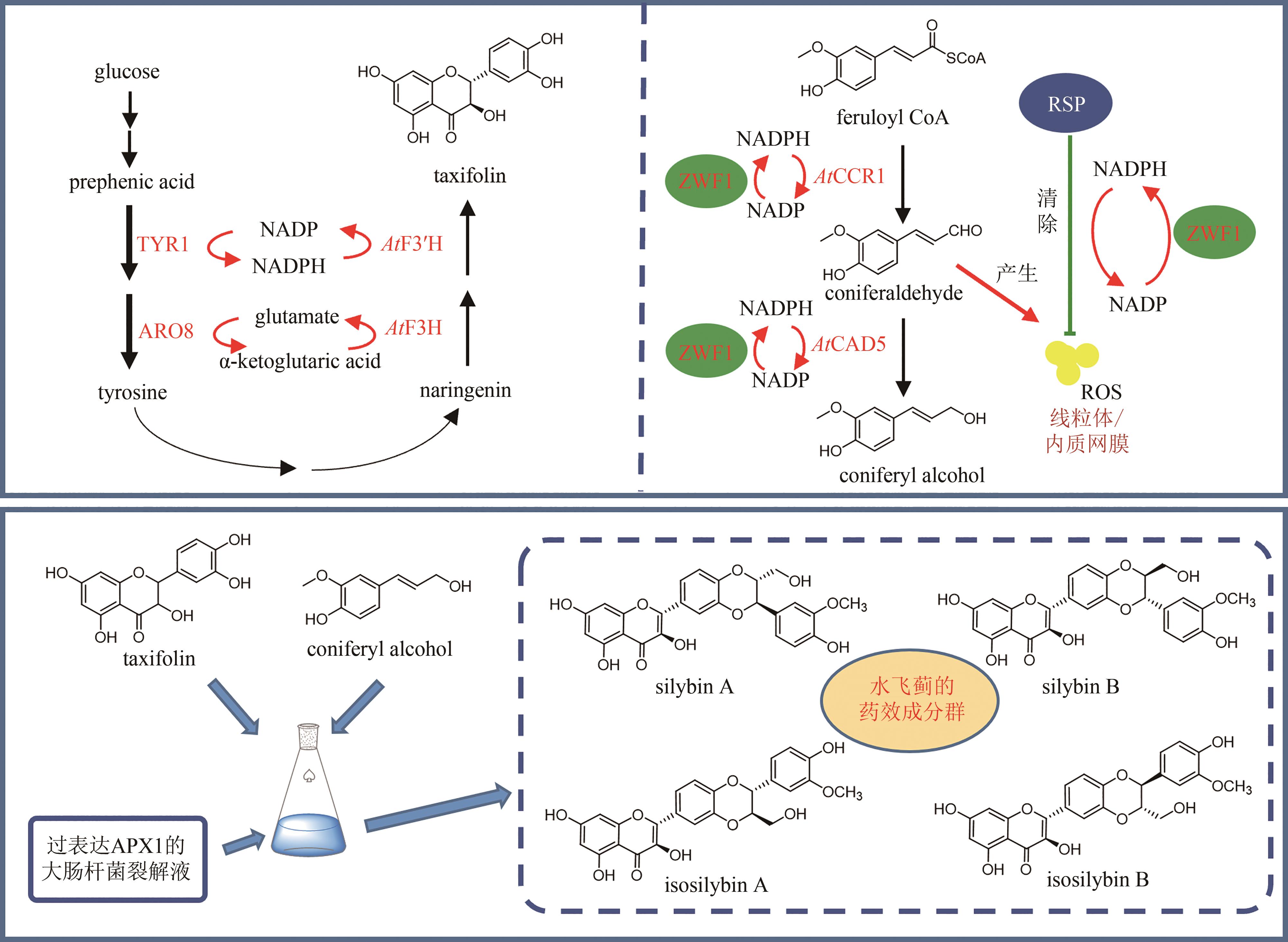
| 82 | TONG Y J, LYU Y B, XU S, et al. Optimum chalcone synthase for flavonoid biosynthesis in microorganisms[J]. Critical Reviews in Biotechnology, 2021, 41(8): 1194-1208. |
| 83 | FANG D N, ZHENG C W, MA Y L. Effectiveness of Scutellaria baicalensis Georgi root in pregnancy-related diseases: a review[J]. Journal of Integrative Medicine, 2023, 21(1): 17-25. |
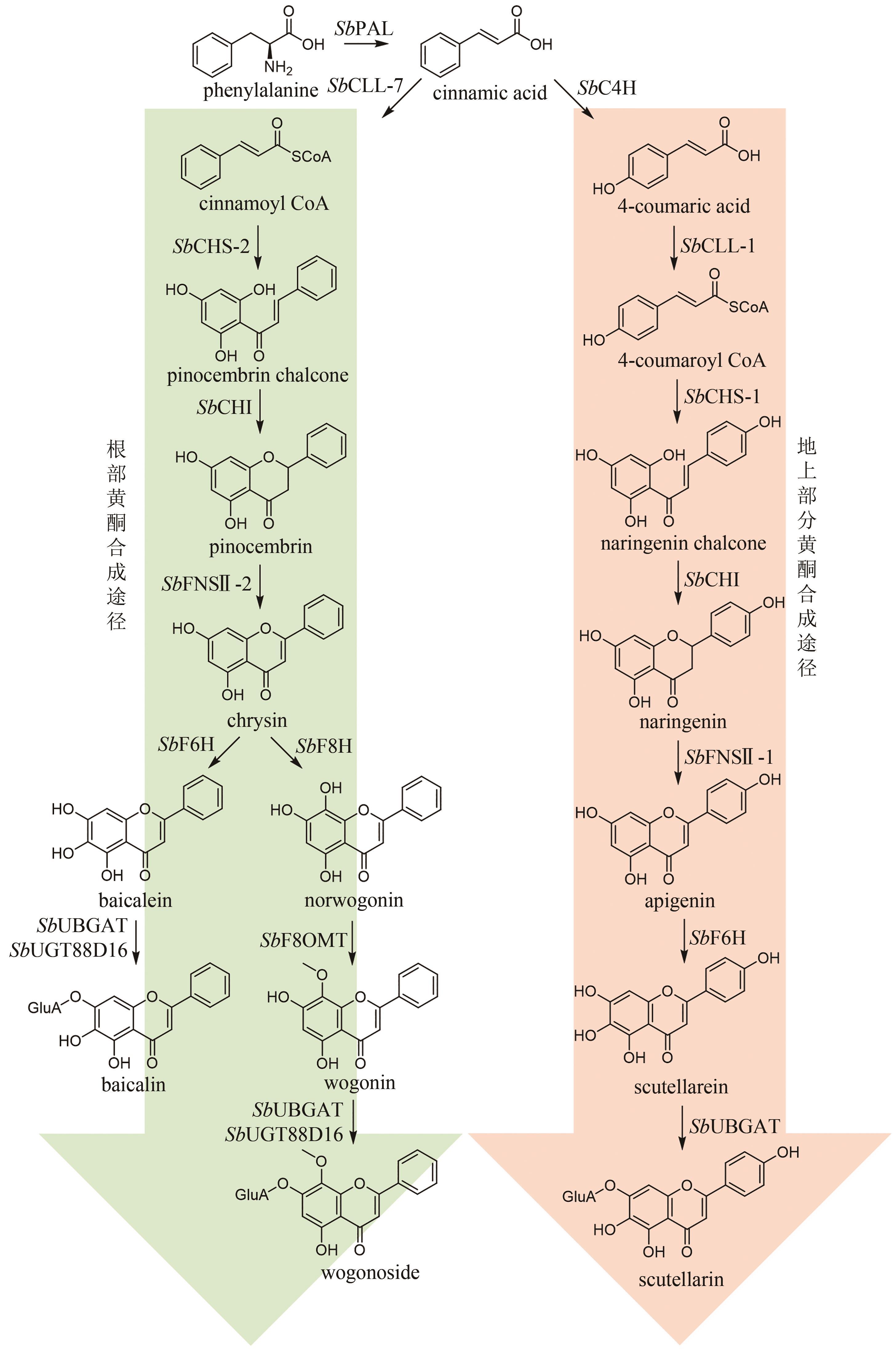
| 84 | WANG Z L, WANG S, KUANG Y, et al. A comprehensive review on phytochemistry, pharmacology, and flavonoid biosynthesis of Scutellaria baicalensis [J]. Pharmaceutical Biology, 2018, 56(1): 465-484. |
| 85 | PEI T L, YAN M X, HUANG Y B, et al. Specific flavonoids and their biosynthetic pathway in Scutellaria baicalensis [J]. Frontiers in Plant Science, 2022, 13: 866282. |
| 86 | ZHAO Q, YANG J, CUI M Y, et al. The reference genome sequence of Scutellaria baicalensis provides insights into the evolution of wogonin biosynthesis[J]. Molecular Plant, 2019, 12(7): 935-950. |
| 87 | XU H, PARK N I, LI X H, et al. Molecular cloning and characterization of phenylalanine ammonia-lyase, cinnamate 4-hydroxylase and genes involved in flavone biosynthesis in Scutellaria baicalensis [J]. Bioresource Technology, 2010, 101(24): 9715-9722. |
| 88 | ZHAO Q, ZHANG Y, WANG G, et al. A specialized flavone biosynthetic pathway has evolved in the medicinal plant, Scutellaria baicalensis [J]. Science Advances, 2016, 2(4): e1501780. |
| 89 | ZHAO Q, CUI M Y, LEVSH O, et al. Two CYP82D enzymes function as flavone hydroxylases in the biosynthesis of root-specific 4'-deoxyflavones in Scutellaria baicalensis [J]. Molecular Plant, 2018, 11(1): 135-148. |
| 90 | LIU X N, CHENG J, ZHU X X, et al. De novo biosynthesis of multiple pinocembrin derivatives in Saccharomyces cerevisiae [J]. ACS Synthetic Biology, 2020, 9(11): 3042-3051. |
| 91 | 韩搏云, 王子龙, 王双, 等. 黄芩中黄酮O-糖基转移酶的发现及功能表征[J]. 药学学报, 2021, 56(12): 3345-3352. |
| HAN B Y, WANG Z L, WANG S, et al. Discovery and functional characterization of flavone O-glycosyltransferases in Scutellaria baicalensis [J]. Acta Pharmaceutica Sinica, 2021, 56(12): 3345-3352. | |
| 92 | NAGASHIMA S, HIROTANI M, YOSHIKAWA T. Purification and characterization of UDP-glucuronate: baicalein 7-O-glucuronosyltransferase from Scutellaria baicalensis Georgi. cell suspension cultures[J]. Phytochemistry, 2000, 53(5): 533-538. |
| 93 | ZHANG H F, YANG T S, LI Z Z, et al. Simultaneous extraction of epimedin A, B, C and icariin from Herba Epimedii by ultrasonic technique[J]. Ultrasonics Sonochemistry, 2008, 15(4): 376-385. |
| 94 | 张云风, 郭宝林. 拟巫山淫羊藿生长地土壤理化性质调查分析[J]. 中国现代中药, 2017, 19(4): 543-547. |
| ZHANG Y F, GUO B L. Investigation and analysis of soil physicochemical properties of Epimedium pseudowushanense growing regions[J]. Modern Chinese Medicine, 2017, 19(4): 543-547. | |
| 95 | HUANG W J, ZENG S H, XIAO G, et al. Elucidating the biosynthetic and regulatory mechanisms of flavonoid-derived bioactive components in Epimedium sagittatum [J]. Frontiers in Plant Science, 2015, 6: 689. |
| 96 | AN T, LIN G Y, LIU Y, et al. De novo biosynthesis of anticarcinogenic icariin in engineered yeast[J]. Metabolic Engineering, 2023, 80: 207-215. |
| 97 | WANG P P, LI C J, LI X D, et al. Complete biosynthesis of the potential medicine icaritin by engineered Saccharomyces cerevisiae and Escherichia coli [J]. Science Bulletin, 2021, 66(18): 1906-1916. |
| 98 | FENG K P, CHEN R D, XIE K B, et al. A regiospecific rhamnosyltransferase from Epimedium pseudowushanense catalyzes the 3-O-rhamnosylation of prenylflavonols[J]. Organic & Biomolecular Chemistry, 2018, 16(3): 452-458. |
| 99 | FENG K P, CHEN R D, XIE K B, et al. Ep7GT, a glycosyltransferase with sugar donor flexibility from Epimedium pseudowushanense, catalyzes the 7-O-glycosylation of baohuoside[J]. Organic & Biomolecular Chemistry, 2019, 17(35): 8106-8114. |
| 100 | 虞沂, 曹应龙, 刘亚婷. 朝藿定合成用糖苷糖基转移酶及其编码基因和应用: CN114350634B[P]. 2022-04-15. |
| YU Y, CAO Y L, LIU Y T. Glycoside glycosyltransferase and its coding gene for the synthesis of epimedins and its application: CN114350634B[P]. 2022-04-15. | |
| 101 | TEPONNO R B, KUSARI S, SPITELLER M. Recent advances in research on lignans and neolignans[J]. Natural Product Reports, 2016, 33(9): 1044-1092. |
| 102 | CHEN Q, LAN H Y, PENG W, et al. Isatis indigotica: a review of phytochemistry, pharmacological activities and clinical applications[J]. The Journal of Pharmacy and Pharmacology, 2021, 73(9): 1137-1150. |
| 103 | FENG J X, HUANG D D, YANG Y B, et al. Isatis indigotica: from (ethno) botany, biochemistry to synthetic biology[J]. Molecular Horticulture, 2021, 1(1): 17. |
| 104 | BOERJAN W, RALPH J, BAUCHER M. Lignin biosynthesis[J]. Annual Review of Plant Biology, 2003, 54: 519-546. |
| 105 | PANIAGUA C, BILKOVA A, JACKSON P, et al. Dirigent proteins in plants: modulating cell wall metabolism during abiotic and biotic stress exposure[J]. Journal of Experimental Botany, 2017, 68(13): 3287-3301. |
| 106 | CHEN R B, YU J, YU L Y, et al. The ERF transcription factor LTF1 activates DIR1 to control stereoselective synthesis of antiviral lignans and stress defense in Isatis indigotica roots[J]. Acta Pharmaceutica Sinica B, 2024, 14(1): 405-420. |
| 107 | XIAO Y, SHAO K, ZHOU J W, et al. Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases[J]. Nature Communications, 2021, 12(1): 2828. |
| 108 | TAN Y P, YANG J, JIANG Y Y, et al. Functional characterization of UDP-glycosyltransferases involved in anti-viral lignan glycosides biosynthesis in Isatis indigotica [J]. Frontiers in Plant Science, 2022, 13: 921815. |
| 109 | CHEN X, CHEN J F, FENG J X, et al. Tandem UGT71B5s catalyze lignan glycosylation in Isatis indigotica with substrates promiscuity[J]. Frontiers in Plant Science, 2021, 12: 637695. |
| 110 | RENNER U D, OERTEL R, KIRCH W. Pharmacokinetics and pharmacodynamics in clinical use of scopolamine[J]. Therapeutic Drug Monitoring, 2005, 27(5): 655-665. |
| 111 | ROGALLA P, LEMBCKE A, RÜCKERT J C, et al. Spasmolysis at CT colonography: butyl scopolamine versus glucagon[J]. Radiology, 2005, 236(1): 184-188. |
| 112 | BEDEWITZ M A, GÓNGORA-CASTILLO E, UEBLER J B, et al. A root-expressed L-phenylalanine: 4-hydroxyphenylpyruvate aminotransferase is required for tropane alkaloid biosynthesis in Atropa belladonna [J]. The Plant Cell, 2014, 26(9): 3745-3762. |
| 113 | QIU F, YANG C X, YUAN L N, et al. A phenylpyruvic acid reductase is required for biosynthesis of tropane alkaloids[J]. Organic Letters, 2018, 20(24): 7807-7810. |
| 114 | QIU F, ZENG J L, WANG J, et al. Functional genomics analysis reveals two novel genes required for littorine biosynthesis[J]. The New Phytologist, 2020, 225(5): 1906-1914. |
| 115 | LI R, REED D W, LIU E W, et al. Functional genomic analysis of alkaloid biosynthesis in Hyoscyamus niger reveals a cytochrome P450 involved in littorine rearrangement[J]. Chemistry & Biology, 2006, 13(5): 513-520. |
| 116 | NASOMJAI P, REED D W, TOZER D J, et al. Mechanistic insights into the cytochrome P450-mediated oxidation and rearrangement of littorine in tropane alkaloid biosynthesis[J]. ChemBioChem, 2009, 10(14): 2382-2393. |
| 117 | LI J, VAN BELKUM M J, VEDERAS J C. Functional characterization of recombinant hyoscyamine 6β-hydroxylase from Atropa belladonna [J]. Bioorganic & Medicinal Chemistry, 2012, 20(14): 4356-4363. |
| 118 | SRINIVASAN P, SMOLKE C D. Biosynthesis of medicinal tropane alkaloids in yeast[J]. Nature, 2020, 585(7826): 614-619. |
| 119 | QIU F, YAN Y J, ZENG J L, et al. Biochemical and metabolic insights into hyoscyamine dehydrogenase[J]. ACS Catalysis, 2021, 11(5): 2912-2924. |
| 120 | YANG J Z, LIANG J C, SHAO L, et al. Green production of silybin and isosilybin by merging metabolic engineering approaches and enzymatic catalysis[J]. Metabolic Engineering, 2020, 59: 44-52. |
| 121 | ARUNKUMAR A N, DHYANI A, JOSHI G. Santalum album[EB/OL]// IUCN red list of threatened speciesThe. 2019: e.T31852A2807668[2023-11-01]. . |
| 122 | ZHA W L, AN T Y, LI T, et al. Reconstruction of the biosynthetic pathway of santalols under control of the GAL regulatory system in yeast[J]. ACS Synthetic Biology, 2020, 9(2): 449-456. |
| 123 | ZHU J X, AN T Y, ZHA W L, et al. Manipulation of IME4 expression, a global regulation strategy for metabolic engineering in Saccharomyces cerevisiae [J]. Acta Pharmaceutica Sinica B, 2023, 13(6): 2795-2806. |
| 124 | LV Y K, GAO S, XU S, et al. Spatial organization of silybin biosynthesis in milk thistle[Silybum marianum (L.) Gaertn][J]. The Plant Journal, 2017, 92(6): 995-1004. |
| 125 | GALANIE S, THODEY K, TRENCHARD I J, et al. Complete biosynthesis of opioids in yeast[J]. Science, 2015, 349(6252): 1095-1100. |
| 126 | LIU Y F, WANG B, SHU S H, et al. Analysis of the Coptis chinensis genome reveals the diversification of protoberberine-type alkaloids[J]. Nature Communications, 2021, 12(1): 3276. |
| 127 | LI M J, KILDEGAARD K R, CHEN Y, et al. De novo production of resveratrol from glucose or ethanol by engineered Saccharomyces cerevisiae [J]. Metabolic Engineering, 2015, 32: 1-11. |
| 128 | ALPER H, FISCHER C, NEVOIGT E, et al. Tuning genetic control through promoter engineering[J]. Proceedings of the National Academy of Sciences of the United States of America, 2005, 102(36): 12678-12683. |
| 129 | CURRAN K A, KARIM A S, GUPTA A, et al. Use of expression-enhancing terminators in Saccharomyces cerevisiae to increase mRNA half-life and improve gene expression control for metabolic engineering applications[J]. Metabolic Engineering, 2013, 19: 88-97. |
| 130 | ALPER H, MOXLEY J, NEVOIGT E, et al. Engineering yeast transcription machinery for improved ethanol tolerance and production[J]. Science, 2006, 314(5805): 1565-1568. |
| 131 | HU Y T, ZHU Z W, NIELSEN J, et al. Heterologous transporter expression for improved fatty alcohol secretion in yeast[J]. Metabolic Engineering, 2018, 45: 51-58. |
| 132 | CHEN R B, GAO J Q, YU W, et al. Engineering cofactor supply and recycling to drive phenolic acid biosynthesis in yeast[J]. Nature Chemical Biology, 2022, 18(5): 520-529. |
| 133 | HAMMER S K, AVALOS J L. Harnessing yeast organelles for metabolic engineering[J]. Nature Chemical Biology, 2017, 13(8): 823-832. |
| 134 | NIELSEN J, KEASLING J D. Engineering cellular metabolism[J]. Cell, 2016, 164(6): 1185-1197. |
| 135 | WANG Y, YU L Y, SHAO J, et al. Structure-driven protein engineering for production of valuable natural products[J]. Trends in Plant Science, 2023, 28(4): 460-470. |
| 136 | WANG Y J, XUE P, CAO M F, et al. Directed evolution: methodologies and applications[J]. Chemical Reviews, 2021, 121(20): 12384-12444. |
| 137 | LOVELOCK S L, CRAWSHAW R, BASLER S, et al. The road to fully programmable protein catalysis[J]. Nature, 2022, 606(7912): 49-58. |
| 138 | DE ROND T, GAO J, ZARGAR A, et al. A high-throughput mass spectrometric enzyme activity assay enabling the discovery of cytochrome P450 biocatalysts[J]. Angewandte Chemie International Edition, 2019, 58(30): 10114-10119. |
| [1] | 高歌, 边旗, 王宝俊. 合成基因线路的工程化设计研究进展与展望[J]. 合成生物学, 2025, 6(1): 45-64. |
| [2] | 李冀渊, 吴国盛. 合成生物学视域下有机体的两种隐喻[J]. 合成生物学, 2025, 6(1): 190-202. |
| [3] | 焦洪涛, 齐蒙, 邵滨, 蒋劲松. DNA数据存储技术的法律治理议题[J]. 合成生物学, 2025, 6(1): 177-189. |
| [4] | 唐兴华, 陆钱能, 胡翌霖. 人类世中对合成生物学的哲学反思[J]. 合成生物学, 2025, 6(1): 203-212. |
| [5] | 徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176. |
| [6] | 石婷, 宋展, 宋世怡, 张以恒. 体外生物转化(ivBT):生物制造的新前沿[J]. 合成生物学, 2024, 5(6): 1437-1460. |
| [7] | 柴猛, 王风清, 魏东芝. 综合利用木质纤维素生物转化合成有机酸[J]. 合成生物学, 2024, 5(6): 1242-1263. |
| [8] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [9] | 陈雨, 张康, 邱以婧, 程彩云, 殷晶晶, 宋天顺, 谢婧婧. 微生物电合成技术转化二氧化碳研究进展[J]. 合成生物学, 2024, 5(5): 1142-1168. |
| [10] | 郑皓天, 李朝风, 刘良叙, 王嘉伟, 李恒润, 倪俊. 负碳人工光合群落的设计、优化与应用[J]. 合成生物学, 2024, 5(5): 1189-1210. |
| [11] | 夏孔晨, 徐维华, 吴起. 光酶催化混乱性反应的研究进展[J]. 合成生物学, 2024, 5(5): 997-1020. |
| [12] | 陈子苓, 向阳飞. 类器官技术与合成生物学协同研究进展[J]. 合成生物学, 2024, 5(4): 795-812. |
| [13] | 蔡冰玉, 谭象天, 李伟. 合成生物学在干细胞工程化改造中的研究进展[J]. 合成生物学, 2024, 5(4): 782-794. |
| [14] | 谢皇, 郑义蕾, 苏依婷, 阮静怡, 李永泉. 放线菌聚酮类化合物生物合成体系重构研究进展[J]. 合成生物学, 2024, 5(3): 612-630. |
| [15] | 惠真, 唐啸宇. CRISPR/Cas9编辑系统在微生物天然产物研究中的应用[J]. 合成生物学, 2024, 5(3): 658-671. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||