Synthetic Biology Journal ›› 2023, Vol. 4 ›› Issue (4): 756-778.DOI: 10.12211/2096-8280.2023-032
• Invited Review • Previous Articles Next Articles
Progress in metabolic engineering of microorganisms for the utilization of formate
CHENG Zhenzhen1,2, ZHANG Jian1,2, GAO Cong1,2, LIU Liming1,2, CHEN Xiulai1,2
- 1.State Key Laboratory of Food Science and Technology,Jiangnan University,Wuxi 214122,Jiangsu,China
2.International Joint Laboratory on Food Safety,Jiangnan University,Wuxi 214122,Jiangsu,China
-
Received:2023-04-17Revised:2023-05-22Online:2023-09-14Published:2023-08-31 -
Contact:CHEN Xiulai
代谢工程改造微生物利用甲酸研究进展
程真真1,2, 张健1,2, 高聪1,2, 刘立明1,2, 陈修来1,2
- 1.江南大学,食品科学与技术国家重点实验室,江苏 无锡 214122
2.江南大学,食品安全国际合作联合实验室,江苏 无锡 214122
-
通讯作者:陈修来 -
作者简介:程真真 (2001—),女,学士。研究方向为微生物固碳途径的设计、构建与应用。E-mail:zzch0929@163.com陈修来 (1985—),男,博士,教授。研究方向为微生物代谢工程与合成生物学。E-mail:xlchen@jiangnan.edu.cn -
基金资助:国家重点研发计划(2021YFC2103500);国家自然科学基金(22122806);江苏省自然科学基金(BK20211529);江南大学基本科研计划面上培育项目(JUSRP22031)
CLC Number:
Cite this article
CHENG Zhenzhen, ZHANG Jian, GAO Cong, LIU Liming, CHEN Xiulai. Progress in metabolic engineering of microorganisms for the utilization of formate[J]. Synthetic Biology Journal, 2023, 4(4): 756-778.
程真真, 张健, 高聪, 刘立明, 陈修来. 代谢工程改造微生物利用甲酸研究进展[J]. 合成生物学, 2023, 4(4): 756-778.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2023-032
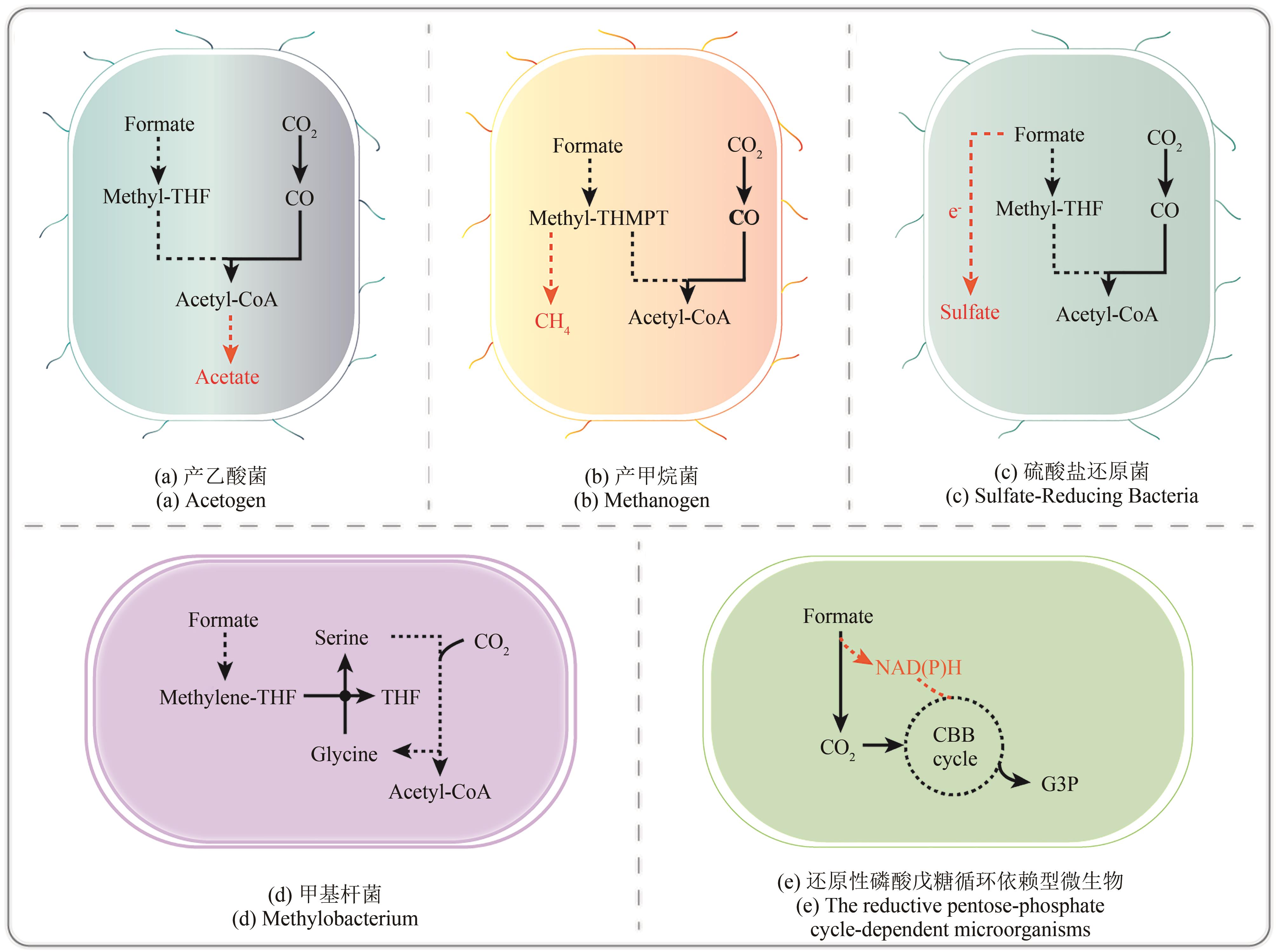
| 微生物种类 | 需氧类型 | 模式菌株 | 天然甲酸 代谢路径 | 应用优劣势 | 参考 文献 |
|---|---|---|---|---|---|
| 产乙酸菌 | 专性厌氧 | 伍氏醋酸杆菌 | 还原性乙酰辅酶A途径 | 优势:能够天然利用甲酸和CO2合成乙酸、乙醇等化合物 劣势:在厌氧条件下生长,不利于工业应用;代谢研究尚不完善,代谢改造工具少 | [ |
| 产甲烷菌 | 专性厌氧 | 氢营养型海沼甲烷球菌 | 还原性乙酰辅酶A途径 | 优势:能够天然利用甲酸和CO2合成甲烷,在清洁能源产业方面具有应用潜能 劣势:在厌氧条件下生长,不利于工业应用;代谢研究尚不完善,代谢改造工具少 | [ |
| 硫酸盐还原菌 | 专型厌氧 | 普通脱硫弧菌 | 还原性乙酰辅酶A途径 | 优势:具有非常高的氢化酶活性,能够天然利用甲酸产氢,在清洁能源产业方面具有应用潜能 劣势:厌氧条件下生长,不利于工业应用;代谢研究尚不完善,代谢改造工具少 | [ |
| 甲基杆菌 | 好氧或 兼性厌氧 | 扭脱甲基杆菌AM1 | 丝氨酸循环 | 优势:能够天然利用甲酸、CO2与甲醇等一碳底物进行细胞生长;模式菌株的研究与改造相对充分,具有一定的改造潜能 劣势:所具有的甲酸代谢路径的能量利用效率偏低,不利于工业应用 | [ |
| CBB循环 依赖型微生物 | 好氧 | 钩虫贪铜菌H16 | 还原性磷酸戊糖循环 | 优势:具有甲酸脱氢酶,能够利用甲酸作为唯一碳源和能源;模式菌株的研究与改造相对充分,具有一定的应用潜能 劣势:甲酸的氧化供能存在能量浪费,因此微生物的细胞生长速率与工业生产潜能均偏低 | [ |
Table 1 Natural formate-utilizing microorganisms
| 微生物种类 | 需氧类型 | 模式菌株 | 天然甲酸 代谢路径 | 应用优劣势 | 参考 文献 |
|---|---|---|---|---|---|
| 产乙酸菌 | 专性厌氧 | 伍氏醋酸杆菌 | 还原性乙酰辅酶A途径 | 优势:能够天然利用甲酸和CO2合成乙酸、乙醇等化合物 劣势:在厌氧条件下生长,不利于工业应用;代谢研究尚不完善,代谢改造工具少 | [ |
| 产甲烷菌 | 专性厌氧 | 氢营养型海沼甲烷球菌 | 还原性乙酰辅酶A途径 | 优势:能够天然利用甲酸和CO2合成甲烷,在清洁能源产业方面具有应用潜能 劣势:在厌氧条件下生长,不利于工业应用;代谢研究尚不完善,代谢改造工具少 | [ |
| 硫酸盐还原菌 | 专型厌氧 | 普通脱硫弧菌 | 还原性乙酰辅酶A途径 | 优势:具有非常高的氢化酶活性,能够天然利用甲酸产氢,在清洁能源产业方面具有应用潜能 劣势:厌氧条件下生长,不利于工业应用;代谢研究尚不完善,代谢改造工具少 | [ |
| 甲基杆菌 | 好氧或 兼性厌氧 | 扭脱甲基杆菌AM1 | 丝氨酸循环 | 优势:能够天然利用甲酸、CO2与甲醇等一碳底物进行细胞生长;模式菌株的研究与改造相对充分,具有一定的改造潜能 劣势:所具有的甲酸代谢路径的能量利用效率偏低,不利于工业应用 | [ |
| CBB循环 依赖型微生物 | 好氧 | 钩虫贪铜菌H16 | 还原性磷酸戊糖循环 | 优势:具有甲酸脱氢酶,能够利用甲酸作为唯一碳源和能源;模式菌株的研究与改造相对充分,具有一定的应用潜能 劣势:甲酸的氧化供能存在能量浪费,因此微生物的细胞生长速率与工业生产潜能均偏低 | [ |
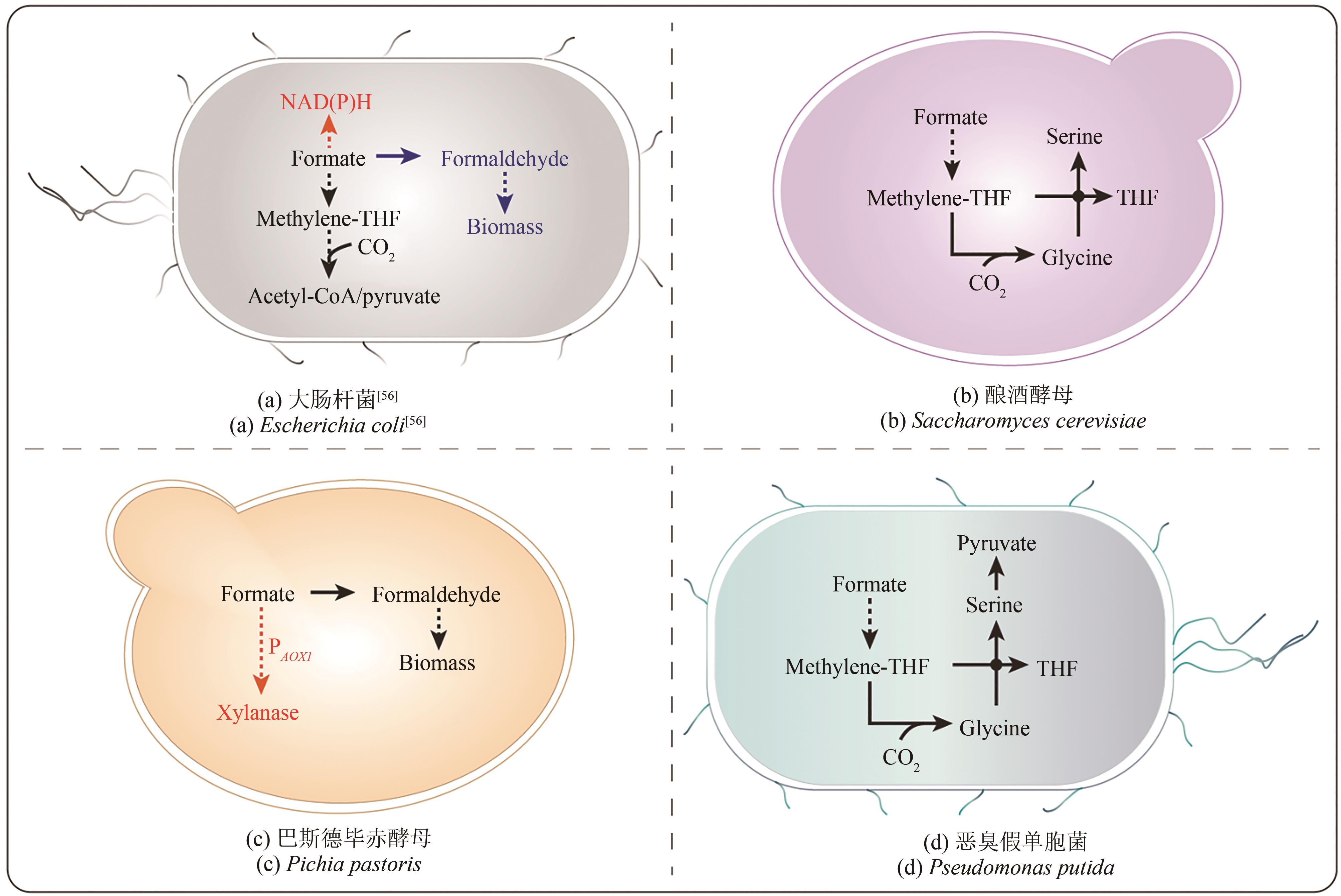
| 微生物 | 非天然甲酸代谢路径 | 主要的应用优势 | 改造潜能 | 参考文献 |
|---|---|---|---|---|
| 大肠杆菌 | 重构的卡尔文循环; 改良的丝氨酸循环; 高丝氨酸循环; rTHF-rgcv途径及关键模块 合成乙酰辅酶A途径等 | 生长周期短; 遗传背景清晰; 代谢工程改造工具种类丰富且高效 | 设计并构建更加高效的甲酸利用路径; 通过实验室适应性进化与培养条件优化等策略提高菌株对甲酸的耐受性 | [ |
| 酿酒酵母 | rTHF-rgcv途径 的关键模块 | 遗传背景清晰; 分子遗传操作工具与技术成熟且高效; 具有内源性甲酸脱氢酶,对甲酸的耐受性较高 | 通过代谢改造进一步提高rTHF-rgcv途径对甲酸的利用效率,开发以甲酸为唯一碳源和能源的工程菌株 | [ |
| 毕赤酵母 | 以甲酸作为P AOX1 的诱导物 | 能够利用甲醇作为唯一碳源和能源进行细胞生长; 在生物医药产业和工业酶生产方面具有巨大潜力 | 通过代谢改造进一步提高菌株对甲酸的利用效率; 基于菌株的代谢网络,设计并构建更加高效的甲酸利用路径 | [ |
| 恶臭假单胞菌 | rTHF-rgcv途径 | 能够编码多种天然甲酸脱氢酶; 具有灵活的代谢机制,能够抵抗氧化应激和多种有毒化合物 | 开发更高效的分子遗传操作工具,进一步完善菌株的甲酸代谢网络 | [ |
Table 2 Metabolically engineered microorganisms
| 微生物 | 非天然甲酸代谢路径 | 主要的应用优势 | 改造潜能 | 参考文献 |
|---|---|---|---|---|
| 大肠杆菌 | 重构的卡尔文循环; 改良的丝氨酸循环; 高丝氨酸循环; rTHF-rgcv途径及关键模块 合成乙酰辅酶A途径等 | 生长周期短; 遗传背景清晰; 代谢工程改造工具种类丰富且高效 | 设计并构建更加高效的甲酸利用路径; 通过实验室适应性进化与培养条件优化等策略提高菌株对甲酸的耐受性 | [ |
| 酿酒酵母 | rTHF-rgcv途径 的关键模块 | 遗传背景清晰; 分子遗传操作工具与技术成熟且高效; 具有内源性甲酸脱氢酶,对甲酸的耐受性较高 | 通过代谢改造进一步提高rTHF-rgcv途径对甲酸的利用效率,开发以甲酸为唯一碳源和能源的工程菌株 | [ |
| 毕赤酵母 | 以甲酸作为P AOX1 的诱导物 | 能够利用甲醇作为唯一碳源和能源进行细胞生长; 在生物医药产业和工业酶生产方面具有巨大潜力 | 通过代谢改造进一步提高菌株对甲酸的利用效率; 基于菌株的代谢网络,设计并构建更加高效的甲酸利用路径 | [ |
| 恶臭假单胞菌 | rTHF-rgcv途径 | 能够编码多种天然甲酸脱氢酶; 具有灵活的代谢机制,能够抵抗氧化应激和多种有毒化合物 | 开发更高效的分子遗传操作工具,进一步完善菌株的甲酸代谢网络 | [ |
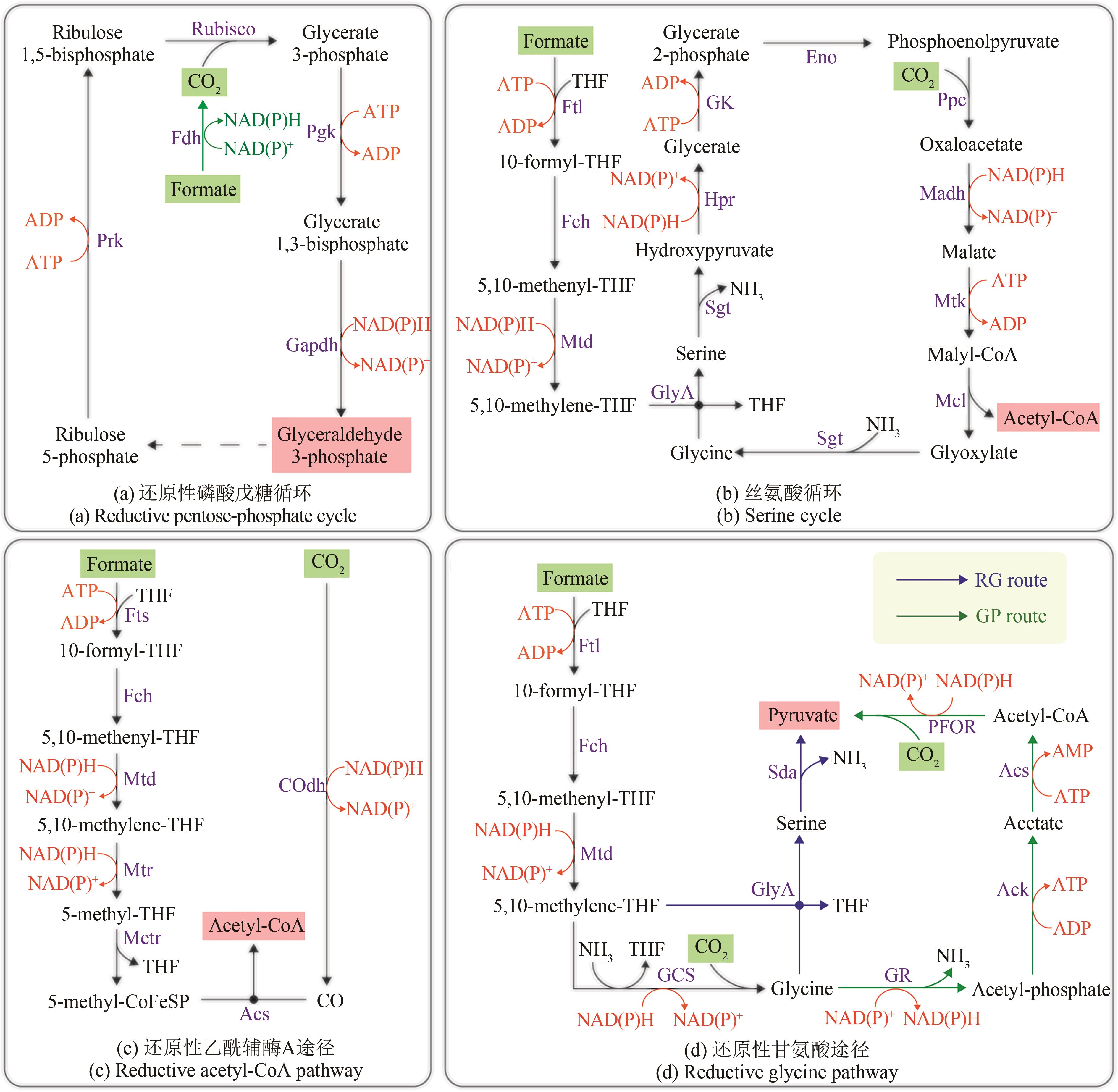
Fig. 3 Natural formate-utilizing pathways(compounds with a green background are pathway substrates; compounds with a pink background are pathway products) Ack—Acetate kinase; Acs—acetyl-CoA synthase; COdh—CO dehydrogenase; CoFeSP—corrinoid ironsulfur protein; Eno—enolase; Fch—methenyl-tetrahydrofolate cyclohydrolase; Fdh—formate dehydrogenase; Ftl—formate-tetrahydrofolate ligase; Fts—formyl-tetrahydrofolate synthetase; Gapdh—glyceraldehyde-3-phosphate dehydrogenase; GCS—glycine cleavage system; Gk—glycerate kinase; GlyA—serine hydroxymethyltransferase; GR—Glycine reductase complex; Hpr—hydroxypyruvate reductase; Madh—malate dehydrogenase; Mcl—malyl-CoA lyase; Metr—methyltransferase; Mtd—methylene-tetrahydrofolate dehydrogenase; Mtk—malate thiokinase; Mtr—methylene-tetrahydrofolate reductase; PFOR—Pyruvate ferredoxin oxidoreductase; Pgk—Phosphoglycerate kinase; Ppc—phosphoenolpyruvate carboxylase; Prk—phosphoribulo kinase; Rubisco—Ribulose-1,5-bisphosphate carboxylase/oxygenase; Sda—serine deaminase; Sgt—serine-glyoxylate transaminase; THF—tetrahydrofolate
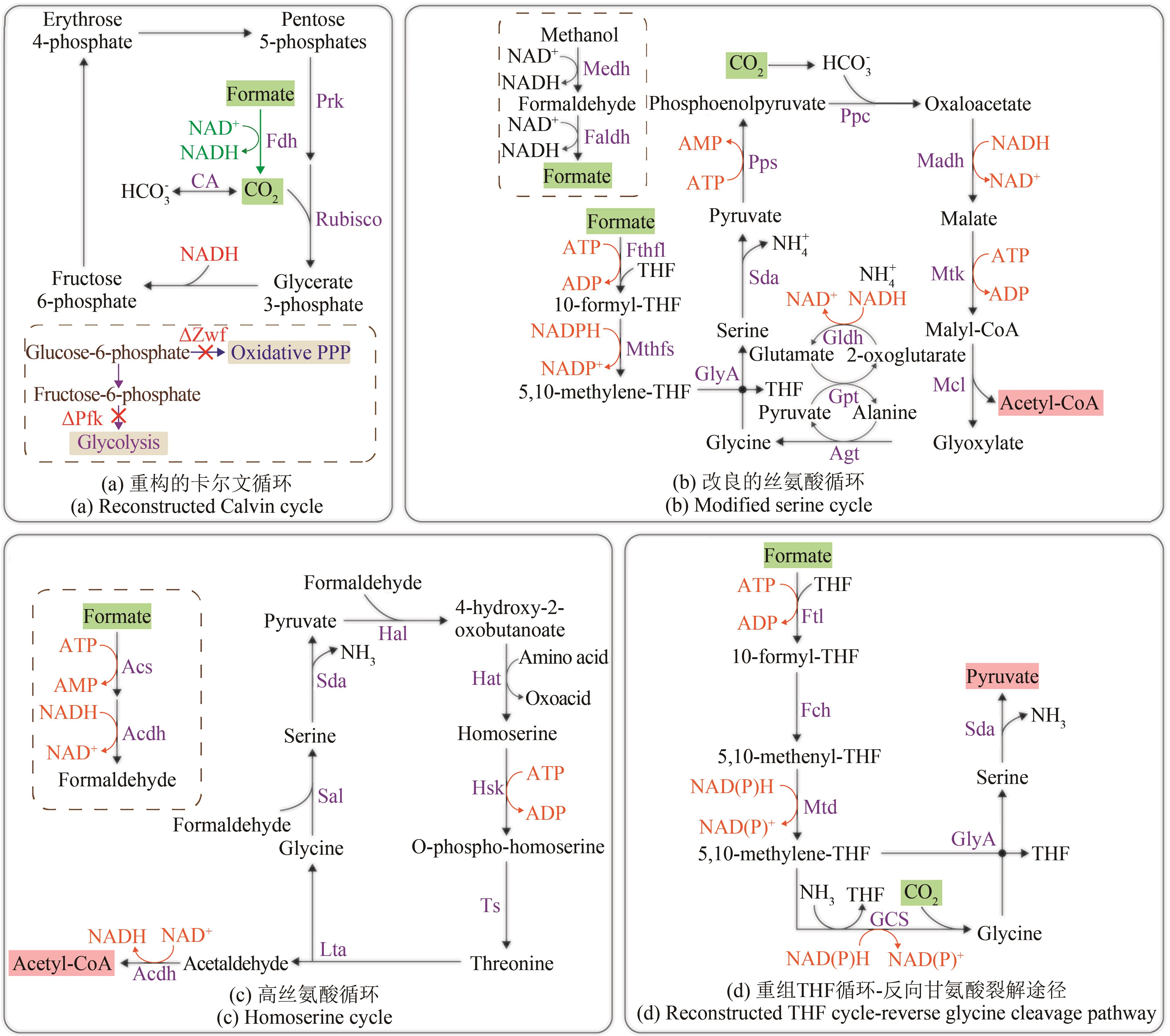
Fig. 4 Reconstruction and optimization of formate-utilizing pathways(compounds with a green background are pathway substrates; compounds with a pink background are pathway products) Acdh—acetaldehyde dehydrogenase; Acs—acetyl-CoA synthase; Agt—alanine-glyoxylate transaminase; CA—carbonic anhydrase; Faldh—formaldehyde dehydrogenase; Fch—methenyl-tetrahydrofolate cyclohydrolase; Fdh—formate dehydrogenase; Fthfl—formate-tetrahydrofolate ligase; Ftl—formate-tetrahydrofolate ligase; GCS—glycine cleavage system; Gldh—glutamate dehydrogenase; GlyA—serine hydroxymethyltransferase; Gpt—glutamate-pyruvate transaminase; Hal—4-hydroxy-2-oxobutanoate aldolase; Hat—4-hydroxy-2-oxobutanoate aminotransferase; Hsk—homoserine kinase; Lta—threonine aldolase; Madh—malate dehydrogenase; Mcl—malyl-CoA lyase; Medh—methanol dehydrogenase; Mtd—methylene-tetrahydrofolate dehydrogenase; Mthfs—5,10-methylene-tetrahydrofolate synthase; Mtk—malate thiokinase; Oxidative PPP—Oxidative pentose-phosphate pathway; Pfk—phosphofructokinase; Ppc—phosphoenolpyruvate carboxylase; Pps—phosphoenolpyruvate synthase; Prk—phosphoribulo kinase; Rubisco—ribulose-1,5-bisphosphate carboxylase; Sal—serine aldolase; Sda—serine deaminase; THF—tetrahydrofolate; Ts—threonine synthase; Zwf—glucose-6-phosphate dehydrogenase
| 菌株 | 路径酶来源 | 参考文献 |
|---|---|---|
| Escherichia coli | Ftl、Fch、Mtd(源自Clostridium ljungdahalii) GCS、GlyA、Sda(内源酶) | [ |
| Escherichia coli | Ftl、Fch、Mtd(源自Methylobacterium extorquens AM1) GCS、GlyA(内源酶) | [ |
| Escherichia coli | Ftl、Fch、Mtd(源自Methylobacterium extorquens CM4) GCS、GlyA、Sda(内源酶) Fdh(源自Candida boidinii) | [ |
| Escherichia coli | Ftl(源自Clostridium kluyveri) FolD(Fch/ Mtd)、GCS、GlyA(内源酶) | [ |
| Escherichia coli | Ftl、Fch、Mtd(源自Methylobacterium extorquens AM1) GCS、GlyA、Sda(内源酶) Fdh(源自Pseudomonas sp.) | [ |
Escherichia coli (Bang等[ | Ftl、Fch、Mtd(源自Methylobacterium extorquens CM4) GCS、GlyA、Sda(内源酶) Fdh(源自Candida boidinii、Arabidopsis thaliana) | [ |
Escherichia coli (Döring等[ | Ftl(源自Clostridium kluyveri) FolD(Fch/ Mtd)、GCS、GlyA(内源酶) | [ |
Escherichia coli (Kim等[ | Ftl、Fch、Mtd(源自Methylobacterium extorquens AM1) GCS、GlyA、Sda(内源酶) Fdh(源自Pseudomonas sp. ) | [ |
| Saccharomyces cerevisiae | MIS1(Ftl/ Fch/ Mtd)、GCS(内源酶) Fdh(内源酶) | [ |
| Cupriavidus necator | Ftl、Fch、Mtd(源自Methylobacterium extorquens AM1) GCS、GlyA、Sda(内源酶) Fdh(内源酶) | [ |
| Clostridium pasteurianum | GCS(源自Gottschalkia acidurici) Ftl、Fch、Mtd、GlyA、Sda(内源酶) | [ |
| Pseudomonas putida | Ftl、Fch、Mtd(源自Methylobacterium extorquens AM1) GCS、GlyA、Sda(内源酶) Fdh(内源酶) | [ |
Table 3 Source of pathway enzymes of the rTHF-rgcv pathway
| 菌株 | 路径酶来源 | 参考文献 |
|---|---|---|
| Escherichia coli | Ftl、Fch、Mtd(源自Clostridium ljungdahalii) GCS、GlyA、Sda(内源酶) | [ |
| Escherichia coli | Ftl、Fch、Mtd(源自Methylobacterium extorquens AM1) GCS、GlyA(内源酶) | [ |
| Escherichia coli | Ftl、Fch、Mtd(源自Methylobacterium extorquens CM4) GCS、GlyA、Sda(内源酶) Fdh(源自Candida boidinii) | [ |
| Escherichia coli | Ftl(源自Clostridium kluyveri) FolD(Fch/ Mtd)、GCS、GlyA(内源酶) | [ |
| Escherichia coli | Ftl、Fch、Mtd(源自Methylobacterium extorquens AM1) GCS、GlyA、Sda(内源酶) Fdh(源自Pseudomonas sp.) | [ |
Escherichia coli (Bang等[ | Ftl、Fch、Mtd(源自Methylobacterium extorquens CM4) GCS、GlyA、Sda(内源酶) Fdh(源自Candida boidinii、Arabidopsis thaliana) | [ |
Escherichia coli (Döring等[ | Ftl(源自Clostridium kluyveri) FolD(Fch/ Mtd)、GCS、GlyA(内源酶) | [ |
Escherichia coli (Kim等[ | Ftl、Fch、Mtd(源自Methylobacterium extorquens AM1) GCS、GlyA、Sda(内源酶) Fdh(源自Pseudomonas sp. ) | [ |
| Saccharomyces cerevisiae | MIS1(Ftl/ Fch/ Mtd)、GCS(内源酶) Fdh(内源酶) | [ |
| Cupriavidus necator | Ftl、Fch、Mtd(源自Methylobacterium extorquens AM1) GCS、GlyA、Sda(内源酶) Fdh(内源酶) | [ |
| Clostridium pasteurianum | GCS(源自Gottschalkia acidurici) Ftl、Fch、Mtd、GlyA、Sda(内源酶) | [ |
| Pseudomonas putida | Ftl、Fch、Mtd(源自Methylobacterium extorquens AM1) GCS、GlyA、Sda(内源酶) Fdh(内源酶) | [ |
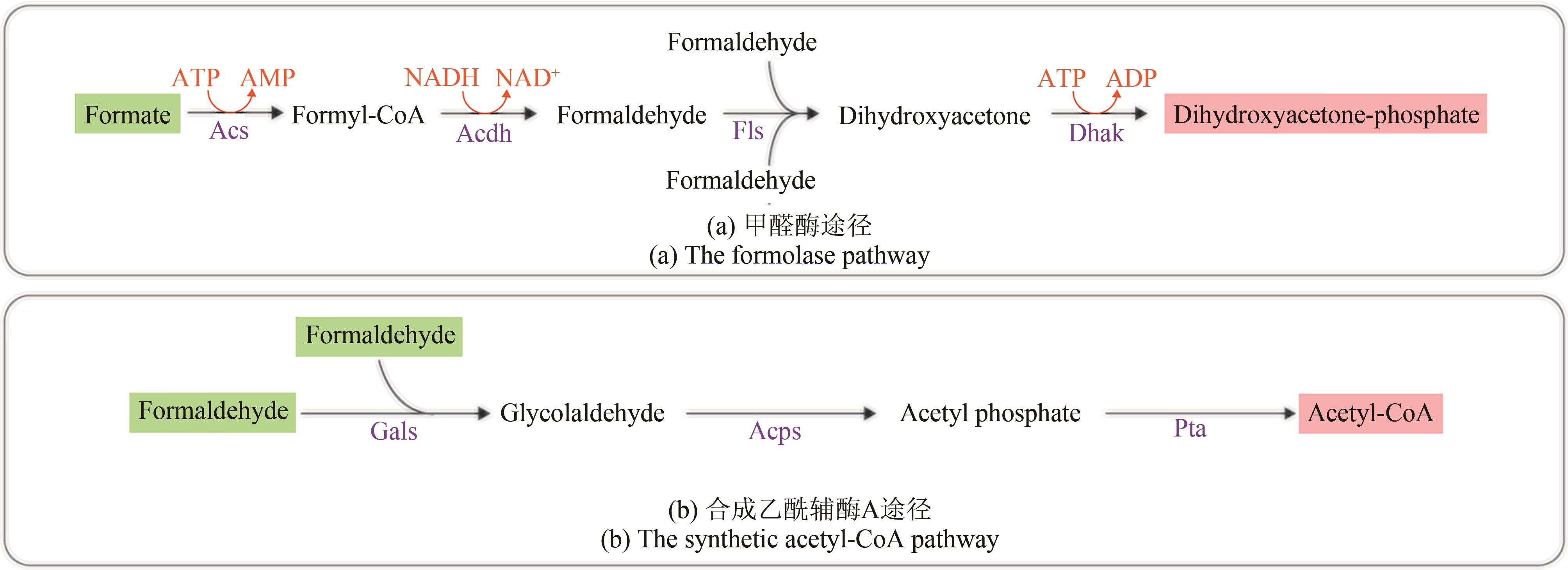
Fig. 5 Artificial formate-utilizing pathwaysAcdh—acetaldehyde dehydrogenase; Acs—acetyl-CoA synthase; Acps—acetyl phosphate synthase; Dhak—dihydroxyacetone kinase; Fls—formolase; Gals—glycolaldehyde synthase; Pta—phosphate acetyltransferase (compounds with a green background are pathway substrates, compounds with a pink background are pathway products)

Fig. 6 Key methods to improve the efficiency of formate assimilationFDH—formate dehydrogenase; Hint—aminomethylated form of H protein; Hox—oxidized form of H protein; Hred—reduced form of H protein; TCA cycle—tricarboxylic acid cycle; THF—tetrahydrofolate;
| 1 | ESCOBAR J C, LORA E S, VENTURINI O J, et al. Biofuels: environment, technology and food security[J]. Renewable & Sustainable Energy Reviews, 2009, 13(6/7): 1275-1287. |
| 2 | ZHOU Y J, KERKHOVEN E J, NIELSEN J. Barriers and opportunities in bio-based production of hydrocarbons[J]. Nature Energy, 2018, 3(11): 925-935. |
| 3 | LV X Q, YU W W, ZHANG C Y, et al. C1-based biomanufacturing: advances, challenges and perspectives[J]. Bioresource Technology, 2023, 367: 128259. |
| 4 | ZHANG C Q, OTTENHEIM C, WEINGARTEN M, et al. Microbial utilization of next-generation feedstocks for the biomanufacturing of value-added chemicals and food ingredients[J]. Frontiers in Bioengineering and Biotechnology, 2022, 10: 874612. |
| 5 | SAKARIKA M, GANIGUÉ R, RABAEY K. Methylotrophs: from C1 compounds to food[J]. Current Opinion in Biotechnology, 2022, 75: 102685. |
| 6 | FLAIZ M, LUDWIG G, BENGELSDORF F R, et al. Production of the biocommodities butanol and acetone from methanol with fluorescent FAST-tagged proteins using metabolically engineered strains of Eubacterium limosum [J]. Biotechnology for Biofuels, 2021, 14(1): 117. |
| 7 | OKOYE-CHINE C G, OTUN K, SHIBA N, et al. Conversion of carbon dioxide into fuels—a review[J]. Journal of CO2 Utilization, 2022, 62: 102099. |
| 8 | YOON J H, CHANG W J, OH S H, et al. Metabolic engineering of Methylorubrum extorquens AM1 for poly (3-hydroxybutyrate-co-3-hydroxyvalerate) production using formate[J]. International Journal of Biological Macromolecules, 2021, 177: 284-293. |
| 9 | RAY S, JIN J O, CHOI I, et al. Recent trends of biotechnological production of polyhydroxyalkanoates from C1 carbon sources[J]. Frontiers in Bioengineering and Biotechnology, 2023, 10: 907500. |
| 10 | LIU Y Q, BAI C X, XU Q, et al. Improved methanol-derived lovastatin production through enhancement of the biosynthetic pathway and intracellular lovastatin efflux in methylotrophic yeast[J].Bioresources and Bioprocessing, 2018, 5(1): 22. |
| 11 | LIU Y Q, TU X H, XU Q, et al. Engineered monoculture and co-culture of methylotrophic yeast for de novo production of monacolin J and lovastatin from methanol[J]. Metabolic Engineering, 2018, 45: 189-199. |
| 12 | YASIN M, JEONG Y S, PARK S Y, et al. Microbial synthesis gas utilization and ways to resolve kinetic and mass-transfer limitations[J]. Bioresource Technology, 2015, 177: 361-374. |
| 13 | COTTON C A, CLAASSENS N J, BENITO-VAQUERIZO S, et al. Renewable methanol and formate as microbial feedstocks[J]. Current Opinion in Biotechnology, 2020, 62: 168-180. |
| 14 | JIANG W, HERNÁNDEZ VILLAMOR D, PENG H D, et al. Metabolic engineering strategies to enable microbial utilization of C1 feedstocks[J]. Nature Chemical Biology, 2021, 17(8): 845-855. |
| 15 | THIJS B, RONGÉ J, MARTENS J A. Matching emerging formic acid synthesis processes with application requirements[J]. Green Chemistry, 2022, 24(6): 2287-2295. |
| 16 | CLAASSENS N J, BORDANABA-FLORIT G, COTTON C A R, et al. Replacing the Calvin cycle with the reductive glycine pathway in Cupriavidus necator [J]. Metabolic Engineering, 2020, 62: 30-41. |
| 17 | KIM S, LINDNER S N, ASLAN S, et al. Growth of E. coli on formate and methanol via the reductive glycine pathway[J]. Nature Chemical Biology, 2020, 16(5): 538-545. |
| 18 | BANG J, HWANG C H, AHN J H, et al. Escherichia coli is engineered to grow on CO2 and formic acid[J]. Nature Microbiology, 2020, 5(12): 1459-1463. |
| 19 | GONZALEZ DE LA CRUZ J, MACHENS F, MESSERSCHMIDT K, et al. Core catalysis of the reductive glycine pathway demonstrated in yeast[J]. ACS Synthetic Biology, 2019, 8(5): 911-917. |
| 20 | DRAKE H L, KÜSEL K, MATTHIES C. Acetogenic prokaryotes[M/OL]//The Prokaryotes. Berlin, Heidelberg: Springer Berlin Heidelberg, 2013: 3-60 [2023-03-01]. . |
| 21 | MOON J, DÖNIG J, KRAMER S, et al. Formate metabolism in the acetogenic bacterium Acetobacterium woodii [J]. Environmental Microbiology, 2021, 23(8): 4214-4227. |
| 22 | NEUENDORF C S, VIGNOLLE G A, DERNTL C, et al. A quantitative metabolic analysis reveals Acetobacterium woodii as a flexible and robust host for formate-based bioproduction[J]. Metabolic Engineering, 2021, 68: 68-85. |
| 23 | REEVE J N. Molecular biology of methanogens[J]. Annual Review of Microbiology, 1992, 46: 165-191. |
| 24 | LUPA B, HENDRICKSON E L, LEIGH J A, et al. Formate-dependent H2 production by the mesophilic methanogen Methanococcus maripaludis [J]. Applied and Environmental Microbiology, 2008, 74(21): 6584-6590. |
| 25 | SARMIENTO F B, LEIGH J A, WHITMAN W B. Genetic systems for hydrogenotrophic methanogens[M/OL]//Methods in Methane Metabolism, Part A: Methods in Enzymology. Amsterdam: Elsevier, 2011: 43-73 [2023-03-01]. . |
| 26 | GOYAL N, ZHOU Z, KARIMI I A. Metabolic processes of Methanococcus maripaludis and potential applications[J]. Microbial Cell Factories, 2016, 15(1): 107. |
| 27 | MARTINS M, PEREIRA I A C. Sulfate-reducing bacteria as new microorganisms for biological hydrogen production[J]. International Journal of Hydrogen Energy, 2013, 38(28): 12294-12301. |
| 28 | RITTMANN S K M R, LEE H S, LIM J K, et al. One-carbon substrate-based biohydrogen production: microbes, mechanism, and productivity[J]. Biotechnology Advances, 2015, 33(1): 165-177. |
| 29 | MARTINS M, MOURATO C, PEREIRA I A C. Desulfovibrio vulgaris growth coupled to formate-driven H2 production[J]. Environmental Science & Technology, 2015, 49(24): 14655-14662. |
| 30 | SINGH R P, SINGH R N, SRIVASTAVA M K, et al. Structure prediction and analysis of MxaF from obligate, facultative and restricted facultative methylobacterium[J]. Bioinformation, 2012, 8(21): 1042-1046. |
| 31 | CROWTHER G J, KOSÁLY G, LIDSTROM M E. Formate as the main branch point for methylotrophic metabolism in Methylobacterium extorquens AM1[J]. Journal of Bacteriology, 2008, 190(14): 5057-5062. |
| 32 | CUI L Y, YANG J, LIANG W F, et al. Sodium formate redirects carbon flux and enhances heterologous mevalonate production in Methylobacterium extorquens AM1[J]. Biotechnology Journal, 2023, 18(2): 2200402. |
| 33 | POHLMANN A, FRICKE W F, REINECKE F, et al. Genome sequence of the bioplastic-producing "Knallgas" bacterium Ralstonia eutropha H16[J]. Nature Biotechnology, 2006, 24(10): 1257-1262. |
| 34 | CRAMM R. Genomic view of energy metabolism in Ralstonia eutropha H16[J]. Journal of Molecular Microbiology and Biotechnology, 2008, 16(1/2): 38-52. |
| 35 | PAN H J, WANG J, WU H L, et al. Synthetic biology toolkit for engineering Cupriviadus necator H16 as a platform for CO2 valorization[J].Biotechnology for Biofuels, 2021, 14(1): 212. |
| 36 | CALVEY C H, SANCHEZ I N V, WHITE A M, et al. Improving growth of Cupriavidus necator H16 on formate using adaptive laboratory evolution-informed engineering[J]. Metabolic Engineering, 2023, 75: 78-90. |
| 37 | SCHUCHMANN K, MÜLLER V. Autotrophy at the thermodynamic limit of life: a model for energy conservation in acetogenic bacteria[J]. Nature Reviews Microbiology, 2014, 12(12): 809-821. |
| 38 | LIU Y C, WHITMAN W B. Metabolic, phylogenetic, and ecological diversity of the methanogenic Archaea[J]. Annals of the New York Academy of Sciences, 2008, 1125(1): 171-189. |
| 39 | 冷欢, 杨清, 黄钢锋, 等. 氢营养型产甲烷代谢途径研究进展[J]. 微生物学报, 2020, 60(10): 2136-2160. |
| LENG H, YANG Q, HUANG G F, et al. Recent advances in hydrogenotrophic methanogenesis[J]. Acta Microbiologica Sinica, 2020, 60(10): 2136-2160. | |
| 40 | THAUER R K, KASTER A K, SEEDORF H, et al. Methanogenic Archaea: ecologically relevant differences in energy conservation[J]. Nature Reviews Microbiology, 2008, 6(8): 579-591. |
| 41 | COSTA K C, WONG P M, WANG T S, et al. Protein complexing in a methanogen suggests electron bifurcation and electron delivery from formate to heterodisulfide reductase[J]. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(24): 11050-11055. |
| 42 | LI J, ZHANG L Y, XU Q, et al. CRISPR-Cas9 toolkit for genome editing in an autotrophic CO2-fixing methanogenic archaeon[J]. Microbiology Spectrum, 2022, 10(4): 01165-22. |
| 43 | BAO J C, DE DIOS MATEOS E, SCHELLER S. Efficient CRISPR/Cas12a-based genome-editing toolbox for metabolic engineering in Methanococcus maripaludis [J]. ACS Synthetic Biology, 2022, 11(7): 2496-2503. |
| 44 | HEIDELBERG J F, SESHADRI R, HAVEMAN S A, et al. The genome sequence of the anaerobic, sulfate-reducing bacterium Desulfovibrio vulgaris hildenborough[J]. Nature Biotechnology, 2004, 22(5): 554-559. |
| 45 | JANSEN K, THAUER R K, WIDDEL F, et al. Carbon assimilation pathways in sulfate reducing bacteria. Formate, carbon dioxide, carbon monoxide, and acetate assimilation by Desulfovibrio baarsii [J]. Archives of Microbiology, 1984, 138(3): 257-262. |
| 46 | SÁNCHEZ-ANDREA I, ALVES GUEDES I, HORNUNG B, et al. The reductive glycine pathway allows autotrophic growth of Desulfovibrio desulfuricans [J]. Nature Communications, 2020, 11: 5090. |
| 47 | SUN H, SPRING S, LAPIDUS A, et al. Complete genome sequence of Desulfarculus baarsii type strain (2st14T)[J]. Standards in Genomic Sciences, 2010, 3(3): 276-284. |
| 48 | MARTINS M, MOURATO C, MORAIS-SILVA F O, et al. Electron transfer pathways of formate-driven H2 production in Desulfovibrio [J]. Applied Microbiology and Biotechnology, 2016, 100(18): 8135-8146. |
| 49 | ZHANG W M, SONG M, YANG Q, et al. Current advance in bioconversion of methanol to chemicals[J]. Biotechnology for Biofuels, 2018, 11: 260. |
| 50 | KIM S M, LEE S H, KIM I K, et al. Structural insight into a molecular mechanism of methenyltetrahydrofolate cyclohydrolase from Methylobacterium extorquens AM1[J]. International Journal of Biological Macromolecules, 2022, 202: 234-240. |
| 51 | TONG S, ZHAO L Z, ZHU D L, et al. From formic acid to single-cell protein: genome-scale revealing the metabolic network of Paracoccus communis MA5[J].Bioresources and Bioprocessing, 2022, 9(1): 55. |
| 52 | PICKERING B S, ORESNIK I J. Formate-dependent autotrophic growth in Sinorhizobium meliloti [J]. Journal of Bacteriology, 2008, 190(19): 6409-6418. |
| 53 | SOROKIN D Y, LÜCKER S, VEJMELKOVA D, et al. Nitrification expanded: discovery, physiology and genomics of a nitrite-oxidizing bacterium from the Phylum chloroflexi [J]. The ISME Journal, 2012, 6(12): 2245-2256. |
| 54 | GROSTERN A, ALVAREZ-COHEN L. RubisCO-based CO2 fixation and C1 metabolism in the actinobacterium Pseudonocardia dioxanivorans CB1190[J]. Environmental Microbiology, 2013, 15(11): 3040-3053. |
| 55 | ORLOVA M V, TARLACHKOV S V, DUBININA G A, et al. Genomic insights into metabolic versatility of a lithotrophic sulfur-oxidizing diazotrophic Alphaproteobacterium Azospirillum thiophilum [J]. FEMS Microbiology Ecology, 2016, 92(12): fiw199. |
| 56 | BANG J, AHN J H, LEE J A, et al. Synthetic formatotrophs for one-carbon biorefinery[J]. Advanced Science, 2021, 8(12): 2100199. |
| 57 | GLEIZER S, BEN-NISSAN R, BAR-ON Y M, et al. Conversion of Escherichia coli to generate all biomass carbon from CO2 [J]. Cell, 2019, 179(6): 1255-1263.e12. |
| 58 | YU H, LIAO J C. A modified serine cycle in Escherichia coli coverts methanol and CO2 to two-carbon compounds[J]. Nature Communications, 2018, 9: 3992. |
| 59 | HE H, HOPER R, DODENHOFT M, et al. An optimized methanol assimilation pathway relying on promiscuous formaldehyde-condensing aldolases in E. coli [J]. Metabolic Engineering, 2020, 60: 1-13. |
| 60 | YISHAI O, GOLDBACH L, TENENBOIM H, et al. Engineered assimilation of exogenous and endogenous formate in Escherichia coli [J]. ACS Synthetic Biology, 2017, 6(9): 1722-1731. |
| 61 | TASHIRO Y, HIRANO S, MATSON M M, et al. Electrical-biological hybrid system for CO2 reduction[J]. Metabolic Engineering, 2018, 47: 211-218. |
| 62 | YISHAI O, BOUZON M, DÖRING V, et al. In vivo assimilation of one-carbon via a synthetic reductive glycine pathway in Escherichia coli [J]. ACS Synthetic Biology, 2018, 7(9): 2023-2028. |
| 63 | BANG J, LEE S Y. Assimilation of formic acid and CO2 by engineered Escherichia coli equipped with reconstructed one-carbon assimilation pathways[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(40): E9271-E9279. |
| 64 | DÖRING V, DARII E, YISHAI O, et al. Implementation of a reductive route of one-carbon assimilation in Escherichia coli through directed evolution[J]. ACS Synthetic Biology, 2018, 7(9): 2029-2036. |
| 65 | DELMAS V A, PERCHAT N, MONET O, et al. Genetic and biocatalytic basis of formate dependent growth of Escherichia coli strains evolved in continuous culture[J]. Metabolic Engineering, 2022, 72: 200-214. |
| 66 | KIM S, GIRALDO N, RAINALDI V, et al. Optimizing E. coli as a formatotrophic platform for bioproduction via the reductive glycine pathway[J]. Frontiers in Bioengineering and Biotechnology, 2023, 11: 1091899. |
| 67 | LU X Y, LIU Y W, YANG Y Q, et al. Constructing a synthetic pathway for acetyl-coenzyme A from one-carbon through enzyme design[J]. Nature Communications, 2019, 10: 1378. |
| 68 | LIU B, LI H J, ZHOU H L, et al. Enhancing xylanase expression by Komagataella phaffii by formate as carbon source and inducer[J].Applied Microbiology and Biotechnology, 2022, 106(23): 7819-7829. |
| 69 | LIU B, ZHAO Y X, ZHOU H L, et al. Enhancing xylanase expression of Komagataella phaffii induced by formate through Mit1 co-expression[J].Bioprocess and Biosystems Engineering, 2022, 45(9): 1515-1525. |
| 70 | TURLIN J, DRONSELLA B, DE MARIA A, et al. Integrated rational and evolutionary engineering of genome-reduced Pseudomonas putida strains promotes synthetic formate assimilation[J]. Metabolic Engineering, 2022, 74: 191-205. |
| 71 | BAR-EVEN A, NOOR E, FLAMHOLZ A, et al. Design and analysis of metabolic pathways supporting formatotrophic growth for electricity-dependent cultivation of microbes[J]. Biochimica et Biophysica Acta (BBA)-Bioenergetics, 2013, 1827(8/9): 1039-1047. |
| 72 | BAR-EVEN A, FLAMHOLZ A, NOOR E, et al. Thermodynamic constraints shape the structure of carbon fixation pathways[J]. Biochimica et Biophysica Acta (BBA) - Bioenergetics, 2012, 1817(9): 1646-1659. |
| 73 | KIM S J, YOON J, IM D K, et al. Adaptively evolved Escherichia coli for improved ability of formate utilization as a carbon source in sugar-free conditions[J].Biotechnology for Biofuels, 2019, 12:207. |
| 74 | NITSCHKE W, RUSSELL M J. Beating the acetyl coenzyme A-pathway to the origin of life[J]. Philosophical Transactions of the Royal Society of London Series B, Biological Sciences, 2013, 368(1622): 20120258. |
| 75 | YISHAI O, LINDNER S N, GONZALEZ DE LA CRUZ J, et al. The formate bio-economy[J]. Current Opinion in Chemical Biology, 2016, 35: 1-9. |
| 76 | LIU Z H, WANG K, CHEN Y, et al. Third-generation biorefineries as the means to produce fuels and chemicals from CO2 [J]. Nature Catalysis, 2020, 3(3): 274-288. |
| 77 | SONG Y, LEE J S, SHIN J, et al. Functional cooperation of the glycine synthase-reductase and Wood-Ljungdahl pathways for autotrophic growth of Clostridium drakei [J]. Proceedings of the National Academy of Sciences of the United States of America, 2020, 117(13): 7516-7523. |
| 78 | MAO W, YUAN Q Q, QI H G, et al. Recent progress in metabolic engineering of microbial formate assimilation[J].Applied Microbiology and Biotechnology, 2020, 104(16): 6905-6917. |
| 79 | ZHANG H, LI Y C, NIE J L, et al. Structure-based dynamic analysis of the glycine cleavage system suggests key residues for control of a key reaction step[J]. Communications Biology, 2020, 3: 756. |
| 80 | XU Y Y, REN J, WANG W, et al. Improvement of glycine biosynthesis from one-carbon compounds and ammonia catalyzed by the glycine cleavage system in vitro [J]. Engineering in Life Sciences, 2022, 22(1): 40-53. |
| 81 | ERB T J, KELLER P, VORHOLT J A. Escherichia coli in auto(trophic) mode[J]. Cell, 2019, 179(6): 1244-1245. |
| 82 | ZHANG Y X, LI F L, DONG J, et al. Recent advances in designing efficient electrocatalysts for electrochemical carbon dioxide reduction to formic acid/formate[J]. Journal of Electroanalytical Chemistry, 2023, 928: 117018. |
| 83 | JUNQUEIRA J R C, DAS D, CATHRIN BRIX A, et al. Simultaneous anodic and cathodic formate production in a paired electrolyzer by CO2 reduction and glycerol oxidation[J]. ChemSusChem, 2023: e202300667. |
| 84 | WANG T, CHEN J D, REN X Y, et al. Halogen-incorporated Sn catalysts for selective electrochemical CO2 reduction to formate[J]. Angewandte Chemie International Edition, 2023, 62(10): e202211174. |
| 85 | BAR-EVEN A. Formate assimilation: the metabolic architecture of natural and synthetic pathways[J]. Biochemistry, 2016, 55(28): 3851-3863. |
| 86 | CHEN N H, DJOKO K Y, VEYRIER F J, et al. Formaldehyde stress responses in bacterial pathogens[J]. Frontiers in Microbiology, 2016, 7: 257. |
| 87 | ALKIM C, FARIAS D, FREDONNET J, et al. Toxic effect and inability of L-homoserine to be a nitrogen source for growth of Escherichia coli resolved by a combination of in vivo evolution engineering and omics analyses[J]. Frontiers in Microbiology, 2022, 13: 1051425. |
| 88 | KLEIN V J, IRLA M, GIL LÓPEZ M, et al. Unravelling formaldehyde metabolism in bacteria: road towards synthetic methylotrophy[J]. Microorganisms, 2022, 10(2): 220. |
| 89 | HONG Y, ARBTER P, WANG W, et al. Introduction of glycine synthase enables uptake of exogenous formate and strongly impacts the metabolism in Clostridium pasteurianum [J]. Biotechnology and Bioengineering, 2021, 118(3): 1366-1380. |
| 90 | KHANA D B, CALLAGHAN M M, AMADOR-NOGUEZ D. Novel computational and experimental approaches for investigating the thermodynamics of metabolic networks[J]. Current Opinion in Microbiology, 2022, 66: 21-31. |
| 91 | CHEN K, ARNOLD F H. Engineering new catalytic activities in enzymes[J]. Nature Catalysis, 2020, 3(3): 203-213. |
| 92 | KIM D H, NOH M H, PARK M H, et al. Enzyme activity engineering based on sequence co-evolution analysis[J]. Metabolic Engineering, 2022, 74: 49-60. |
| 93 | SIEGEL J B, SMITH A L, POUST S, et al. Computational protein design enables a novel one-carbon assimilation pathway[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(12): 3704-3709. |
| 94 | HU G P, LI Z H, MA D L, et al. Light-driven CO2 sequestration in Escherichia coli to achieve theoretical yield of chemicals[J]. Nature Catalysis, 2021, 4(5): 395-406. |
| 95 | HU G P, GUO L, GAO C, et al. Synergistic metabolism of glucose and formate increases the yield of short-chain organic acids in Escherichia coli [J]. ACS Synthetic Biology, 2022, 11(1): 135-143. |
| 96 | CLAASSENS N J, HE H, BAR-EVEN A. Synthetic methanol and formate assimilation via modular engineering and selection strategies[J]. Current Issues in Molecular Biology, 2019, 33(1): 237-248. |
| 97 | DRAGOSITS M, MATTANOVICH D. Adaptive laboratory evolution—principles and applications for biotechnology[J]. Microbial Cell Factories, 2013, 12: 64. |
| 98 | AHN J H, BANG J, KIM W J, et al. Formic acid as a secondary substrate for succinic acid production by metabolically engineered Mannheimia succiniciproducens [J]. Biotechnology and Bioengineering, 2017, 114(12): 2837-2847. |
| 99 | SINGH A, MOESTEDT J, BERG A, et al. Microbiological surveillance of biogas plants: targeting acetogenic community[J]. Frontiers in Microbiology, 2021, 12: 700256. |
| 100 | SUDA K, SAKAMOTO S, IGUCHI A, et al. Novel quantitative method for individual isotopomer of organic acids from 13C tracer experiments determines carbon flow in acetogenesis[J]. Talanta, 2023, 257: 124328. |
| 101 | KIRST H, FERLEZ B H, LINDNER S N, et al. Toward a glycyl radical enzyme containing synthetic bacterial microcompartment to produce pyruvate from formate and acetate[J]. Proceedings of the National Academy of Sciences of the United States of America, 2022, 119(8): e2116871119. |
| 102 | NA D, YOO S M, CHUNG H, et al. Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs[J]. Nature Biotechnology, 2013, 31(2): 170-174. |
| 103 | PONATH F, HÖR J, VOGEL J. An overview of gene regulation in bacteria by small RNAs derived from mRNA 3′ ends[J]. FEMS Microbiology Reviews, 2022, 46(5): fuac017. |
| 104 | WENK S, SCHANN K, HE H, et al. An "energy-auxotroph" Escherichia coli provides an in vivo platform for assessing NADH regeneration systems[J]. Biotechnology and Bioengineering, 2020, 117(11): 3422-3434. |
| [1] | GUO Shuyuan, ZHANG Qiannan, Gulikezi· MAIMAITIREXIATI, YANG Yiqun, YU Tao. Advances in microbial production of liquid biofuels [J]. Synthetic Biology Journal, 2025, 6(1): 18-44. |
| [2] | SHAO Mingwei, SUN Simian, YANG Shimao, CHEN Guoqiang. Bioproduction based on extremophiles [J]. Synthetic Biology Journal, 2024, 5(6): 1419-1436. |
| [3] | ZHAO Liang, LI Zhenshuai, FU Liping, LYU Ming, WANG Shi’an, ZHANG Quan, LIU Licheng, LI Fuli, LIU Ziyong. Progress in biomanufacturing of lipids and single cell protein from one-carbon compounds [J]. Synthetic Biology Journal, 2024, 5(6): 1300-1318. |
| [4] | ZHU Fanghuan, CEN Xuecong, CHEN Zhen. Research progress of diols production by microbes [J]. Synthetic Biology Journal, 2024, 5(6): 1367-1385. |
| [5] | YU Wei, GAO Jiaoqi, ZHOU Yongjin. Bioconversion of one carbon feedstocks for producing organic acids [J]. Synthetic Biology Journal, 2024, 5(5): 1169-1188. |
| [6] | CHEN Xiwei, ZHANG Huaran, ZOU Yi. Biosynthesis and metabolic engineering of fungal non-ribosomal peptides [J]. Synthetic Biology Journal, 2024, 5(3): 571-592. |
| [7] | XIE Huang, ZHENG Yilei, SU Yiting, RUAN Jingyi, LI Yongquan. An overview on reconstructing the biosynthetic system of actinomycetes for polyketides production [J]. Synthetic Biology Journal, 2024, 5(3): 612-630. |
| [8] | HUI Zhen, TANG Xiaoyu. Applications of the CRISPR/Cas9 editing system in the study of microbial natural products [J]. Synthetic Biology Journal, 2024, 5(3): 658-671. |
| [9] | ZHAO Jingyu, ZHANG Jian, QI Qingsheng, WANG Qian. Research progress in biosensors based on bacterial two-component systems [J]. Synthetic Biology Journal, 2024, 5(1): 38-52. |
| [10] | ZHOU Qiang, ZHOU Dawei, SUN Jingxiang, WANG Jingnan, JIANG Wankui, ZHANG Wenming, JIANG Yujia, XIN Fengxue, JIANG Min. Research progress in synthesis of astaxanthin by microbial fermentation [J]. Synthetic Biology Journal, 2024, 5(1): 126-143. |
| [11] | LIU Weisong, ZHANG Kuncheng, CUI Huijuan, ZHU Zhiguang, ZHANG Yiheng, ZHANG Lingling. Electro-assisted carbon dioxide biotransformation [J]. Synthetic Biology Journal, 2023, 4(6): 1191-1222. |
| [12] | SUN Huili, CUI Jinyu, LUAN Guodong, LYU Xuefeng. Progress of cyanobacterial synthetic biotechnology for efficient light-driven carbon fixation and ethanol production [J]. Synthetic Biology Journal, 2023, 4(6): 1161-1177. |
| [13] | YAN Xiongying, WANG Zhen, LOU Jiyun, ZHANG Haoyu, HUANG Xingyu, WANG Xia, YANG Shihui. Progress in the construction of microbial cell factories for efficient biofuel production [J]. Synthetic Biology Journal, 2023, 4(6): 1082-1121. |
| [14] | ZHANG Chenyue, MA Yingqun, WANG Xing, FU Rongzhan, HUANG Jiwei, HUA Xiufu, FAN Daidi, FEI Qiang. Progress in the bioconversion of biogas into sustainable aviation fuel [J]. Synthetic Biology Journal, 2023, 4(6): 1246-1258. |
| [15] | CHEN Yaru, CAO Yingxiu, SONG Hao. Advances and applications of gene editing and transcriptional regulation in electroactive microorganisms [J]. Synthetic Biology Journal, 2023, 4(6): 1281-1299. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||