Synthetic Biology Journal ›› 2021, Vol. 2 ›› Issue (6): 1046-1060.DOI: 10.12211/2096-8280.2021-098
De novo biosynthesis of 3-phenylpropanol in E. coli
GAO Hutao, WANG Jia, SUN Xinxiao, SHEN Xiaolin, YUAN Qipeng
- State Key Laboratory of Effective Utilization of Chemical Resources,Beijing University of Chemical Technology,Beijing 100029,China
-
Received:2021-10-21Revised:2021-11-29Online:2022-01-21Published:2021-12-31 -
Contact:SHEN Xiaolin, YUAN Qipeng
在大肠杆菌中从头生物合成3-苯丙醇
高虎涛, 王佳, 孙新晓, 申晓林, 袁其朋
- 北京化工大学,化工资源有效利用国家重点实验室,北京 100029
-
通讯作者:申晓林,袁其朋 -
作者简介:高虎涛 (1997—),男,硕士研究生。研究方向为芳香氨基酸衍生物的合成生物学和代谢工程。E-mail:2019201106@buct.edu.cn申晓林 (1984—),女,博士,副教授。研究方向为代谢工程及微生物合成生物学。E-mail:shenxl@mail.buct.edu.cn袁其朋 (1969—),男,博士,教授。研究方向为代谢工程及微生物合成生物学。E-mail:yuanqp@mail.buct.edu.cn -
基金资助:国家重点研发计划(2018YFA0901800)
CLC Number:
Cite this article
GAO Hutao, WANG Jia, SUN Xinxiao, SHEN Xiaolin, YUAN Qipeng. De novo biosynthesis of 3-phenylpropanol in E. coli[J]. Synthetic Biology Journal, 2021, 2(6): 1046-1060.
高虎涛, 王佳, 孙新晓, 申晓林, 袁其朋. 在大肠杆菌中从头生物合成3-苯丙醇[J]. 合成生物学, 2021, 2(6): 1046-1060.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2021-098
| Strains | Genotype | Source |
|---|---|---|
| Trans 5α | Lab Stock | |
| BW25113(F′) | Lab Stock | |
| BW25113(F′) ∆pykA ∆pykF | This study | |
| G01 | BW25113(F′) harboring pZE-CCR-4CL1 | This study |
| G02 | BW25113(F′) harboring pCS-ER | This study |
| G03 | BW25113(F′) harboring pZE-CCR-4CL1 and pCS-ER | This study |
| G04 | BW25113(F′) harboring pZE-ER | This study |
| G05 | BW25113(F′) harboring pCS-Carsfp | This study |
| G06 | BW25113(F′) harboring pZE-ER and pCS-Carsfp | This study |
| G07 | BW25113(F′) harboring pZE-RgTAL-ER and pCS-Carsfp | This study |
| G08 | BW25113(F′) harboring pZE-tac-RgTAL-ER-Carsfp | This study |
| G09 | BW25113(F′)∆pykA∆pykF harboring pZE-tac-RgTAL-ER-Carsfp | This study |
| G10 | BW25113(F′) harboring pCS-lac-APTA and pZE-tac-RgTAL-ER-Carsfp | This study |
| G11 | BW25113(F′) harboring pCS-tac-APTA and pZE-tac-RgTAL-ER-Carsfp | This study |
| G12 | BW25113(F′)∆pykA∆pykF harboring pCS-tac-APTA and pZE-tac-RgTAL-ER-Carsfp | This study |
Tab. 1 Strains used in this study
| Strains | Genotype | Source |
|---|---|---|
| Trans 5α | Lab Stock | |
| BW25113(F′) | Lab Stock | |
| BW25113(F′) ∆pykA ∆pykF | This study | |
| G01 | BW25113(F′) harboring pZE-CCR-4CL1 | This study |
| G02 | BW25113(F′) harboring pCS-ER | This study |
| G03 | BW25113(F′) harboring pZE-CCR-4CL1 and pCS-ER | This study |
| G04 | BW25113(F′) harboring pZE-ER | This study |
| G05 | BW25113(F′) harboring pCS-Carsfp | This study |
| G06 | BW25113(F′) harboring pZE-ER and pCS-Carsfp | This study |
| G07 | BW25113(F′) harboring pZE-RgTAL-ER and pCS-Carsfp | This study |
| G08 | BW25113(F′) harboring pZE-tac-RgTAL-ER-Carsfp | This study |
| G09 | BW25113(F′)∆pykA∆pykF harboring pZE-tac-RgTAL-ER-Carsfp | This study |
| G10 | BW25113(F′) harboring pCS-lac-APTA and pZE-tac-RgTAL-ER-Carsfp | This study |
| G11 | BW25113(F′) harboring pCS-tac-APTA and pZE-tac-RgTAL-ER-Carsfp | This study |
| G12 | BW25113(F′)∆pykA∆pykF harboring pCS-tac-APTA and pZE-tac-RgTAL-ER-Carsfp | This study |
| Plasmids | Description | Source |
|---|---|---|
| pZE12-luc | pLlacO-1; luc; ColE1 ori; Ampr | Lab Stock |
| pCS27 | pLlacO-1, P15A ori, Kanr | Lab Stock |
| pZE-CCR-4CL1 | pZE-luc carrying CCR from Leucaena leucocephala, and 4CL1 from Arabidopsis thaliana | This study |
| pZE-ER | pZE-luc carrying ER from Clostridium acetobutylicum | This study |
| pCS-ER | pCS27 carrying ER from C. acetobutylicum | This study |
| pCS-carsfp | pCS27 carrying Car from Mycobacterium marinum and Sfp from Bacillus subtilis | This study |
| pZE-RgTAL-ER | pZE-luc carrying TAL from Rhodobacter glutinis and ER from C.acetobutylicum | This study |
Tab. 2 Plasmids used in this study
| Plasmids | Description | Source |
|---|---|---|
| pZE12-luc | pLlacO-1; luc; ColE1 ori; Ampr | Lab Stock |
| pCS27 | pLlacO-1, P15A ori, Kanr | Lab Stock |
| pZE-CCR-4CL1 | pZE-luc carrying CCR from Leucaena leucocephala, and 4CL1 from Arabidopsis thaliana | This study |
| pZE-ER | pZE-luc carrying ER from Clostridium acetobutylicum | This study |
| pCS-ER | pCS27 carrying ER from C. acetobutylicum | This study |
| pCS-carsfp | pCS27 carrying Car from Mycobacterium marinum and Sfp from Bacillus subtilis | This study |
| pZE-RgTAL-ER | pZE-luc carrying TAL from Rhodobacter glutinis and ER from C.acetobutylicum | This study |
| Primer | Sequence 5′-3′ |
|---|---|
| CCR-4CL1-1-F-KpnI | gggaaaGGTACCatgcctgctgcggctccagc |
| CCR-4CL1-1-R-BamHI | gggaaaGGATCCttatttggtcggcagcggcaggtg |
| CCR-4CL1-2-F-BamHI | gggaaaGGATCCaggagatataccatggcgccacaagaacaagcagt |
| CCR-4CL1-2-R-XbaI | gggaaaTCTAGAttacaatccatttgctagttttgccctc |
| ER-KpnI-F | gggaaaGGTACCatgaacaaatacaagaaattatttgaaccaatcaaaattgg |
| ER-XbaI-F | gggaaaTCTAGAttatatatggtttgcaacttcaaaagcatccc |
| ER框-SpeI-F | gggaaaACTAGTaattgtgagcggataacaattgacattgtga |
| ER框-SacI-R | gggaaaGAGCTCacaacagataaaacgaaaggcccagtc |
| TAL框-SpeI-F | gggaaaACTAGTctcgagaattgtgagcggataacaattga |
| TAL框-SacI-R | gggaaaGAGCTCcgacaaacaacagataaaacgaaaggcc |
| Car-KpnI-F | gggaaaGGTACCatgtcacctatcacccgcgagg |
| Car-BamHI-R | gggaaaGGATCCtcacagcaagcccagcagac |
| sfp-BamHI-F | gggaaaGGATCCaggagatataccatgaagatttacggaatttatatgg |
| sfp-XbaI-R | GGGAAAtctagattataaaagctcttcgtacgagactattgtgat |
| AP-NheI-R | gggaaaGCTAGCttatttcttcagttcagccaggcttaacc |
| TA-NheI-F | gggaaaGCTAGCaggagatataccatgtcctcacgtaaagagcttg |
| APTA-XbaI-F | gggaaaTCTAGAatgacacaacctctttttctgatcggg |
| APTA-BamHI-R | gggaaaGGATCCttacccgcgacgcgctttta |
| pCS-tac-反-BamHI-F | gggaaaGGATCCgtcgccaatcacgcgtgaagagc |
| pCS-tac-反-XbaI-R | gggaaaCATATGttataaaagctcttcgtacgagacta |
| tac-Car框-F-SpeI | gggaaaACTAGTctcgagttgacaattaatcatcggctcg |
| tac-Car框-R-SacI | gggaaaGAGCTCcgacaaacaacagataaaacgaaaggcc |
Tab. 3 Primers used in this study
| Primer | Sequence 5′-3′ |
|---|---|
| CCR-4CL1-1-F-KpnI | gggaaaGGTACCatgcctgctgcggctccagc |
| CCR-4CL1-1-R-BamHI | gggaaaGGATCCttatttggtcggcagcggcaggtg |
| CCR-4CL1-2-F-BamHI | gggaaaGGATCCaggagatataccatggcgccacaagaacaagcagt |
| CCR-4CL1-2-R-XbaI | gggaaaTCTAGAttacaatccatttgctagttttgccctc |
| ER-KpnI-F | gggaaaGGTACCatgaacaaatacaagaaattatttgaaccaatcaaaattgg |
| ER-XbaI-F | gggaaaTCTAGAttatatatggtttgcaacttcaaaagcatccc |
| ER框-SpeI-F | gggaaaACTAGTaattgtgagcggataacaattgacattgtga |
| ER框-SacI-R | gggaaaGAGCTCacaacagataaaacgaaaggcccagtc |
| TAL框-SpeI-F | gggaaaACTAGTctcgagaattgtgagcggataacaattga |
| TAL框-SacI-R | gggaaaGAGCTCcgacaaacaacagataaaacgaaaggcc |
| Car-KpnI-F | gggaaaGGTACCatgtcacctatcacccgcgagg |
| Car-BamHI-R | gggaaaGGATCCtcacagcaagcccagcagac |
| sfp-BamHI-F | gggaaaGGATCCaggagatataccatgaagatttacggaatttatatgg |
| sfp-XbaI-R | GGGAAAtctagattataaaagctcttcgtacgagactattgtgat |
| AP-NheI-R | gggaaaGCTAGCttatttcttcagttcagccaggcttaacc |
| TA-NheI-F | gggaaaGCTAGCaggagatataccatgtcctcacgtaaagagcttg |
| APTA-XbaI-F | gggaaaTCTAGAatgacacaacctctttttctgatcggg |
| APTA-BamHI-R | gggaaaGGATCCttacccgcgacgcgctttta |
| pCS-tac-反-BamHI-F | gggaaaGGATCCgtcgccaatcacgcgtgaagagc |
| pCS-tac-反-XbaI-R | gggaaaCATATGttataaaagctcttcgtacgagacta |
| tac-Car框-F-SpeI | gggaaaACTAGTctcgagttgacaattaatcatcggctcg |
| tac-Car框-R-SacI | gggaaaGAGCTCcgacaaacaacagataaaacgaaaggcc |
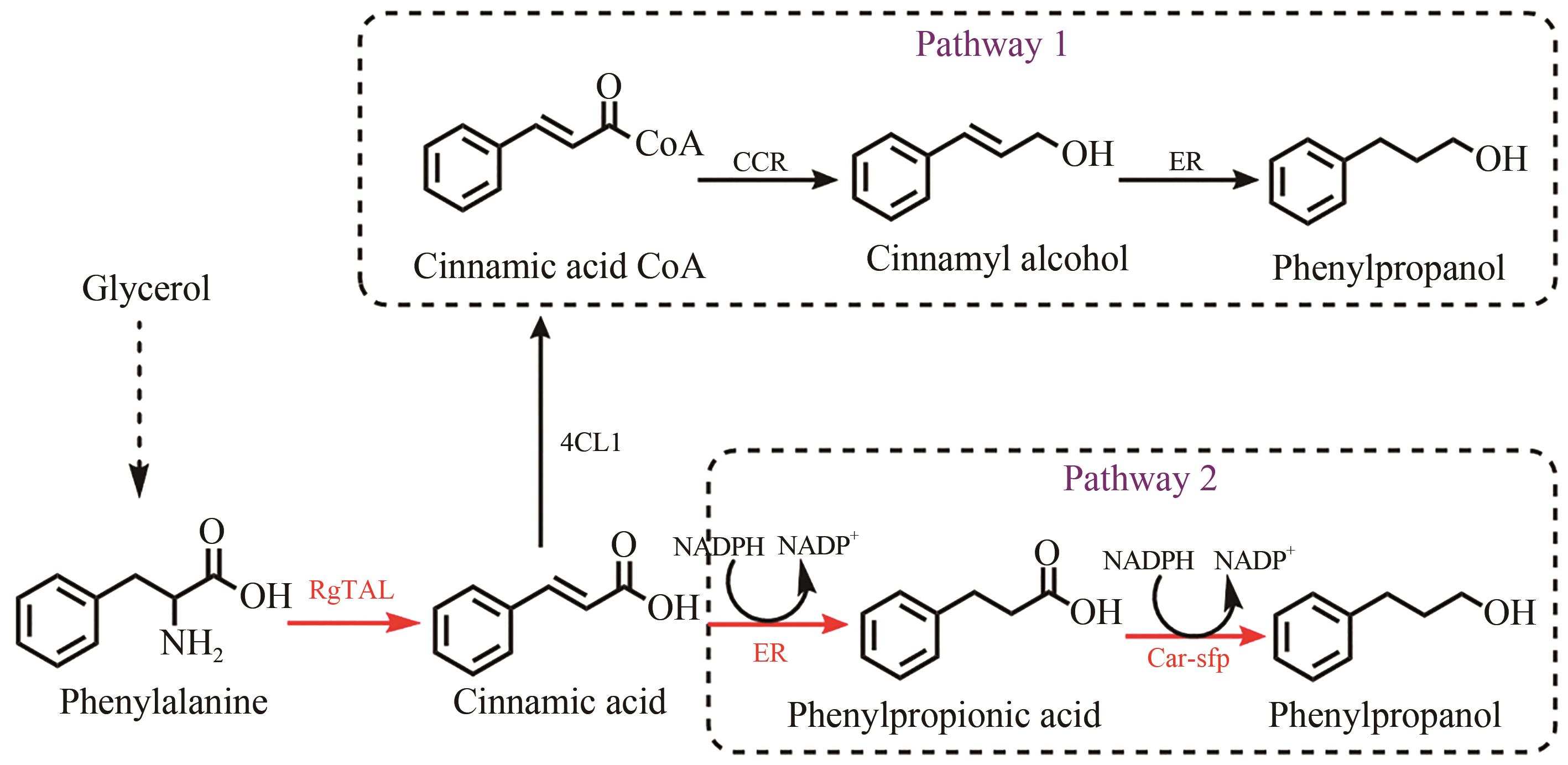
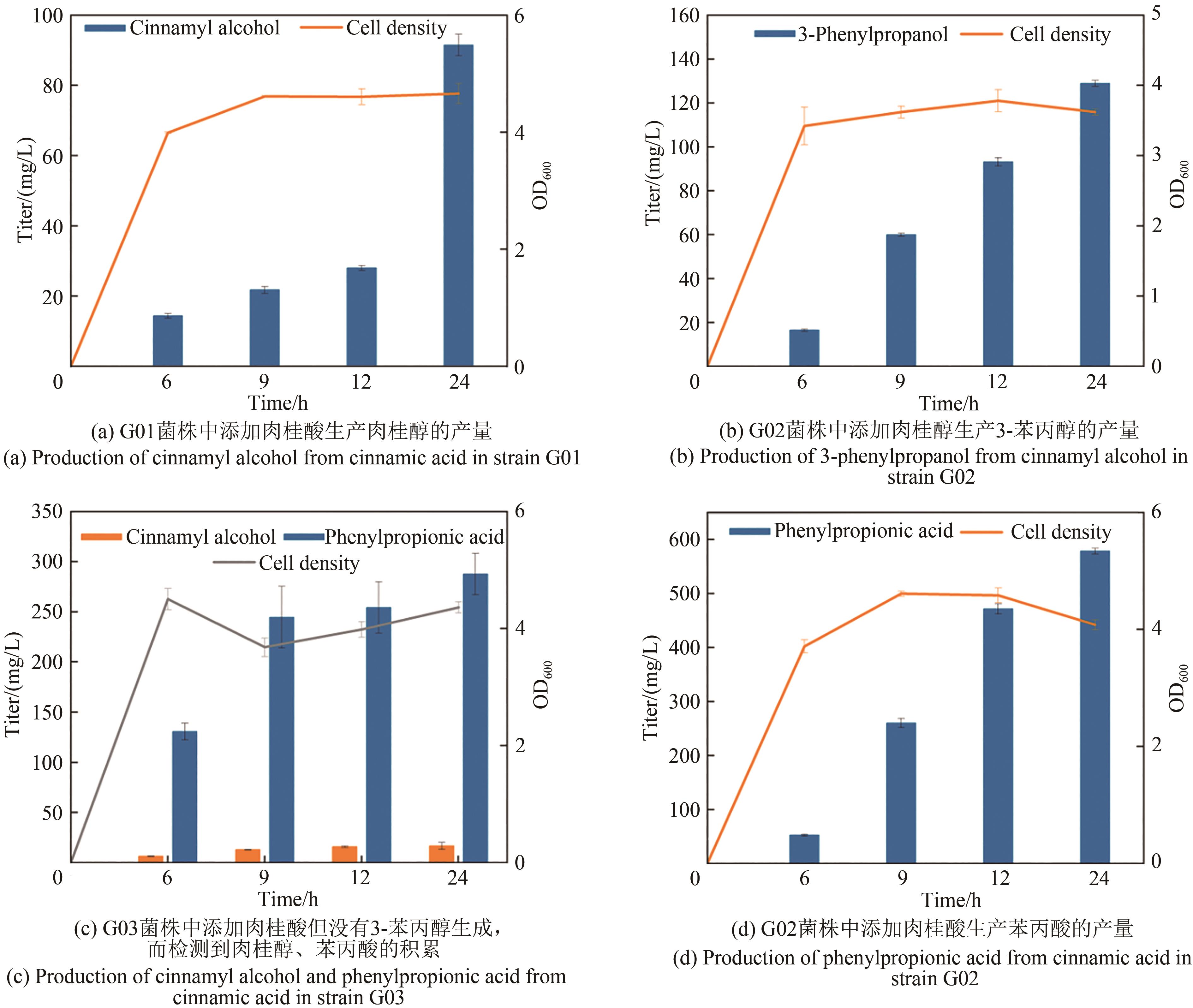
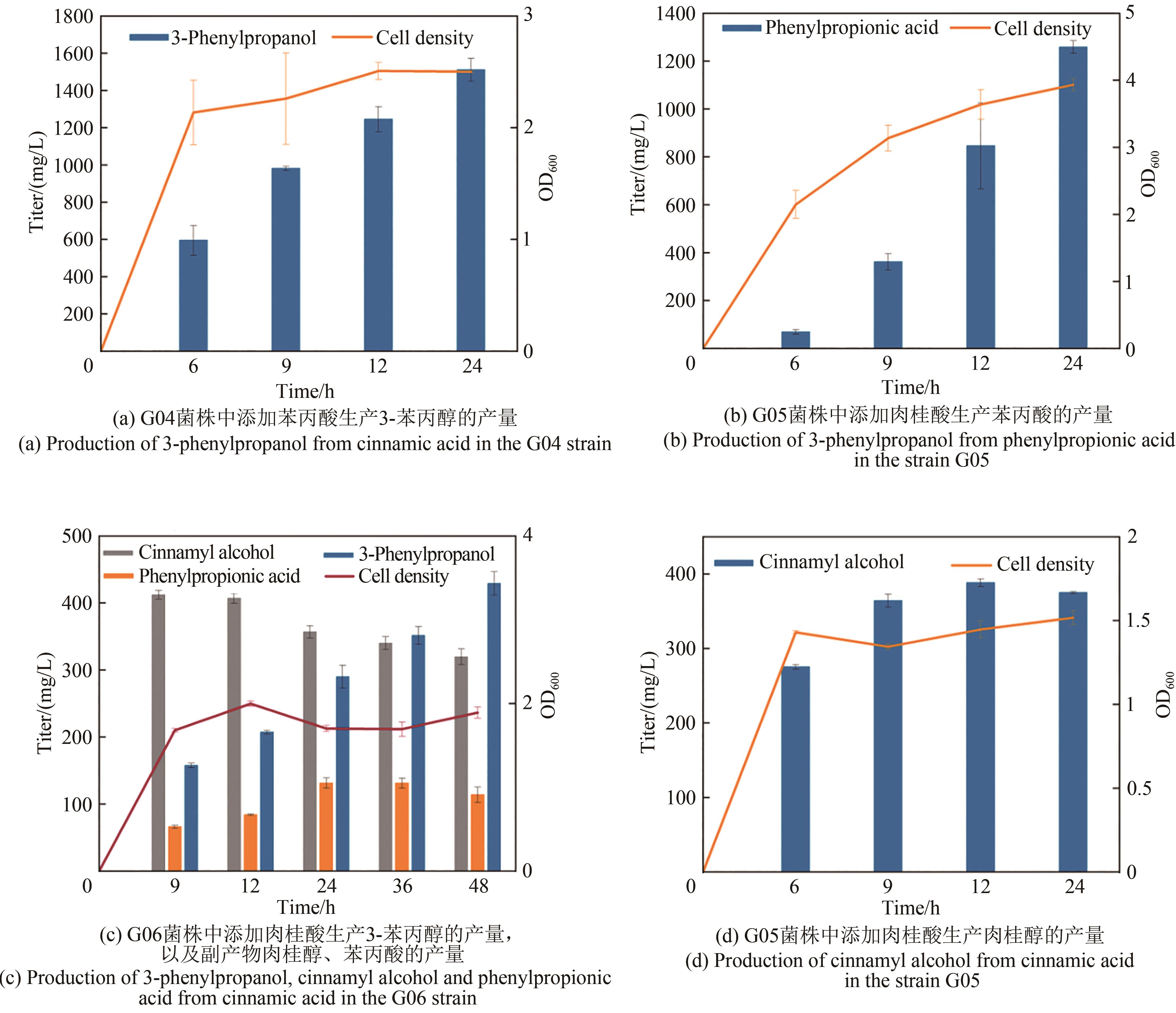
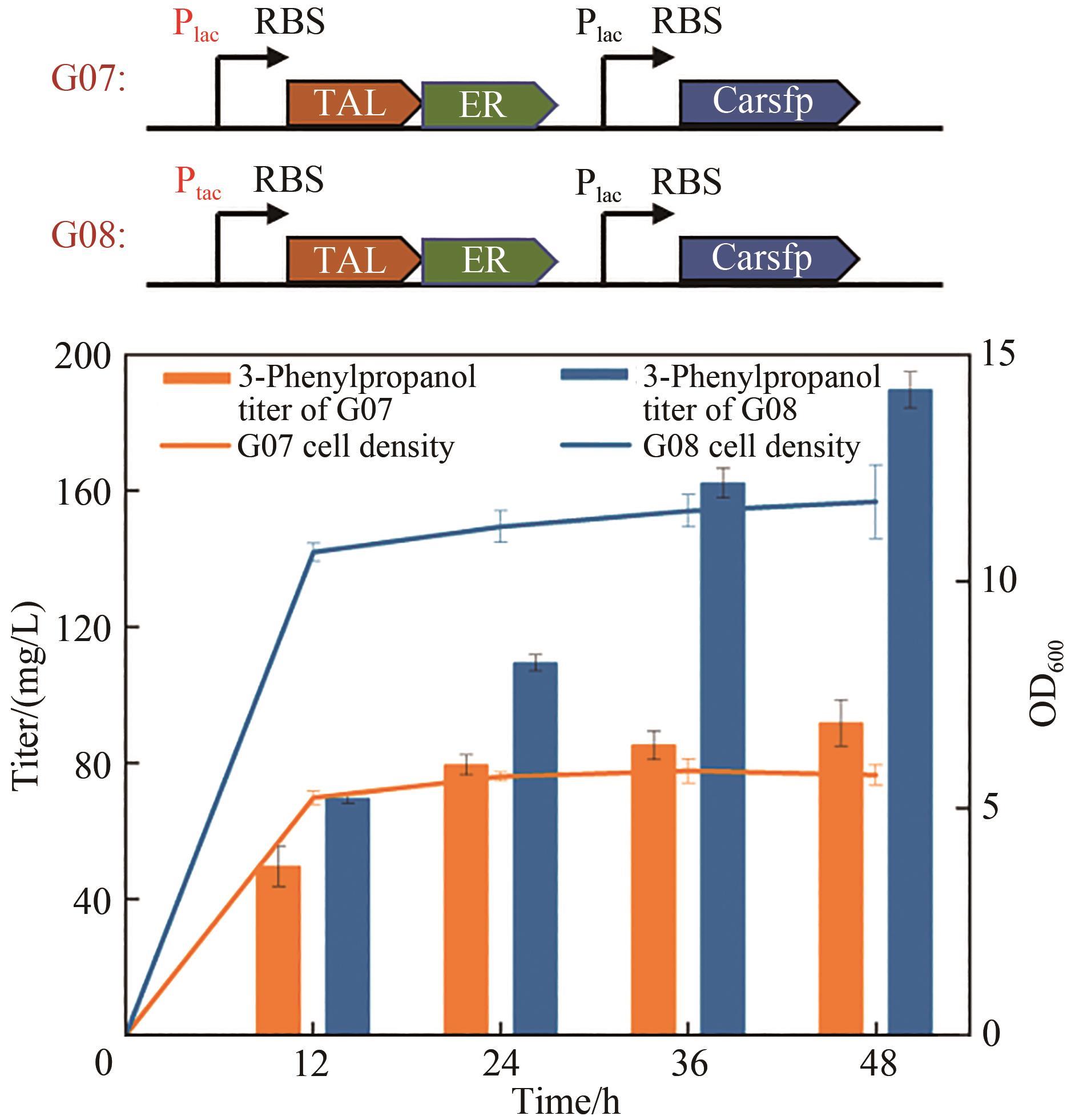
Fig. 1 Design of synthetic routes of 3-phenylpropanolRgTAL—phenylalanine ammonia lyase from Rhodobacter glutinis; 4CL1—from Arabidopsis thaliana coumarate-CoA ligase; CCR— cinnamoyl-CoA reductase from Leucaena leucocephala; ER—enoic acid reduction from Clostridium acetobutylicum Enzyme; Car—carboxylic acid reductase from Marine Mycobacterium; sfp—phosphoubiquitin transferase from Bacillus subtilis
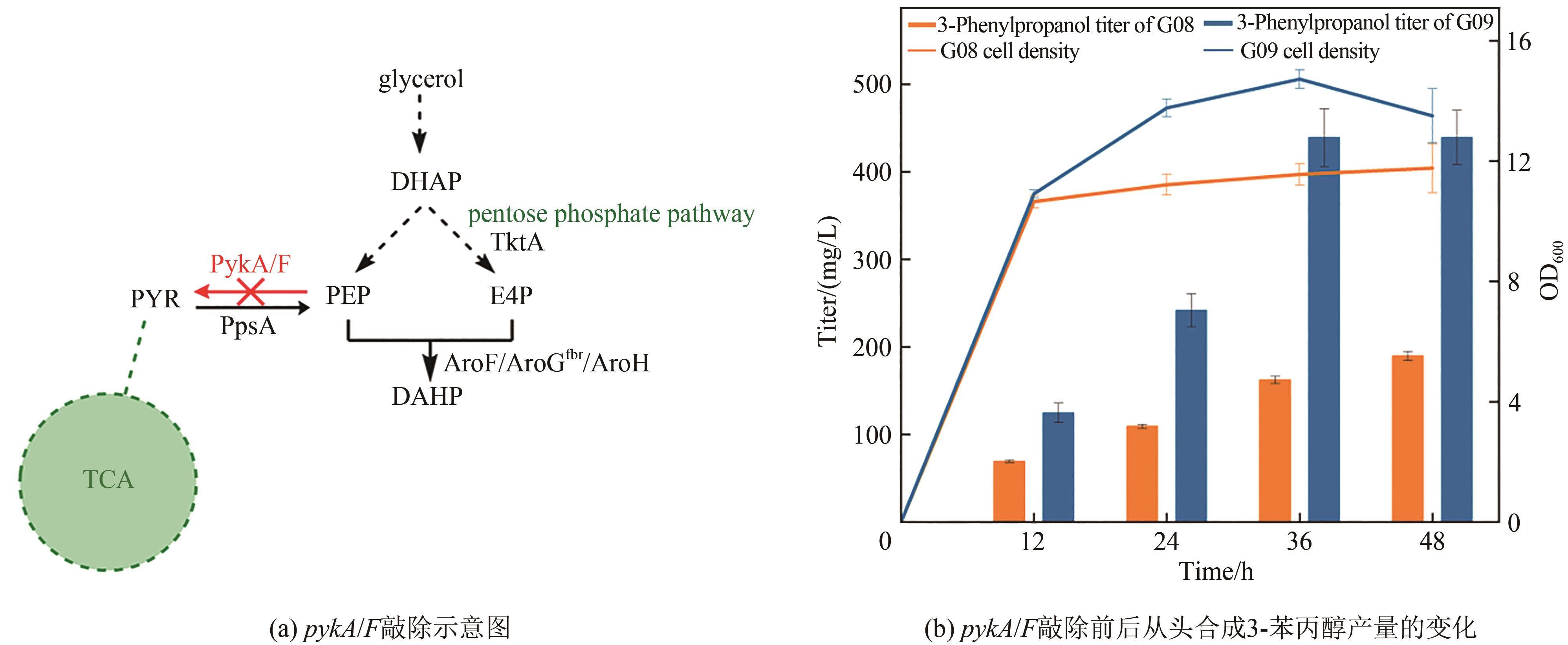
Fig. 5 Production of 3-phenylpropanol in pykA/F knockout strainDHAP—dihydroxyacetone phosphate; PYR—pyruvate; PEP—phosphoenolpyruvate; E4P—D-erythrose-4-phosphate; DAHP—3-deoxy-D-arabinoheptanoate heptaphosphate; TktA—Transketolase; PykA/F—pyruvate kinase; PpsA—phosphoenolpyruvate synthase; AroF/AroGfbr/AroH—3-deoxy-D-arabinoheptanoate heptaphosphate synthase
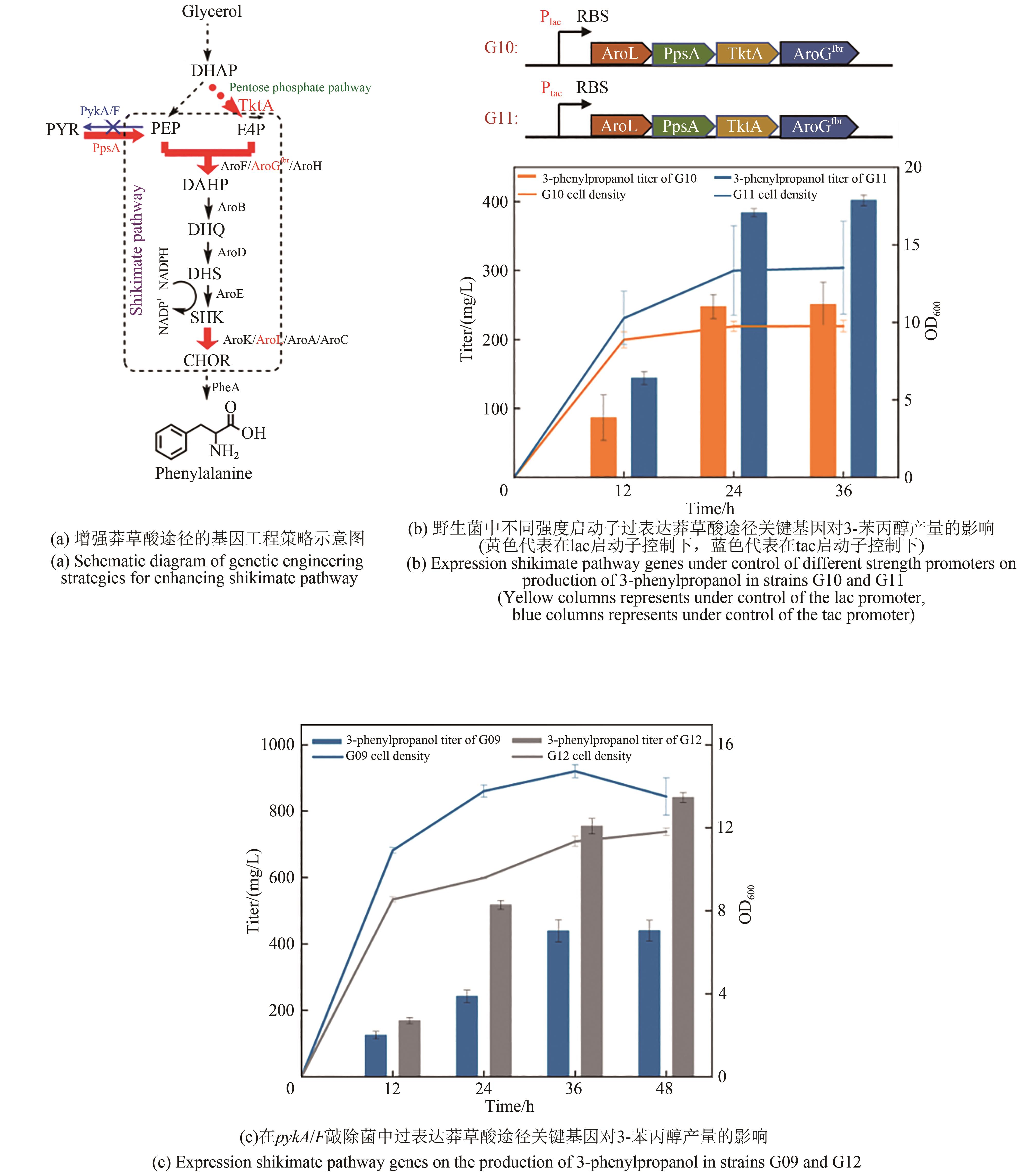
Fig. 6 Strengthening of shikimate pathway on production of 3-phenylpropanolDHQ—3-dehydroquinic acid; DHS—3-dehydroshikimate; SHK—shikimic acid; CHOR—chorismate; AroB—dehydroquinic acid synthase; AroD—dehydroquinic acid dehydratase, AroE—Shikimate dehydrogenase; AroK/AroL/AroA/AroC—dehydroshikimate dehydratase; PheA—prephenate dehydrogenase
| 1 | BHATIA S P, WELLINGTON G A, COCCHIARA J, et al. Fragrance material review on 3-phenyl-1-propanol[J]. Food and Chemical Toxicology, 2011, 49: S246-S51. |
| 2 | SÁ A G A, MENESES A C D, ARAÚJO P H H D, et al. A review on enzymatic synthesis of aromatic esters used as flavor ingredients for food, cosmetics and pharmaceuticals industries[J]. Trends in Food Science & Technology, 2017, 69: 95-105. |
| 3 | LÜ J, YU C Q, GUO Y, et al. Gallstone disease and the risk of type 2 diabetes[J]. Scientific Reports, 2017, 7(1): 15853. |
| 4 | PANTEN J, SURBURG H. Flavors and fragrances (Ⅲ): Aromatic and heterocyclic compounds[J]. Ullmann's Encyclopedia of Industrial Chemistry, 2016: 1-45. |
| 5 | ZHOU Y Y, LI Z H, LIU Y B, et al. Regulating hydrogenation chemoselectivity of α, β-unsaturated aldehydes by combination of transfer and catalytic hydrogenation[J]. ChemSusChem, 2020, 13(7): 1746-1750. |
| 6 | MAO X Y, WANG Y. Cooperative carbon emission reduction through the Belt and Road Initiative[J]. Environmental Science and Pollution Research, 2021: 1-22. |
| 7 | YUAN X L, SHENG X R, CHEN L P, et al. Carbon footprint and embodied carbon transfer at the provincial level of the Yellow River Basin[J]. The Science of the Total Environment, 2022, 803: 149993. |
| 8 | KHALIL A S, COLLINS J J. Synthetic biology: applications come of age[J]. Nature Reviews Genetics, 2010, 11(5): 367-379. |
| 9 | RAN F A, HSU P D, WRIGHT J, et al. Genome engineering using the CRISPR-Cas9 system[J]. Nature Protocols, 2013, 8(11): 2281-308. |
| 10 | FRANGOUL H, ALTSHULER D, CAPPELLINI M D, et al. CRISPR-Cas9 gene editing for sickle cell disease and β-thalassemia[J]. The New England Journal of Medicine, 2021, 384(3): 252-260. |
| 11 | BIAN X Y, HUANG F, STEWART F A, et al. Direct cloning, genetic engineering, and heterologous expression of the syringolin biosynthetic gene cluster in E. coli through Red/ET recombineering[J]. ChemBioChem, 2012, 13(13): 1946-1952. |
| 12 | CORREA A, OPPEZZO P. Tuning different expression parameters to achieve soluble recombinant proteins in E. coli: advantages of high-throughput screening[J]. Biotechnology Journal, 2011, 6(6): 715-730. |
| 13 | WANG Y, LI Q G, ZHENG P, et al. Evolving the L-lysine high-producing strain of Escherichia coli using a newly developed high-throughput screening method[J]. Journal of Industrial Microbiology & Biotechnology, 2016, 43(9): 1227-1235. |
| 14 | LEE S Y, KIM H U, PARK J H, et al. Metabolic engineering of microorganisms: general strategies and drug production[J]. Drug Discovery Today, 2009, 14(1/2): 78-88. |
| 15 | SHEN X L, CHEN X, WANG J, et al. Design and construction of an artificial pathway for biosynthesis of acetaminophen in Escherichia coli [J]. Metabolic Engineering, 2021, 68: 26-33. |
| 16 | FENG J C, YANG C, ZHAO Z H, et al. Application of cell-free protein synthesis system for the biosynthesis of L-theanine[J]. ACS Synthetic Biology, 2021, 10(3): 620-631. |
| 17 | BOO Y C. p-Coumaric acid as an active ingredient in cosmetics: a review focusing on its antimelanogenic effects[J]. Antioxidants, 2019, 8(8): 275. |
| 18 | SHANG Y Z, WEI W P, ZHANG P, et al. Engineering Yarrowia lipolytica for enhanced production of arbutin[J]. Journal of Agricultural and Food Chemistry, 2020, 68(5): 1364-1372. |
| 19 | YIM H, HASELBECK R, NIU W, et al. Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol[J]. Nature Chemical Biology, 2011, 7(7): 445-452. |
| 20 | ATSUMI S, LIAO J C. Metabolic engineering for advanced biofuels production from Escherichia coli [J]. Current Opinion in Biotechnology, 2008, 19(5): 414-419. |
| 21 | DZIGA D, LISZNIANSKA M, WLADYKA B. Bioreactor study employing bacteria with enhanced activity toward cyanobacterial toxins microcystins[J]. Toxins, 2014, 6(8): 2379-2392. |
| 22 | CHEN Z Y, SUN X X, LI Y, et al. Metabolic engineering of Escherichia coli for microbial synthesis of monolignols[J]. Metabolic Engineering, 2017, 39: 102-109. |
| 23 | SUN J, LIN Y H, SHEN X L, et al. Aerobic biosynthesis of hydrocinnamic acids in Escherichia coli with a strictly oxygen-sensitive enoate reductase[J]. Metabolic Engineering, 2016, 35: 75-82. |
| 24 | AKHTAR M K, TURNER N J, JONES P R. Carboxylic acid reductase is a versatile enzyme for the conversion of fatty acids into fuels and chemical commodities[J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(1): 87-92. |
| 25 | WANG J, LI C Y, ZOU Y S, et al. Bacterial synthesis of C3-C5 diols via extending amino acid catabolism[J]. Proceedings of the National Academy of Sciences of the United States of America, 2020, 117(32): 19159-19167. |
| 26 | VENKITASUBRAMANIAN P, DANIELS L, ROSAZZA J P. Reduction of carboxylic acids by Nocardia aldehyde oxidoreductase requires a phosphopantetheinylated enzyme[J]. JBC, 2007, 282(1): 478-485. |
| 27 | HALL M, STUECKLER C, HAUER B, et al. Asymmetric bioreduction of activated C=C bonds using Zymomonas mobilis NCR enoate reductase and old yellow enzymes OYE 1-3 from yeasts[J]. European Journal of Organic Chemistry, 2008, 2008(9): 1511-1516. |
| 28 | WANG S Y, ZHANG S W, XIAO A F, et al. Metabolic engineering of Escherichia coli for the biosynthesis of various phenylpropanoid derivatives[J]. Metabolic Engineering, 2015, 29: 153-159. |
| 29 | MAEDA H, DUDAREVA N. The shikimate pathway and aromatic amino acid biosynthesis in plants[J]. Annual Review of Plant Biology, 2012, 63: 73-105. |
| 30 | ROBERTS C W, ROBERTS F, LYONS R E, et al. The shikimate pathway and its branches in apicomplexan parasites[J]. The Journal of Infectious Diseases, 2002, 185(S1): S25-S36. |
| 31 | XU J Z, YU H B, HAN M, et al. Metabolic engineering of glucose uptake systems in Corynebacterium glutamicum for improving the efficiency of L-lysine production[J]. Journal of Industrial Microbiology & Biotechnology, 2019, 46(7): 937-949. |
| 32 | LINDNER S N, KNEBEL S, PALLERLA S R, et al. Cg2091 encodes a polyphosphate/ATP-dependent glucokinase of Corynebacterium glutamicum [J]. Applied Microbiology and Biotechnology, 2010, 87(2): 703-713. |
| 33 | LINDNER S N, SEIBOLD G M, HENRICH A, et al. Phosphotransferase system-independent glucose utilization in Corynebacterium glutamicum by inositol permeases and glucokinases[J]. Applied and Environmental Microbiology, 2011, 77(11): 3571-3581. |
| 34 | VOGT M, HAAS S, KLAFFL S, et al. Pushing product formation to its limit: metabolic engineering of Corynebacterium glutamicum for L-leucine overproduction[J]. Metabolic Engineering, 2014, 22: 40-52. |
| 35 | KHAMDUANG M, PACKDIBAMRUNG K, CHUTMANOP J, et al. Production of l-phenylalanine from glycerol by a recombinant Escherichia coli [J]. Journal of Industrial Microbiology & Biotechnology, 2009, 36(10): 1267-1274. |
| 36 | AHN J O, LEE H W, SAHA R, et al. Exploring the effects of carbon sources on the metabolic capacity for shikimic acid production in Escherichia coli using in silico metabolic predictions[J]. Journal of Microbiology and Biotechnology, 2008, 18(11): 1773-1784. |
| 37 | LEE S J, LEE D Y, KIM T Y, et al. Metabolic engineering of Escherichia coli for enhanced production of succinic acid, based on genome comparison and in silico gene knockout simulation[J]. Applied and Environmental Microbiology, 2005, 71(12): 7880-7887. |
| 38 | CHAVADI S, WOOFF E, COLDHAM N G, et al. Global effects of inactivation of the pyruvate kinase gene in the Mycobacterium tuberculosis complex[J]. Journal of Bacteriology, 2009, 191(24): 7545-7553. |
| 39 | POTTS A H, VAKULSKAS C A, PANNURI A, et al. Global role of the bacterial post-transcriptional regulator CsrA revealed by integrated transcriptomics[J]. Nature Communications, 2017, 8(1): 1596. |
| 40 | TATARKO M, ROMEO T. Disruption of a global regulatory gene to enhance central carbon flux into phenylalanine biosynthesis in Escherichia coli [J]. Current Microbiology, 2001, 43(1): 26-32. |
| 41 | SUZUKI K, BABITZKE P, KUSHNER S R, et al. Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E[J]. Genes & Development, 2006, 20(18): 2605-2617. |
| 42 | YAKANDAWALA N, ROMEO T, FRIESEN A D, et al. Metabolic engineering of Escherichia coli to enhance phenylalanine production[J]. Applied Microbiology and Biotechnology, 2008, 78(2): 283-291. |
| 43 | JIANG M, ZHANG H R. Engineering the shikimate pathway for biosynthesis of molecules with pharmaceutical activities in E. coli [J]. Current Opinion in Biotechnology, 2016, 42: 1-6. |
| 44 | LI Z, DING D Q, WANG H Y, et al. Engineering Escherichia coli to improve tryptophan production via genetic manipulation of precursor and cofactor pathways[J]. Synthetic and Systems Biotechnology, 2020, 5(3): 200-205. |
| 45 | KNOP D R, DRATHS K M, CHANDRAN S S, et al. Hydroaromatic equilibration during biosynthesis of shikimic acid[J]. Journal of the American Chemical Society, 2001, 123(42): 10173-10182. |
| 46 | LU J-L, LIAO J C. Metabolic engineering and control analysis for production of aromatics: role of transaldolase[J]. Biotechnology and Bioengineering, 1997, 53(2): 132-138. |
| 47 | CHANDRAN S S, YI J, DRATHS K M, et al. Phosphoenolpyruvate availability and the biosynthesis of shikimic acid[J]. Biotechnology Progress, 2003, 19(3): 808-814. |
| 48 | ZHOU H Y, LIAO X Y, WANG T W, et al. Enhanced L-phenylalanine biosynthesis by co-expression of pheAfbr and aroFwt [J]. Bioresource Technology, 2010, 101(11): 4151-4156. |
| 49 | MCCANDLISS R J, POLING M D, HERRMANN K M. 3-Deoxy-D-arabino-heptulosonate 7-phosphate synthase. Purification and molecular characterization of the phenylalanine-sensitive isoenzyme from Escherichia coli [J]. JBC, 1978, 253(12): 4259-4265. |
| 50 | ELY B, PITTARD J. Aromatic amino acid biosynthesis: regulation of shikimate kinase in Escherichia coli K-12[J]. Journal of Bacteriology, 1979, 138(3): 933-43. |
| 51 | DEFEYTER R C, PITTARD J. Purification and properties of shikimate kinase Ⅱ from Escherichia coli K-12[J]. Journal of Bacteriology, 1986, 165(1): 331-333. |
| 52 | KIM B, BINKLEY R, KIM H U, et al. Metabolic engineering of Escherichia coli for the enhanced production of L-tyrosine[J]. Biotechnology and Bioengineering, 2018, 115(10): 2554-2564. |
| 53 | PARSONS C V, HARRIS D M M, PATTEN C L. Regulation of indole-3-acetic acid biosynthesis by branched-chain amino acids in Enterobacter cloacae UW5 [J]. FEMS Microbiology Letters, 2015, 362(18): fnv153. |
| 54 | GOTTARDI M, KNUDSEN J D, PRADO L, et al. De novo biosynthesis of trans-cinnamic acid derivatives in Saccharomyces cerevisiae [J]. Applied Microbiology and Biotechnology, 2017, 101(12): 4883-4893. |
| 55 | LIU Z N, ZHANG X, LEI D W, et al. Metabolic engineering of Escherichia coli for de novo production of 3-phenylpropanol via retrobiosynthesis approach[J]. Microbial Cell Factories, 2021, 20(1): 121. |
| 56 | KAUP B, BRINGER-MEYER S, SAHM H. Metabolic engineering of Escherichia coli: construction of an efficient biocatalyst for D-mannitol formation in a whole-cell biotransformation[J]. Applied Microbiology and Biotechnology, 2004, 64(3): 333-339. |
| 57 | WANG J, YANG Y P, ZHANG R H, et al. Microbial production of branched-chain dicarboxylate 2-methylsuccinic acid via enoate reductase-mediated bioreduction[J]. Metabolic Engineering, 2018, 45: 1-10. |
| [1] | GUO Shuyuan, ZHANG Qiannan, Gulikezi· MAIMAITIREXIATI, YANG Yiqun, YU Tao. Advances in microbial production of liquid biofuels [J]. Synthetic Biology Journal, 2025, 6(1): 18-44. |
| [2] | SHAO Mingwei, SUN Simian, YANG Shimao, CHEN Guoqiang. Bioproduction based on extremophiles [J]. Synthetic Biology Journal, 2024, 5(6): 1419-1436. |
| [3] | ZHAO Liang, LI Zhenshuai, FU Liping, LYU Ming, WANG Shi’an, ZHANG Quan, LIU Licheng, LI Fuli, LIU Ziyong. Progress in biomanufacturing of lipids and single cell protein from one-carbon compounds [J]. Synthetic Biology Journal, 2024, 5(6): 1300-1318. |
| [4] | ZHU Fanghuan, CEN Xuecong, CHEN Zhen. Research progress of diols production by microbes [J]. Synthetic Biology Journal, 2024, 5(6): 1367-1385. |
| [5] | YU Wei, GAO Jiaoqi, ZHOU Yongjin. Bioconversion of one carbon feedstocks for producing organic acids [J]. Synthetic Biology Journal, 2024, 5(5): 1169-1188. |
| [6] | CHEN Xiwei, ZHANG Huaran, ZOU Yi. Biosynthesis and metabolic engineering of fungal non-ribosomal peptides [J]. Synthetic Biology Journal, 2024, 5(3): 571-592. |
| [7] | XIE Huang, ZHENG Yilei, SU Yiting, RUAN Jingyi, LI Yongquan. An overview on reconstructing the biosynthetic system of actinomycetes for polyketides production [J]. Synthetic Biology Journal, 2024, 5(3): 612-630. |
| [8] | ZHAO Jingyu, ZHANG Jian, QI Qingsheng, WANG Qian. Research progress in biosensors based on bacterial two-component systems [J]. Synthetic Biology Journal, 2024, 5(1): 38-52. |
| [9] | SUN Huili, CUI Jinyu, LUAN Guodong, LYU Xuefeng. Progress of cyanobacterial synthetic biotechnology for efficient light-driven carbon fixation and ethanol production [J]. Synthetic Biology Journal, 2023, 4(6): 1161-1177. |
| [10] | YAN Xiongying, WANG Zhen, LOU Jiyun, ZHANG Haoyu, HUANG Xingyu, WANG Xia, YANG Shihui. Progress in the construction of microbial cell factories for efficient biofuel production [J]. Synthetic Biology Journal, 2023, 4(6): 1082-1121. |
| [11] | CHENG Zhenzhen, ZHANG Jian, GAO Cong, LIU Liming, CHEN Xiulai. Progress in metabolic engineering of microorganisms for the utilization of formate [J]. Synthetic Biology Journal, 2023, 4(4): 756-778. |
| [12] | LIU Jiayu, YANG Zhihan, YANG Lei, ZHU Liying, ZHU Zhengming, JIANG Ling. Advances in the development of Clostridium tyrobutyricum cell factories driven by synthetic biotechnology [J]. Synthetic Biology Journal, 2022, 3(6): 1174-1200. |
| [13] | GUO Shuyuan, WU Lianghuan, LIU Xiangjian, WANG Bo, YU Tao. Developing C1-based metabolic network in methylotrophy for biotransformation [J]. Synthetic Biology Journal, 2022, 3(1): 116-137. |
| [14] | CHEN Jiuzhou, WANG Yu, PU Wei, ZHENG Ping, SUN Jibin. Advances and perspective on bioproduction of 5-aminolevulinic acid [J]. Synthetic Biology Journal, 2021, 2(6): 1000-1016. |
| [15] | Qingzhuo WANG, Ping SONG, He HUANG. Synthetic biotechnology drives the development of natural eukaryotic lipid cell factories [J]. Synthetic Biology Journal, 2021, 2(6): 920-941. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||