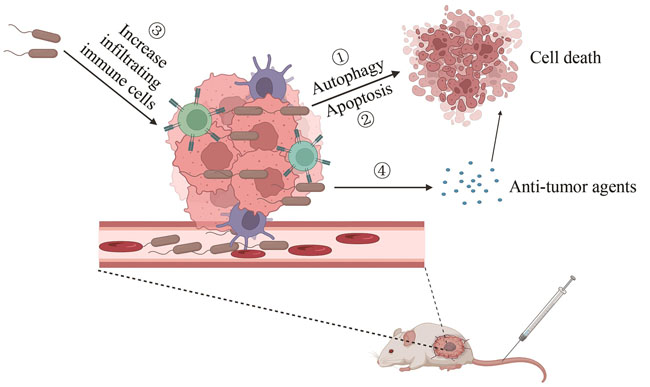
Bacterial therapy has a long history, which originated at the end of the 19th century when bacteria were used for the first time to treat cancer. William B. Coley, a bone sarcoma surgeon, used heat-killed Streptococcus pyogenes to treat patients with unresectable sarcomas and found that the cancer had disappeared spontaneously. Over the next 40 years, he injected more than 1000 cancer patients with heat-killed bacteria Streptococcus pyogenes and Serratia marcescens, which are now known as Coley's toxins. However, owing to criticism from the medical community, rare reproducibility of the results and the emergence of radiotherapy and chemotherapy, bacterial therapy gradually disappeared from medical practice. With the development of immunology and synthetic biology, bacterial therapy has once again gained importance. Over the past 20 years, bacterial therapy has become a major focus area for research, and several types of bacteria have been used in clinical and preclinical studies. Therefore, bacterial therapy is considered an innovative and ideal strategy to treat cancer. In this review, we summarized the recent progress in the treatment of tumours with genetically engineered bacteria. Various bacteria include Salmonella, Escherichia coli, Bifidobacterium and Streptococcus pyogenes, which can accumulate, especially in tumours, and suppress their growth. However, in order to improve bacterial tumour-targeting ability and reduce bacterial virulence, various bacterial species have been genetically engineered through molecular biology techniques, thus forming different genetically engineered strains by using Clostridium novyi-NT, Listeria monocytogenes and Salmonella typhimurium. These genetically engineered bacteria can be selectively colonized in tumours and can inhibit cancer growth. In addition, they can also be used as live delivery carriers for cancer treatment, which can overcome the limitations of conventional antitumour therapies, such as high toxicity to normal tissue cells and the inability to treat deep tumour tissues. Potential targets including cytokines, cytotoxic agents, regulatory factors, prodrug-converting enzymes and small interfering RNAs (siRNAs) can be delivered via genetically engineered bacteria to treat cancer. However, various problems remain to be overcome before bacterial therapy can be used for clinical cancer treatment. The balance between bacterial virulence and anti-tumor ability is a key point, and more sophisticated genetic circuits need to be designed to modify the bacteria. While reducing the virulence of the bacteria, it is also important to enhance the ability of the bacteria to target to the tumor tissue to reduce the impact on other normal tissues. Genetic instability of bacteria is also a potential problem, as mutations may produce ineffective or deleterious phenotypes. However, with the development of synthetic biology, the above problems would be solved in the future, and bacterial therapy will be an approach with great potential for tumor treatment.