Synthetic Biology Journal ›› 2020, Vol. 1 ›› Issue (6): 621-634.DOI: 10.12211/2096-8280.2020-012
• Invited Review • Previous Articles Next Articles
Research advances in synthetic microbial communities
QU Zepeng, CHEN Moxian, CAO Zhaohui, ZUO Wenlong, CHEN Ye, DAI Lei
- Shenzhen Institute of Synthetic Biology,Shenzhen Institutes of Advanced Technology,Chinese Academy of Sciences,Shenzhen 518055,Guangdong,China
-
Received:2020-02-09Revised:2020-06-02Online:2021-01-15Published:2020-12-31 -
Contact:DAI Lei
合成微生物群落研究进展
曲泽鹏, 陈沫先, 曹朝辉, 左文龙, 陈业, 戴磊
- 中国科学院深圳先进技术研究院,深圳合成生物学创新研究院,中国科学院定量工程生物学重点实验室,广东 深圳 518055
-
通讯作者:戴磊 -
作者简介:曲泽鹏(1993—),男,硕士,研究助理。研究方向为合成微生物组学。E-mail:zp.qu@siat.ac.cn
戴磊(1987—),男,博士,研究员。研究方向为合成微生物组学。E-mail:lei.dai@siat.ac.cn -
基金资助:国家自然科学基金(31971513);深圳合成生物学创新研究院科研基金(ZTXM20190003)
CLC Number:
Cite this article
QU Zepeng, CHEN Moxian, CAO Zhaohui, ZUO Wenlong, CHEN Ye, DAI Lei. Research advances in synthetic microbial communities[J]. Synthetic Biology Journal, 2020, 1(6): 621-634.
曲泽鹏, 陈沫先, 曹朝辉, 左文龙, 陈业, 戴磊. 合成微生物群落研究进展[J]. 合成生物学, 2020, 1(6): 621-634.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2020-012
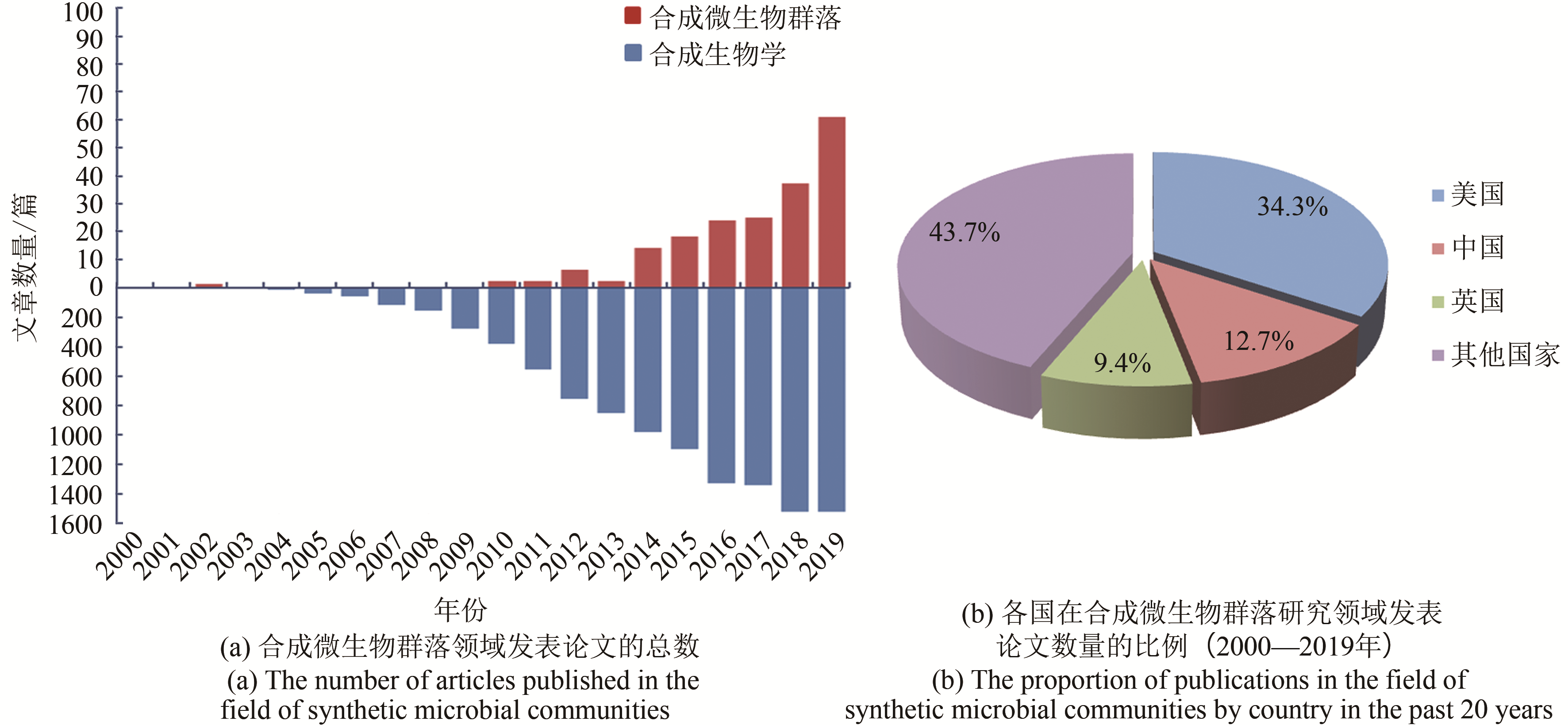
Fig. 1 Research trends in synthetic microbial communities(Based on the Web of Science database, search the publications in the past 20 years, in the field of synthetic microbial communities with‘synthetic microbial communities/consortia, synthetic microbiota/microbiome, microbiome engineering’ and in the field of synthetic biology with ‘synthetic biology’)
| 相互作用类型 | 研究案例 |
|---|---|
| 互利互惠(+/+) | Shou等[ |
| Summers等[ | |
| 偏利共生(+/0) | Freilich等[ |
| Kedia等[ | |
| Lu等[ | |
| 寄生或捕食(+/-) | Contreras-Moreno等[ |
| Zhang等[ | |
| 竞争(-/-) | Gadaga等[ |
| Gause[ |
Tab. 1 Examples of different types of microbial interactions
| 相互作用类型 | 研究案例 |
|---|---|
| 互利互惠(+/+) | Shou等[ |
| Summers等[ | |
| 偏利共生(+/0) | Freilich等[ |
| Kedia等[ | |
| Lu等[ | |
| 寄生或捕食(+/-) | Contreras-Moreno等[ |
| Zhang等[ | |
| 竞争(-/-) | Gadaga等[ |
| Gause[ |
| 1 | GROSSKOPF T, SOYER O S. Synthetic microbial communities[J]. Current Opinion in Microbiology, 2014, 18:72-77. |
| 2 | KAREN D R, MASSIMO M, PIETER V A, et al. Synthetic microbial ecosystems: an exciting tool to understand and apply microbial communities[J]. Environmental Microbiology, 2014, 16(6):1472-1481. |
| 3 | BERNSTEIN H C, CARLSON R P. Microbial consortia engineering for cellular factories: in vitro to in silico systems[J]. Computational and Structural Biotechnology Journal, 2012, 3(4):e201210017. |
| 4 | FALKOWSKI P G, FENCHEL T, DELONG E F. The microbial engines that drive Earth's biogeochemical cycles[J]. Science, 2008, 320(5879):1034-1039. |
| 5 | LEY R E, TURNBAUGH P J, KLEIN S, et al. Microbial ecology - human gut microbes associated with obesity[J]. Nature, 2006, 444(7122):1022-1023. |
| 6 | LUKENS J R, GURUNG P, VOGEL P, et al. Dietary modulation of the microbiome affects autoinflammatory disease[J]. Nature, 2014, 516(7530):246-249. |
| 7 | BERG G. Plant-microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture[J]. Applied Microbiology and Biotechnology, 2009, 84(1):11-18. |
| 8 | PATLE S, LAL B. Ethanol production from hydrolysed agricultural wastes using mixed culture of Zymomonas mobilis and Candida tropicalis [J]. Biotechnology Letters, 2007, 29(12):1839-1843. |
| 9 | PAWELCZYK S, ABRAHAM W R, HARMS H, et al. Community-based degradation of 4-chorosalicylate tracked on the single cell level[J]. Journal of Microbiological Methods, 2008, 75(1):117-126. |
| 10 | RAES J, BORK P. Systems microbiology - timeline - molecular eco-systems biology: towards an understanding of community function[J]. Nature Reviews Microbiology, 2008, 6(9):693-699. |
| 11 | PROSSER J I, BOHANNAN B J M, CURTIS T P, et al. The role of ecological theory in microbial ecology[J]. Nature Reviews Microbiology, 2007, 5(5):384-392. |
| 12 | VENTURELLI O S, CARR A V, FISHER G, et al. Deciphering microbial interactions in synthetic human gut microbiome communities[J]. Molecular Systems Biology, 2018, 14(6):e8157. |
| 13 | LIU F, MAO J W, KONG W T, et al. Interaction variability shapes succession of synthetic microbial ecosystems[J]. Nature Communications, 2020, 11(1):e309. |
| 14 | COYTE K Z, SCHLUTER J, FOSTER K R. The ecology of the microbiome: networks, competition, and stability[J]. Science, 2015, 350(6261):663-666. |
| 15 | BANERJEE S, SCHLAEPPI K, VAN DER H M G. A. Keystone taxa as drivers of microbiome structure and functioning[J]. Nature Reviews Microbiology, 2018, 16(9):567-576. |
| 16 | SONG H, DING M Z, JIA X Q, et al. Synthetic microbial consortia: from systematic analysis to construction and applications[J]. Chemical Society Reviews, 2014, 43(20):6954-6981. |
| 17 | FAUST K, RAES J. Microbial interactions: from networks to models[J]. Nature Reviews Microbiology, 2012, 10(8):538-550. |
| 18 | SHOU W Y, RAM S, VILAR J M G. Synthetic cooperation in engineered yeast populations[J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(6):1877-1882. |
| 19 | SUMMERS Z M, FOGARTY H E, LEANG C, et al. Direct exchange of electrons within aggregates of an evolved syntrophic coculture of anaerobic bacteria[J]. Science, 2010, 330(6009):1413-1415. |
| 20 | FREILICH S, ZARECKI R, EILAM O, et al. Competitive and cooperative metabolic interactions in bacterial communities[J]. Nature Communications, 2011, 2:e589. |
| 21 | KEDIA G, WANG R, PATEL H, et al. Use of mixed cultures for the fermentation of cereal-based substrates with potential probiotic properties[J]. Process Biochemistry, 2007, 42(1):65-70. |
| 22 | LU Y Y, PUTRA S D, LIU S Q. A novel non-dairy beverage from durian pulp fermented with selected probiotics and yeast[J]. International Journal of Food Microbiology, 2018, 265:1-8. |
| 23 | FRANCISCO J C, JOSE M, NATALIA I J, et al. Copper and melanin play a role in Myxococcus xanthus predation on Sinorhizobium meliloti[J]. Frontiers in Microbiology, 2020, 11:e94. |
| 24 | ZHANG W C, WANG Y, LU H N, et al. Dynamics of solitary predation by Myxococcus xanthus on Escherichia coli observed at the Single-Cell Level[J]. Applied and Environmental Microbiology, 2020, 86(3):e02286-19. |
| 25 | GADAGA T H, MUTUKUMIRA A N, NARVHUS J A. The growth and interaction of yeasts and lactic acid bacteria isolated from zimbabwean naturally fermented milk in UHT milk[J]. International Journal of Food Microbiology, 2001, 68(1):21-32. |
| 26 | GAUSE G F. The struggle for existence[J]. Williams &Wilkins, 1935, 16(4): 656-657. |
| 27 | NIU B, PAULSON J N, ZHENG X, et al. Simplified and representative bacterial community of maize roots[J]. Proceedings of the National Academy of Sciences of the United States of America, 2017, 114(12):2450-2459. |
| 28 | WINTERMUTE E H, SILVER P A. Dynamics in the mixed microbial concourse[J]. Genes & Development, 2010, 24(23):2603-2614. |
| 29 | BALAGADDE F K, SONG H, OZAKI J, et al. A synthetic Escherichia coli predator-prey ecosystem[J]. Molecular Systems Biology, 2008, 4:e187. |
| 30 | HU B, DU J, ZOU R Y, et al. An environment-sensitive synthetic microbial ecosystem[J]. PLoS One, 2010, 5(5):e10619. |
| 31 | HONG S H, HEGDE M, KIM J, et al. Synthetic quorum-sensing circuit to control consortial biofilm formation and dispersal in a microfluidic device[J]. Nature Communications, 2012, 3:e613. |
| 32 | CHEN Y, KIM J K, HIRNING A J, et al. Emergent genetic oscillations in a synthetic microbial consortium[J]. Science, 2015, 349(6251):986-989. |
| 33 | KONG W T, MELDGIN D R, COLLINS J J, et al. Designing microbial consortia with defined social interactions[J]. Nature Chemical Biology, 2018, 14(8):821-829. |
| 34 | KERNER A, PARK J, WILLIAMS A, et al. A programmable Escherichia coli consortium via tunable symbiosis[J]. PLoS One, 2012, 7(3):e34032. |
| 35 | KUBO I, HOSODA K, SUZUKI S, et al. Construction of bacteria-eukaryote synthetic mutualism[J]. Biosystems, 2013, 113(2):66-71. |
| 36 | HUANG A C, JIANG T, LIU Y X, et al. A specialized metabolic network selectively modulates Arabidopsis root microbiota[J]. Science, 2019, 364(6440):eaau6389. |
| 37 | TEIXEIRA P, COLAIANNI N R, FITZPATRICK C R, et al. Beyond pathogens: microbiota interactions with the plant immune system[J]. Current Opinion in Microbiology, 2019, 49:7-17. |
| 38 | RAWLS J F, MAHOWALD M A, LEY R E, et al. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection[J]. Cell, 2006, 127(2):423-433. |
| 39 | ROESELERS G, MITTGE E K, STEPHENS W Z, et al. Evidence for a core gut microbiota in the zebrafish[J]. ISME Journal, 2011, 5(10):1595-1608. |
| 40 | RATZKE C, GORE J. Modifying and reacting to the environmental pH can drive bacterial interactions[J]. PLoS Biology, 2018, 16(3):e2004248. |
| 41 | FOERSTNER K U, VON M C, BORK P. Comparative analysis of environmental sequences: potential and challenges[J]. Philosophical Transactions of the Royal Society B-Biological Sciences, 2006, 361(1467):519-523. |
| 42 | RAMETTE A. Multivariate analyses in microbial ecology[J]. FEMS Microbiology Ecology, 2007, 62(2):142-160. |
| 43 | BERTIN P N, MEDIGUE C, NORMAND P. Advances in environmental genomics: towards an integrated view of micro-organisms and ecosystems[J]. Microbiology-(UK), 2008, 154:347-359. |
| 44 | SHONG J, JIMENEZ D M R, COLLINS C H. Towards synthetic microbial consortia for bioprocessing[J]. Current Opinion in Microbiology, 2012, 23(5):798-802. |
| 45 | GARCIA F C, BESTION E, WARFIELD R, et al. Changes in temperature alter the relationship between biodiversity and ecosystem functioning[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(43):10989-10999. |
| 46 | WANG X F, WEI Z, YANG K M, et al. Phage combination therapies for bacterial wilt disease in tomato[J]. Nature Biotechnology, 2019, 37(12):1513-1520. |
| 47 | HSU B B, GIBSON T E, YELISEYEV V, et al. Dynamic modulation of the gut microbiota and metabolome by bacteriophages in a mouse model[J]. Cell Host & Microbe, 2019, 25(6):803-814. |
| 48 | LAWSON C E, HARCOMBE W R, HATZENPICHLER R, et al. Common principles and best practices for engineering microbiomes[J]. Nature Reviews Microbiology, 2019, 17(12):725-741. |
| 49 | STEIN R R, TANOUE T, SZABADY R L, et al. Computer-guided design of optimal microbial consortia for immune system modulation [J]. eLife, 2018, 7:e30916. |
| 50 | ZHAO C H, SINUMVAYO J P, ZHANG Y P, et al. Design and development of a ‘Y-shaped’ microbial consortium capable of simultaneously utilizing biomass sugars for efficient production of butanol [J]. Metabolic Engineering, 2019, 55:111-119. |
| 51 | PAPENFORT K, BASSLER B L. Quorum sensing signal-response systems in Gram-negative bacteria [J]. Nature Reviews Microbiology, 2016, 14(9):576-588. |
| 52 | MARTELLACCI L, QUARANTA G, PATINI R, et al. A Literature review of metagenomics and culturomics of the peri-implant microbiome: current evidence and future perspectives[J]. Materials, 2019, 12(18):e3010. |
| 53 | LAGIER J C, DUBOURG G, MILLION M, et al. Culturing the human microbiota and culturomics[J]. Nature Reviews Microbiology, 2018, 16(9):540-550. |
| 54 | SARHAN M S, HAMZA M A, YOUSSEF H H, et al. Culturomics of the plant prokaryotic microbiome and the dawn of plant-based culture media - a review[J]. Journal of Advanced Research, 2019, 19:15-27. |
| 55 | LAGIER J C, ARMOUGOM F, MILLION M, et al. Microbial culturomics: paradigm shift in the human gut microbiome study[J]. Clinical Microbiology and Infection, 2012, 18(12):1185-1193. |
| 56 | BAI Y, MULLER D B, SRINIVAS G, et al. Functional overlap of the Arabidopsis leaf and root microbiota [J]. Nature, 2015, 528(7582):364-369. |
| 57 | OBERHARDT M A, ZARECKI R, GRONOW S, et al. Harnessing the landscape of microbial culture media to predict new organism-media pairings[J]. Nature Communications, 2015, 6:8493. |
| 58 | WARD T L, LARSON J, MEULEMANS J, et al. BugBase predicts organism-level microbiome phenotypes [EB/OL]. [2017-05-02]. . |
| 59 | KAMINSKI T S, SCHELER O, GARSTECKI P. Droplet microfluidics for microbiology: techniques, applications and challenges[J]. Lab on a Chip, 2016, 16(12):2168-2187. |
| 60 | BEIN A, SHIN W, JALILI F S, et al. Microfluidic organ-on-a-chip models of human Intestine[J]. Cellular and Molecular Gastroenterology and Hepatology, 2018, 5(4):659-668. |
| 61 | ALEKLETT K, KIERS E T, OHLSSON P, et al. Build your own soil: exploring microfluidics to create microbial habitat structures[J]. Isme Journal, 2018, 12(2):312-319. |
| 62 | KEHE J, KULESA A, ORTIZ A, et al. Massively parallel screening of synthetic microbial communities[J]. Proceedings of the National Academy of Sciences of the United States of America, 2019, 116(26):12804-12809. |
| 63 | KEHE J, ORTIZ A, KULESA A, et al. Positive interactions are common among culturable bacteria [EB/OL]. [2020-06-25]. . |
| 64 | ALBERTSEN M, HUGENHOLTZ P, SKARSHEWSKI A, et al. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes[J]. Nature Biotechnology, 2013, 31(6):533-538. |
| 65 | HAWLEY A K, BREWER H M, NORBECK A D, et al. Metaproteomics reveals differential modes of metabolic coupling among ubiquitous oxygen minimum zone microbes[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(31):11395-11400. |
| 66 | SWENSON T L, KARAOZ U, SWENSON J M, et al. Linking soil biology and chemistry in biological soil crust using isolate exometabolomics[J]. Nature Communications, 2018, 9(1):19. |
| 67 | PERNTHALER J, GLOCKNER F O, SCHONHUBER W, et al. Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes[J]. Methods in Microbiology, 2001, 30:207-226. |
| 68 | GHOSH A, NILMEIER J, WEAVER D, et al. A peptide-based method for C-13 metabolic flux analysis in microbial communities[J]. PLoS Computational Biology, 2014, 10(9):e1003827. |
| 69 | GEBRESELASSIE N A, ANTONIEWICZ M R. C-13-metabolic flux analysis of co-cultures: a novel approach[J]. Metabolic Engineering, 2015, 31:132-139. |
| 70 | GILMORE I S, HEILES S, PIETERSE C L. Metabolic imaging at the single-cell scale: recent advances in mass spectrometry imaging[J]. Annual Review of Analytical Chemistry, 2019, 12(1):201-224. |
| 71 | DUNHAM S J B, ELLIS J F, LI B, et al. Mass spectrometry imaging of complex microbial communities[J]. Accounts of Chemical Research, 2017, 50(1):96-104. |
| 72 | NUNEZ J, RENSLOW R, CLIFF J B, et al. NanoSIMS for biological applications: current practices and analyses[J]. Biointerphases, 2018, 13(3):03B301. |
| 73 | VRANCKEN G, GREGORY A C, HUYS G R B, et al. Synthetic ecology of the human gut microbiota[J]. Nature Reviews Microbiology, 2019, 17(12)754-763. |
| 74 | JO J, OH J, PARK C. Microbial community analysis using high-throughput sequencing technology: a beginner's guide for microbiologists[J]. Journal of Microbiology, 2020, 58(3):176-192. |
| 75 | ZHANG X, DEEKE S A, NING Z B, et al. Metaproteomics reveals associations between microbiome and intestinal extracellular vesicle proteins in pediatric inflammatory bowel disease[J]. Nature Communications, 2018, 9(1):2873. |
| 76 | HEYER R, SCHALLERT K, SIEWERT C, et al. Metaproteome analysis reveals that syntrophy, competition, and phage-host interaction shape microbial communities in biogas plants[J]. Microbiome, 2019, 7(1):69. |
| 77 | FRANZOSA E A, MCIVER L J, RAHNAVARD Gholamali, et al. Species-level functional profiling of metagenomes and metatranscriptomes[J]. Nature Methods, 2018, 15(11):962-968. |
| 78 | KNIEF C, DELMOTTE N, CHAFFRON S, et al. Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice[J]. ISME Journal, 2012, 6(7):1378-1390. |
| 79 | BROBERG M, DOONAN J, MUNDT F, et al. Integrated multi-omic analysis of host-microbiota interactions in acute oak decline[J]. Microbiome, 2018, 6(1):21. |
| 80 | PERNTHALER A, DEKAS A E, BROWN C T, et al. Diverse syntrophic partnerships from-deep-sea methane vents revealed by direct cell capture and metagenomics[J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(19):7052-7057. |
| 81 | SEKIGUCHI Y, KAMAGATA Y, NAKAMURA K, et al. Fluorescence in situ hybridization using 16S rRNA-targeted oligonucleotides reveals localization of methanogens and selected uncultured bacteria in mesophilic and thermophilic sludge granules[J]. Applied and Environmental Microbiology, 1999, 65(3):1280-1288. |
| 82 | NAVA G M, FRIEDRICHSEN H J, STAPPENBECK T S. Spatial organization of intestinal microbiota in the mouse ascending colon[J]. ISME Journal, 2011, 5(4):627-638. |
| 83 | SWIDSINSKI A, WEBER J, LOENING-BAUCKE V, et al. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease[J]. Journal of Clinical Microbiology, 2005, 43(7):3380-3389. |
| 84 | WHITAKER W R, SHEPHERD E S, SONNENBURG J L. Tunable expression tools enable single-cell strain distinction in the gut microbiome[J]. Cell, 2017, 169(3):538-546. |
| 85 | GEVA-ZATORSKY N, ALVAREZ D, HUDAK J E, et al. In vivo imaging and tracking of host-microbiota interactions via metabolic labeling of gut anaerobic bacteria[J]. Nature Medicine, 2015, 21(9):1091-1100. |
| 86 | PRAVESCHOTINUNT P, DORVAL C N M, DEN H I, et al. Tracking of engineered bacteria in vivo using nonstandard amino acid incorporation[J]. ACS Synthetic Biology, 2018, 7(6):1640-1650. |
| 87 | EARLE K A, BILLINGS G, SIGAL M, et al. Quantitative imaging of gut microbiota spatial organization[J]. Cell Host Microbe, 2015, 18(4):478-488. |
| 88 | MARK WELCH J. L., HASEGAWA Y., MCNULTY N. P., et al. Spatial organization of a model 15-member human gut microbiota established in gnotobiotic mice[J]. Proceedings of the National Academy of Sciences of the United States of America, 2017, 114(43):9105-9114. |
| 89 | WELCH J L M, ROSSETTI Blair J, RIEKEN C W, et al. Biogeography of a human oral microbiome at the micron scale[J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(6):791-800. |
| 90 | WILBERT S A, WELCH J L M, BORISY G G. Spatial ecology of the human tongue dorsum microbiome[J]. Cell Host & Microbe, 2020, 30(12):4003-4015. |
| 91 | PINTO F, MEDINA D A, PEREZ-CORREA J R, et al. Modeling metabolic interactions in a consortium of the infant gut microbiome[J]. Frontiers in Microbiology, 2017, 8:e2507. |
| 92 | THIELE I, PALSSON B O. A protocol for generating a high-quality genome-scale metabolic reconstruction[J]. Nature Protocols, 2010, 5(1):93-121. |
| 93 | GONZE D, LAHTI L, RAES J, et al. Multi-stability and the origin of microbial community types[J]. ISME Journal, 2017, 11(10):2159-2166. |
| 94 | GOYAL A, DUBINKINA V, MASLOV S. Multiple stable states in microbial communities explained by the stable marriage problem[J]. ISME Journal, 2018, 12(12):2823-2834. |
| 95 | GIBSON T E, BASHAN A, CAO H T, et al. On the origins and control of community types in the human microbiome[J]. PLoS Computational Biology, 2016, 12(2):e1004688. |
| 96 | CARLSTRÖM C I, FIELD C M, BORTFELD-MILLER M, et al. Synthetic microbiota reveal priority effects and keystone strains in the Arabidopsis phyllosphere[J]. Nature Ecology & Evolution, 2019, 3(10):1445-1454. |
| 97 | GONZE D, COYTE Katharine Z, LAHTI L, et al. Microbial communities as dynamical systems[J]. Current Opinion in Microbiology, 2018, 44:41-49. |
| 98 | CAO H T, GIBSON T E, BASHAN A, et al. Inferring human microbial dynamics from temporal metagenomics data: pitfalls and lessons[J]. Bioessays, 2017, 39(2):e1600188. |
| 99 | XIAO Y D, TULIO A M, FRIEDMAN J, et al. Mapping the ecological networks of microbial communities[J]. Nature Communications, 2017, 8(1):2042. |
| 100 | FRIEDMAN J, HIGGINS L M, GORE J. Community structure follows simple assembly rules in microbial microcosms [J]. Nature Ecology & Evolution, 2017, 1(5):0109. |
| 101 | LI C, CHNG K R, KWAH J S, et al. An expectation-maximization algorithm enables accurate ecological modeling using longitudinal microbiome sequencing data [J]. Microbiome, 2019, 7(1):118. |
| 102 | BUCCI V, TZEN B, LI N, et al. MDSINE: Microbial dynamical systems inference engine for microbiome time-series analyses[J]. Genome Biology, 2016, 17(1):e121. |
| 103 | GOTTSTEIN W, OLIVIER B G, BRUGGEMAN F J, et al. Constraint-based stoichiometric modelling from single organisms to microbial communities[J]. Journal of the Royal Society Interface, 2016, 13(124):e20160627. |
| 104 | KETTLE H, LOUIS P, HOLTROP G, et al. Modelling the emergent dynamics and major metabolites of the human colonic microbiota[J]. Environmental Microbiology, 2015, 17(5):1615-1630. |
| 105 | PACHECO Alan R, MOEL Mauricio, SEGRE Daniel. Costless metabolic secretions as drivers of interspecies interactions in microbial ecosystems[J]. Nature Communications, 2019, 10(1):103. |
| 106 | GOLDFORD J E, LU N X, BAJIC D, et al. Emergent simplicity in microbial community assembly[J]. Science, 2018, 361(6401):469-474. |
| 107 | HUTTENHOWER C. Structure, function and diversity of the healthy human microbiome[J]. Nature, 2012, 486:207-214. |
| 108 | LLOYD-PRICE J, ABU-ALI, G, HUTTENHOWER C. The healthy human microbiome[J]. Genome Medicine, 2016, 8:51-62. |
| 109 | GENSOLLEN T, IYER S S, KASPER D L, et al. How colonization by microbiota in early life shapes the immune system[J]. Science, 2016, 352:539-544. |
| 110 | HONDA K, LITTMAN D R. The microbiota in adaptive immune homeostasis and disease[J]. Nature, 2016, 535:75-84. |
| 111 | CLEMENTE J C, URSELL L K, PARFREY L W, et al. The impact of the gut microbiota on human health: an integrative view[J]. Cell, 2012, 148(6):1258-1270. |
| 112 | BÄCKHED F, DING H, WANG T, et al. The gut microbiota as an environmental factor that regulates fat storage[J]. Proceedings of the National Academy of Sciences of the United States of America, 2004, 101:15718-15723. |
| 113 | LEY R E, BÄCKHED F, TURNBAUGH P, et al. Obesity alters gut microbial ecology[J]. Proceedings of the National Academy of Sciences of the United States of America, 2005, 102:11070-11075. |
| 114 | RIDAURA V K. Gut microbiota from twins discordant for obesity modulate metabolism in mice[J]. Science, 2013, 341(6150):e1241214. |
| 115 | TURNBAUGH P J. A core gut microbiome in obese and lean twins[J]. Nature, 2009, 457:480-484. |
| 116 | KARLSSON F H. Gut metagenome in European women with normal impaired and diabetic glucose control[J]. Nature, 2013, 498:99-103. |
| 117 | LARSEN N. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults[J]. PLoS One, 2010, 5:9085. |
| 118 | QIN J J, LI R Q, RAES J, et al. A human gut microbial gene catalogue established by metagenomic sequencing[J]. Nature, 2010, 464:59-65. |
| 119 | TILG H, MOSCHEN A R. Microbiota and diabetes: an evolving relationship[J]. Gut Microbes, 2014, 63:1513-1521. |
| 120 | CANI P D. Human gut microbiome: hopes, threats and promises[J]. Gut Microbes, 2018, 67:1716-1725. |
| 121 | SCHMIDT T S B, RAES J, BORK P. The human gut microbiome: from association to modulation[J]. Cell, 2018, 172:1198-1215. |
| 122 | GIANOTTI R J, MOSS A C. Fecal microbiota transplantation: from Clostridium difficile to inflammatory bowel disease[J]. Gastroenterology & Hepatology, 2017, 13(4):209-213. |
| 123 | ANDERSON J L, EDNEY R J, WHELAN K. Systematic review: faecal microbiota transplantation in the management of inflammatory bowel disease[J]. Alimentary Pharmacology & Therapeutics, 2012, 36(6):503-516. |
| 124 | ZHANG F M, WANG H G, WANG M, et al. Fecal microbiota transplantation for severe enterocolonic fistulizing Crohn's disease[J]. World Journal of Gastroenterology, 2013, 19(41):7213-7216. |
| 125 | WILLEM M V. Fame and future of faecal transplantations - developing next-generation therapies with synthetic microbiomes[J]. Microbial Biotechnology, 2013, 6(4):316-325. |
| 126 | ATARASHI K, TANOUE T, SHIMA T, et al. Induction of colonic regulatory T cells by indigenous clostridium species[J]. Science, 2011, 331(6015):337-341. |
| 127 | CHIBA T, SENO H. Indigenous clostridium species regulate systemic immune responses by induction of colonic regulatory T cells[J]. Gastroenterology, 2011, 141(3):1114-1116. |
| 128 | TANOUE T, MORITA S, PLICHTA D R, et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity[J]. Nature, 2019, 565(7741):600-605. |
| 129 | BERENDSEN R L, PIETERSE C M J, BAKKER P. The rhizosphere microbiome and plant health[J]. Trends in Plant Science, 2012, 17(8):478-486. |
| 130 | ZHANG J Y, LIU Y X, ZHANG N, et al. NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice[J]. Nature Biotechnology, 2019, 37(6):676-684. |
| 131 | FINKEL O M, SALAS-GONZALEZ I, CASTRILLO G, et al. The effects of soil phosphorus content on plant microbiota are driven by the plant phosphate starvation response[J]. PLoS Biology, 2019, 17(11):e3000534. |
| 132 | LEBEIS S L, PAREDES S H, LUNDBERG D S, et al. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa[J]. Science, 2015, 349(6250):860-864. |
| 133 | CASTRILLO G, TEIXEIRA P J P L, PAREDES S H, et al. Root microbiota drive direct integration of phosphate stress and immunity[J]. Nature, 2017, 543(7646):513-518. |
| 134 | WEI Z, GU Y, FRIMAN V P, et al. Initial soil microbiome composition and functioning predetermine future plant health[J]. Science Advances, 2019, 5(9):eaaw0759. |
| 135 | DURÁN P, THIERGART T, GARRIDO-OTER R, et al. Microbial interkingdom interactions in roots promote Arabidopsis survival[J]. Cell, 2018, 175(4):973-983. |
| 136 | LI M, WEI Z, WANG J N, et al. Facilitation promotes invasions in plant-associated microbial communities[J]. Ecology Letters, 2019, 22(1):149-158. |
| 137 | LIU Y X, QIN Y, BAI Y. Reductionist synthetic community approaches in root microbiome research[J]. Current Opinion in Microbiology, 2019, 49:97-102. |
| 138 | SYED AB RAHMAN S F, SINGH E, PIETERSE C M J, et al. Emerging microbial biocontrol strategies for plant pathogens[J]. Plant Science : an International Journal of Experimental Plant Biology, 2018, 267:102-111. |
| 139 | AZIZOGLU U. Bacillus thuringiensis as a biofertilizer and biostimulator: a mini-review of the little-known plant growth-promoting properties of Bt[J]. Current Microbiology, 2019, 76(11):1379-1385. |
| 140 | 刘炜伟, 吴冰, 向梅春, 等. 从微生物组到合成功能菌群[J]. 微生物学通报, 2017, 44(4):881-889. |
| LIU W W, WU B, XIANG M C, et al. From microbiome to synthetic microbial community[J]. Microbiology China, 2017, 44(4):881-889. | |
| 141 | 王丽娟. 产乙偶姻功能微生物群落的分离及其在食醋酿造中的应用 [D]. 无锡:江南大学, 2018. |
| WANG L J. Isolation of acetoin-producing microbial community in vinegar Pei and its application in vinegar brewing process[D]. Wuxi: Jiangnan University, 2018. | |
| 142 | WU Q, LING J, XU Y. Starter culture selection for making Chinese sesame-flavored liquor based on microbial metabolic activity in mixed-culture fermentation[J]. Applied and Environmental Microbiology, 2014, 80(14):4450-4459. |
| 143 | CHEN Y L. Development and application of co-culture for ethanol production by co-fermentation of glucose and xylose: a systematic review[J]. Journal of Industrial Microbiology & Biotechnology, 2011, 38(5):581-597. |
| 144 | ABDALLA M A, SULIEMAN S, MCGAW L J. Microbial communication: a significant approach for new leads[J]. South African Journal of Botany, 2017, 113:461-470. |
| 145 | SCHROECKH V, SCHERLACH K, NUTZMANN H W, et al. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans [J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(34):14558-14563. |
| 146 | ZHOU K, QIAO K J, EDGAR S, et al. Distributing a metabolic pathway among a microbial consortium enhances production of natural products[J]. Nature Biotechnology, 2015, 33(4):377-383. |
| 147 | RICHARDSON M. Pesticides - friend or foe?[J]. Water Science and Technology, 1998, 37(8):19-25. |
| 148 | DEJONGHE W, BERTELOOT E, GORIS J, et al. Synergistic degradation of linuron by a bacterial consortium and isolation of a single linuron-degrading Variovorax strain[J]. Applied and Environmental Microbiology, 2003, 69(3):1532-1541. |
| 149 | HEIJDEN M G A VAN DER, KLIRONOMOS J N, URSIC M, et al. Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity[J]. Nature, 1998, 396(6706):69-72. |
| 150 | BRENNER K, YOU L C, ARNOLD F H. Engineering microbial consortia: a new frontier in synthetic biology[J]. Trends in Biotechnology, 2008, 26(9):483-489. |
| [1] | GAO Ge, BIAN Qi, WANG Baojun. Synthetic genetic circuit engineering: principles, advances and prospects [J]. Synthetic Biology Journal, 2025, 6(1): 45-64. |
| [2] | LI Jiyuan, WU Guosheng. Two hypothesises for the origins of organisms from the synthetic biology perspective [J]. Synthetic Biology Journal, 2025, 6(1): 190-202. |
| [3] | JIAO Hongtao, QI Meng, SHAO Bin, JIANG Jinsong. Legal issues for the storage of DNA data [J]. Synthetic Biology Journal, 2025, 6(1): 177-189. |
| [4] | TANG Xinghua, LU Qianneng, HU Yilin. Philosophical reflections on synthetic biology in the Anthropocene [J]. Synthetic Biology Journal, 2025, 6(1): 203-212. |
| [5] | XU Huaisheng, SHI Xiaolong, LIU Xiaoguang, XU Miaomiao. Key technologies for DNA storage: encoding, error correction, random access, and security [J]. Synthetic Biology Journal, 2025, 6(1): 157-176. |
| [6] | SHI Ting, SONG Zhan, SONG Shiyi, ZHANG Yi-Heng P. Job. In vitro BioTransformation (ivBT): a new frontier of industrial biomanufacturing [J]. Synthetic Biology Journal, 2024, 5(6): 1437-1460. |
| [7] | CHAI Meng, WANG Fengqing, WEI Dongzhi. Synthesis of organic acids from lignocellulose by biotransformation [J]. Synthetic Biology Journal, 2024, 5(6): 1242-1263. |
| [8] | SHAO Mingwei, SUN Simian, YANG Shimao, CHEN Guoqiang. Bioproduction based on extremophiles [J]. Synthetic Biology Journal, 2024, 5(6): 1419-1436. |
| [9] | CHEN Yu, ZHANG Kang, QIU Yijing, CHENG Caiyun, YIN Jingjing, SONG Tianshun, XIE Jingjing. Progress of microbial electrosynthesis for conversion of CO2 [J]. Synthetic Biology Journal, 2024, 5(5): 1142-1168. |
| [10] | ZHENG Haotian, LI Chaofeng, LIU Liangxu, WANG Jiawei, LI Hengrun, NI Jun. Design, optimization and application of synthetic carbon-negative phototrophic community [J]. Synthetic Biology Journal, 2024, 5(5): 1189-1210. |
| [11] | CHEN Ziling, XIANG Yangfei. Integrated development of organoid technology and synthetic biology [J]. Synthetic Biology Journal, 2024, 5(4): 795-812. |
| [12] | CAI Bingyu, TAN Xiangtian, LI Wei. Advances in synthetic biology for engineering stem cell [J]. Synthetic Biology Journal, 2024, 5(4): 782-794. |
| [13] | XIE Huang, ZHENG Yilei, SU Yiting, RUAN Jingyi, LI Yongquan. An overview on reconstructing the biosynthetic system of actinomycetes for polyketides production [J]. Synthetic Biology Journal, 2024, 5(3): 612-630. |
| [14] | ZHA Wenlong, BU Lan, ZI Jiachen. Advances in synthetic biology for producing potent pharmaceutical ingredients of traditional Chinese medicine [J]. Synthetic Biology Journal, 2024, 5(3): 631-657. |
| [15] | HUI Zhen, TANG Xiaoyu. Applications of the CRISPR/Cas9 editing system in the study of microbial natural products [J]. Synthetic Biology Journal, 2024, 5(3): 658-671. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||

