Synthetic Biology Journal ›› 2021, Vol. 2 ›› Issue (2): 194-221.DOI: 10.12211/2096-8280.2020-080
• Invited Review • Previous Articles Next Articles
Recent advances in metabolic engineering of clostridia for n-butanol production
WEN Zhiqiang1, SUN Xiaoman1, WANG Qingzhuo1, LI Yanan1, LIU Wenzheng1, JIANG Yu2, YANG Sheng2,3
- 1.School of Food Science and Pharmaceutical Engineering,Nanjing Normal University,Nanjing 210046,Jiangsu,China
2.Huzhou Center of Industrial Biotechnology,Shanghai Institutes of Biological Sciences,Chinese Academy of Sciences,Huzhou 313000,Zhejiang,China
3.Key Laboratory of Synthetic Biology,CAS Center for Excellence in Molecular Plant Sciences,Shanghai Institute of Plant Physiology and Ecology,Chinese Academy of Sciences,Shanghai 200032,China
-
Received:2020-10-22Revised:2021-02-09Online:2021-04-30Published:2021-04-30 -
Contact:YANG Sheng
梭菌正丁醇代谢工程研究进展
闻志强1, 孙小曼1, 汪庆卓1, 李亚楠1, 刘文正1, 蒋宇2, 杨晟2,3
- 1.南京师范大学食品与制药工程学院,江苏 南京 210046
2.中国科学院上海生命科学研究院湖州工业生物技术研发中心,浙江 湖州 313000
3.中国科学院分子植物科学卓越创新中心,中科院合成生物学重点实验室,上海 200032
-
通讯作者:杨晟 -
作者简介:闻志强 (1985—),男,博士,讲师,研究方向为合成梭菌菌群代谢工程。E-mail:zqwen@njnu.edu.cn
杨晟(1973—),男,博士,研究员,研究方向为合成生物催化剂工程与基因编辑。E-mail:syang@sibs.ac.cn -
基金资助:国家自然科学基金杰出青年科学基金(21825804);国家自然科学基金创新研究群体科学基金(31921006);国家自然科学基金青年项目(21706133)
CLC Number:
Cite this article
WEN Zhiqiang, SUN Xiaoman, WANG Qingzhuo, LI Yanan, LIU Wenzheng, JIANG Yu, YANG Sheng. Recent advances in metabolic engineering of clostridia for n-butanol production[J]. Synthetic Biology Journal, 2021, 2(2): 194-221.
闻志强, 孙小曼, 汪庆卓, 李亚楠, 刘文正, 蒋宇, 杨晟. 梭菌正丁醇代谢工程研究进展[J]. 合成生物学, 2021, 2(2): 194-221.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2020-080

Fig. 1 n-Butanol biosynthesis pathway in microorganism(1—Phosphoenolpyruvate carboxylase; 2—Aspartate transaminase; 3—Citramalate synthase; 4—3-isopropylmalate dehydrogenase/isomerase; 5—Threonine dehydratase; 6—2-isopropylmalate synthase; 7,8—3-isopropylmalate isomerase; 9—3-isopropylmalate dehydrogenase; 10—2-ketoacid decarboxylase 11—pyruvate kinase; 12—pyruvate dehydrogenase complex/pyruvate formate lyase/ pyruvate-ferredoxin oxidoreductase; 13—thiolase/acetyl-CoA transacetylase; 14—acetyl CoA carboxylase; 15—acetoacetyl-CoA synthase; 16—3-hydroxybutyryl-CoA dehydrogenase/hydroxyalkyl-CoA dehydrogenase; 17—crotonase; 18—butyryl-CoA dehydrogenase/trans-2-enoyl-CoA reductase; 19—acetaldehyde/butyraldehyde dehydrogenase; 20— ethanol/butanol dehydrogenase; 21—acetyl-CoA transacylase; 22—malonyl-CoA transacylase; 23—β-ketoacyl-ACP synthase; 24—β-ketoacyl-ACP reductase; 25—β-ketoacyl-ACP dehydratase; 26—enoyl-ACP reductase; 27—acyl-ACP thioesterase; 28—carboxylic acid reductase; 29—glycine oxidase; 30—malate synthase; DHAP—dihydroxyacetone phosphate)
| 菌株 | 底物 | 代谢途径 | 正丁醇/g·L-1 | 文献 |
|---|---|---|---|---|
| E. coli | 甘油 | CoA依赖的途径 | 0.552 | [ |
| E. coli | 葡萄糖 | CoA依赖的途径 | 15 | [ |
| E. coli | 葡萄糖 | 反式β氧化途径 | 14 | [ |
| E. coli | 葡萄糖 | ACP依赖的途径 | 0.3 | [ |
| E. coli | 葡萄糖 | 2-酮酸依赖的途径 | 1 | [ |
| E. coli | 葡萄糖+丁酸 | (截短的)CoA依赖的途径 | 6.2 | [ |
| E. coli | 葡萄糖+甘油 | CoA依赖的途径 | 18.3 | [ |
| E. coli | 葡萄糖 | CoA依赖的途径 | 20 | [ |
| S. cerevisiae | 半乳糖 | CoA依赖的途径 | 0.00 025 | [ |
| S. cerevisiae | 葡萄糖 | CoA依赖的途径 | 0.120 | [ |
| S. cerevisiae | 葡萄糖+甘氨酸 | 2-酮酸依赖的途径 | 0.092 | [ |
| S. cerevisiae | 葡萄糖 | 2-酮酸依赖的途径 | 0.2428 | [ |
| S. cerevisiae | 葡萄糖 | 2-酮酸依赖的途径 | 0.835 | [ |
| Y. lipolytica | 葡萄糖 | CoA依赖的途径 | 0.123 | [ |
| B. subtilis | 甘油 | CoA依赖的途径 | 0.024 | [ |
| L. brevis | 葡萄糖 | CoA依赖的途径 | 0.3 | [ |
| Synechococcus 7942 | CO2 | Malonyl-CoA (CoA)依赖的途径 | 0.404 | [ |
| S.7942 | CO2 | CoA依赖的途径 | 0.030 | [ |
| S.7942 | CO2 | CoA依赖的途径 | 0.404 | [ |
| S.7942 | CO2 | CoA依赖的途径 | 0.4187 | [ |
| Synechocystis PCC 6803 | CO2 | CoA依赖的途径 | 4.8 | [ |
| K. pneumonia | 甘油 | CoA依赖的途径 | 0.0150 | [ |
| K. pneumonia | 甘油 | 2-酮酸依赖的途径 | 0.0287 | [ |
| K. pneumonia | 甘油 | 2-酮酸依赖的途径 | 0.100 | [ |
| P. putida | 甘油 | CoA依赖的途径 | 0.122 | [ |
| Thermoanaerobacterium saccharolyticum | 木糖 | CoA依赖的途径 | 1.05 | [ |
| C. tyrobutyricum | 葡萄糖 | CoA依赖的途径 | 26.2 | [ |
| C. saccharoperbutylacetonicum N1-4 | 葡萄糖+木糖 | CoA依赖的途径 | 16 | [ |
| C. pasteurianum | 甘油+葡萄糖 | CoA依赖的途径 | 21.1 | [ |
| C. acetobutylicum ATCC 824 | 葡萄糖 | CoA依赖的途径 | 11.27 | [ |
| C. beijerinckii BA101 | 葡萄糖 | CoA依赖的途径 | 18.6 | [ |
| C. cellulovorans | 碱处理玉米棒芯 | CoA依赖的途径 | 4.97 | [ |
| C. ljungdahlii | CO2 | CoA依赖的途径 | 0.148 | [ |
| C. carboxidivorans P7 | CO2 | CoA依赖的途径 | 1.67 | [ |
| C. autoethanogenum | CO | CoA依赖的途径 | 1.54 | [ |
| C. cellulovorans/ C. beijerinckii | 碱处理玉米棒芯 | CoA依赖的途径 | 8.3 | [ |
| C. cellulovorans/ C. beijerinckii | 碱处理玉米棒芯 | CoA依赖的途径 | 11.5 | [ |
Tab. 1 Comparison of n-butanol production by various microorganisms
| 菌株 | 底物 | 代谢途径 | 正丁醇/g·L-1 | 文献 |
|---|---|---|---|---|
| E. coli | 甘油 | CoA依赖的途径 | 0.552 | [ |
| E. coli | 葡萄糖 | CoA依赖的途径 | 15 | [ |
| E. coli | 葡萄糖 | 反式β氧化途径 | 14 | [ |
| E. coli | 葡萄糖 | ACP依赖的途径 | 0.3 | [ |
| E. coli | 葡萄糖 | 2-酮酸依赖的途径 | 1 | [ |
| E. coli | 葡萄糖+丁酸 | (截短的)CoA依赖的途径 | 6.2 | [ |
| E. coli | 葡萄糖+甘油 | CoA依赖的途径 | 18.3 | [ |
| E. coli | 葡萄糖 | CoA依赖的途径 | 20 | [ |
| S. cerevisiae | 半乳糖 | CoA依赖的途径 | 0.00 025 | [ |
| S. cerevisiae | 葡萄糖 | CoA依赖的途径 | 0.120 | [ |
| S. cerevisiae | 葡萄糖+甘氨酸 | 2-酮酸依赖的途径 | 0.092 | [ |
| S. cerevisiae | 葡萄糖 | 2-酮酸依赖的途径 | 0.2428 | [ |
| S. cerevisiae | 葡萄糖 | 2-酮酸依赖的途径 | 0.835 | [ |
| Y. lipolytica | 葡萄糖 | CoA依赖的途径 | 0.123 | [ |
| B. subtilis | 甘油 | CoA依赖的途径 | 0.024 | [ |
| L. brevis | 葡萄糖 | CoA依赖的途径 | 0.3 | [ |
| Synechococcus 7942 | CO2 | Malonyl-CoA (CoA)依赖的途径 | 0.404 | [ |
| S.7942 | CO2 | CoA依赖的途径 | 0.030 | [ |
| S.7942 | CO2 | CoA依赖的途径 | 0.404 | [ |
| S.7942 | CO2 | CoA依赖的途径 | 0.4187 | [ |
| Synechocystis PCC 6803 | CO2 | CoA依赖的途径 | 4.8 | [ |
| K. pneumonia | 甘油 | CoA依赖的途径 | 0.0150 | [ |
| K. pneumonia | 甘油 | 2-酮酸依赖的途径 | 0.0287 | [ |
| K. pneumonia | 甘油 | 2-酮酸依赖的途径 | 0.100 | [ |
| P. putida | 甘油 | CoA依赖的途径 | 0.122 | [ |
| Thermoanaerobacterium saccharolyticum | 木糖 | CoA依赖的途径 | 1.05 | [ |
| C. tyrobutyricum | 葡萄糖 | CoA依赖的途径 | 26.2 | [ |
| C. saccharoperbutylacetonicum N1-4 | 葡萄糖+木糖 | CoA依赖的途径 | 16 | [ |
| C. pasteurianum | 甘油+葡萄糖 | CoA依赖的途径 | 21.1 | [ |
| C. acetobutylicum ATCC 824 | 葡萄糖 | CoA依赖的途径 | 11.27 | [ |
| C. beijerinckii BA101 | 葡萄糖 | CoA依赖的途径 | 18.6 | [ |
| C. cellulovorans | 碱处理玉米棒芯 | CoA依赖的途径 | 4.97 | [ |
| C. ljungdahlii | CO2 | CoA依赖的途径 | 0.148 | [ |
| C. carboxidivorans P7 | CO2 | CoA依赖的途径 | 1.67 | [ |
| C. autoethanogenum | CO | CoA依赖的途径 | 1.54 | [ |
| C. cellulovorans/ C. beijerinckii | 碱处理玉米棒芯 | CoA依赖的途径 | 8.3 | [ |
| C. cellulovorans/ C. beijerinckii | 碱处理玉米棒芯 | CoA依赖的途径 | 11.5 | [ |

Fig. 2 Milestones in the development of genetic manipulation tools for clostridia(Blue font represents genetic manipulation tools based on homologous recombination, and red represents genetic manipulation tools independent on homologous recombination)
| 遗传操作工具 | 原理及效果 | 优点 | 缺点 | 文献 |
|---|---|---|---|---|
| RNA干扰 | 利用反义RNA干扰靶标基因转录 | 周期短,见效快 | 设计和操作烦琐,仅在转录水平控制 | [ |
| 二型内含子插入失活 | 二型内含子可重编程靶向插入目的基因的特定位置 | 操作简单,几乎适用于所有梭菌 | 有一定的脱靶概率,可能造成极性效应 | [ |
| 转座 | 利用Mariner转座子在梭菌染色体上随机插入使基因失活 | 随机插入失活,适合建库 | 不可用于靶向敲除 | [ |
| 小RNA介导基因下调 | 小RNA形成的特定结构影响目的mRNA的转录水平 | 周期短,见效快 | 设计和操作烦琐,仅在转录水平上能调控 | [ |
| 整合酶介导的染色体整合 | 噬菌体丝氨酸整合酶将目标基因整合到染色体预先设计的整合位点上 | 可以很方便进行大片段整合 | 需先将整合位点插入染色体靶标位置 | [ |
| 等位基因替换 | 依赖同源重组单交换及之后的二次交换 | 可精确进行基因编辑 | 梭菌同源重效率低,过程耗时长 | [ |
| 反筛标记介导的等位基因替换 | 在等位基因替换基础上,在同源臂内设计pyrE、mazF等反筛标记,方便筛选二次交换突变株 | 相比纯粹的等位基因替换,效率有所提升 | 受制于第一次单交换效率 | [ |
| I-SceI归巢内切酶介导的等位基因替换 | 在等位基因替换基础上,在同源臂内设计I-SceI酶切位点,表达I-SceI切割双键,促进二次交换或帮助筛选二次交换突变株 | 相比纯粹的等位基因替换,效率有所提升 | 受制于第一次单交换效率 | [ |
| 单链寡核苷酸介导的点突变 | 重组蛋白RecT介导单链寡核苷酸更强的侵入和重组 | 可在较短同源臂时实现点突变 | 同源重组效率仍然较低 | [ |
| CRISPR/Cas9系统介导的基因编辑 | Cas9在sgRNA引导下切割靶标DNA,造成双键断裂(平末端),依赖同源重组修复 | 增加第一次单交换效率,可实现多功能编辑 | 双键断裂的毒性大,转化子很难获得 | [ |
| CRISPR/nCas9系统介导的基因编辑 | 失活Cas9蛋白的其中一个切割蛋白结构域,使其仅能单链切割靶标DNA,造成缺刻,然后依赖同源重组修复 | 相比CRISPR/Cas9毒性有所降低,转化子数目增加 | 仍有毒性,转化子依然偏少 | [ |
| CRISPR/Cpf1系统介导的基因编辑 | Cpf1在sgRNA引导下切割靶标DNA,造成双键断裂(有悬端),依赖同源重组修复 | 悬端的存在有利于DNA修复;脱靶概率比CRISPR/Cas9低 | 仍有毒性,转化子依然偏少 | [ |
| CRISPR/dCas9系统介导的基因下调 | 失活Cas9蛋白的全部两个切割蛋白结构域,使其仅能与靶标DNA结合,利用位阻效应降低mRNA转录水平 | 对靶标基因转型转录调控,尤其适用于必需基因的操作 | 依赖于sgRNA和基因 | [ |
| CRISPR/Cas系统辅助的碱基编辑 | 将胞嘧啶脱氨酶和尿嘧啶DNA糖苷酶与Cas蛋白融合表达,在sgRNA引导下对目标碱基实现C-G到T-A的替换 | 理论上可对任何碱基进行编辑,不依赖于同源重组 | 碱基转换能力待拓展,编辑效率有待提升 | [ |
Tab. 2 Comparison of different genetic tools applicable in Clostridium
| 遗传操作工具 | 原理及效果 | 优点 | 缺点 | 文献 |
|---|---|---|---|---|
| RNA干扰 | 利用反义RNA干扰靶标基因转录 | 周期短,见效快 | 设计和操作烦琐,仅在转录水平控制 | [ |
| 二型内含子插入失活 | 二型内含子可重编程靶向插入目的基因的特定位置 | 操作简单,几乎适用于所有梭菌 | 有一定的脱靶概率,可能造成极性效应 | [ |
| 转座 | 利用Mariner转座子在梭菌染色体上随机插入使基因失活 | 随机插入失活,适合建库 | 不可用于靶向敲除 | [ |
| 小RNA介导基因下调 | 小RNA形成的特定结构影响目的mRNA的转录水平 | 周期短,见效快 | 设计和操作烦琐,仅在转录水平上能调控 | [ |
| 整合酶介导的染色体整合 | 噬菌体丝氨酸整合酶将目标基因整合到染色体预先设计的整合位点上 | 可以很方便进行大片段整合 | 需先将整合位点插入染色体靶标位置 | [ |
| 等位基因替换 | 依赖同源重组单交换及之后的二次交换 | 可精确进行基因编辑 | 梭菌同源重效率低,过程耗时长 | [ |
| 反筛标记介导的等位基因替换 | 在等位基因替换基础上,在同源臂内设计pyrE、mazF等反筛标记,方便筛选二次交换突变株 | 相比纯粹的等位基因替换,效率有所提升 | 受制于第一次单交换效率 | [ |
| I-SceI归巢内切酶介导的等位基因替换 | 在等位基因替换基础上,在同源臂内设计I-SceI酶切位点,表达I-SceI切割双键,促进二次交换或帮助筛选二次交换突变株 | 相比纯粹的等位基因替换,效率有所提升 | 受制于第一次单交换效率 | [ |
| 单链寡核苷酸介导的点突变 | 重组蛋白RecT介导单链寡核苷酸更强的侵入和重组 | 可在较短同源臂时实现点突变 | 同源重组效率仍然较低 | [ |
| CRISPR/Cas9系统介导的基因编辑 | Cas9在sgRNA引导下切割靶标DNA,造成双键断裂(平末端),依赖同源重组修复 | 增加第一次单交换效率,可实现多功能编辑 | 双键断裂的毒性大,转化子很难获得 | [ |
| CRISPR/nCas9系统介导的基因编辑 | 失活Cas9蛋白的其中一个切割蛋白结构域,使其仅能单链切割靶标DNA,造成缺刻,然后依赖同源重组修复 | 相比CRISPR/Cas9毒性有所降低,转化子数目增加 | 仍有毒性,转化子依然偏少 | [ |
| CRISPR/Cpf1系统介导的基因编辑 | Cpf1在sgRNA引导下切割靶标DNA,造成双键断裂(有悬端),依赖同源重组修复 | 悬端的存在有利于DNA修复;脱靶概率比CRISPR/Cas9低 | 仍有毒性,转化子依然偏少 | [ |
| CRISPR/dCas9系统介导的基因下调 | 失活Cas9蛋白的全部两个切割蛋白结构域,使其仅能与靶标DNA结合,利用位阻效应降低mRNA转录水平 | 对靶标基因转型转录调控,尤其适用于必需基因的操作 | 依赖于sgRNA和基因 | [ |
| CRISPR/Cas系统辅助的碱基编辑 | 将胞嘧啶脱氨酶和尿嘧啶DNA糖苷酶与Cas蛋白融合表达,在sgRNA引导下对目标碱基实现C-G到T-A的替换 | 理论上可对任何碱基进行编辑,不依赖于同源重组 | 碱基转换能力待拓展,编辑效率有待提升 | [ |

Fig. 3 Metabolic engineering of clostridiafor n-butanol production(pfk—6-phosphofructokinase; fba—fructose-bisphosphate aldolase; pyk—pyruvate kinase; ilvB—acetolactate synthase; aldc—acetolactate decarboxylase; acr—acetoin reductase; pfor—pyruvate-ferredoxin oxidoreductase; hyd—hydrogenase; pta—phosphate acyltransferase; ack—acetate kinase; thl—thiolase; hbd—3-hydroxybutyryl-CoA dehydrogenase; crt—crotonase; bcd—butyryl-CoA dehydrogenase; ter—trans-enoyl-CoA reductase; ptb—phosphobutyryl transferase; buk—butyrate kinase; ctfAB—acetoacetyl-CoA: acetate/butyrate: CoA transferase; adc—acetoacetate decarboxylase; sadh—isopropanol dehydrogenase; adh/edh—alcohol/ehanol dehydrogenase; bdh/edh—butanol/ehanol dehydrogenase; adhE—alcohol/aldehyde dehydrogenase)
| 出发菌株 | 基因型 | 增产策略及效果描述 | 正丁醇/g·L-1 | 文献 |
|---|---|---|---|---|
| C. acetobutylicum ATCC 824 | +pykA+pfkA | 过表达EMP途径的6-磷酸果糖激酶和丙酮酸激酶基因,增强碳源供应,正丁醇增加44.3% | 19.12 | [ |
| C. acetobutylicum ATCC 824 | +thl(R133G,H156N;G222V) | 过表达点突变的thl(3处点突变减少辅酶A对硫酯酶的反馈抑制),增强主途径代谢,乙醇和正丁醇产量分别增加46%和18%,丙酮几乎没有变化 | 12.4 | [ |
| C. diolis DSM15140 | +ter | 过表达不可逆的反式烯酰辅酶A还原酶(来自齿垢密螺旋体),正丁醇产量提升50.5% | 10.1 | [ |
| C. acetobutylicum ATCC 824 | △adc,+gshAB+adhE+ctfAB,+thl+hbd+crt+bcd | 正丁醇14.86g/L提升187%;乙醇产量3.25 g/L,提升278%;丙酮0.15 g/L,下降94.3% | 14.86 | [ |
| C. acetobutylicum ATCC 824 | △solR | 敲除产溶剂操纵子的负调控因子,正丁醇产量增加 | 17.8 | [ |
| C. acetobutylicum ATCC 824 | 野生型 | — | 11.71 | [ |
| C. acetobutylicum ATCC 824 | △solR | 敲除产溶剂操纵子的负调控因子,正丁醇产量小幅提升 | 14.6 | [ |
| C. acetobutylicum ATCC 824 | △solR+aad | 敲除产溶剂操纵子的负调控因子后过表达醇醛脱氢酶基因(aad) | 17.6 | [ |
| C. acetobutylicum ATCC 824 | 野生型 | — | 11.27 | [ |
| C. acetobutylicum ATCC 824 | +aad+thl | 过表达aad和thl增强主途径,但正丁醇产量和比例未见明显改变 | 11.34 | [ |
| C. acetobutylicum ATCC 824 | +aad | 过表达thl增强主途径,正丁醇产量和比例未见明显改变 | 11.86 | [ |
| C. acetobutylicum ATCC 824 | +aad-ctfB | 过表达aad及辅酶A转移酶基因ctfB的反义RNA。正丁醇产量小幅增加,但乙醇增加明显,导致正丁醇比例下降 | 13.19 | [ |
| C. acetobutylicum ATCC 824 | +aad D458G | 过表达点突变(485位天冬氨酸突变为甘氨酸)aad(对NADH和NADPH均有较好亲和力),正丁醇仅增加11.3%;乙醇大幅增加294% | 14.8 | [ |
| C. acetobutylicum ATCC 824 | +aad D458G +(Cb ald-Cl bdh) | 过表达aad D458G,以及拜氏梭菌来源的醛脱氢酶基因(ald),杨氏梭菌来源的正丁醇脱氢酶基因(bdh),试图增强主途径,正丁醇产量增加27.1%,乙醇和丙酮均未大幅增加 | 16.9 | [ |
| C. acetobutylicum ATCC 824 | △ack | TargeTron中断乙酸激酶基因(ack),试图阻断产乙酸途径,正丁醇产量提升22.9%,乙醇产量提升305% | 8.6 | [ |
| C. acetobutylicum ATCC 824 | △ack | TargeTron中断ack,试图阻断产乙酸途径,正丁醇产量几乎不变 | 11.3 | [ |
| C. acetobutylicum ATCC 824 | △pta | TargeTron中断乙酰磷酸转移酶基因pta,试图阻断产乙酸途径,溶剂产量大幅增加,其中正丁醇增加45.8% | 17.2 | [ |
| C. acetobutylicum ATCC 824 | △ptb | TargeTron中断丁酰磷酸转移酶基因ptb,试图阻断产丁酸途径,正丁醇减少32.8%;乙醇产量12.1 g/L,增加505%;丙酮5.3 g/L | 8 | [ |
| C. acetobutylicum ATCC 824 | △ptb | TargeTron中断丁ptb,试图阻断产丁酸途径,正丁醇增加17.8%,乙醇0.9 g/L,几乎不变 | 13.9 | [ |
| C. acetobutylicum ATCC 824 | △buk | TargeTron中断丁酸激酶基因buk,试图阻断产丁酸途径,正丁醇增加28.8%;乙醇1.9 g/L,增加111%;丙酮8 g/L,增加48.1% | 15.2 | [ |
| C. acetobutylicum ATCC 824 | △pta△buk | TargeTron连续中断pta和buk,正丁醇增加35.6% | 16 | [ |
| C. acetobutylicum ATCC 824 | △pta△buk +adhE1 | TargeTron连续中断pta和buk后,过表达醇醛脱氢酶基因adhE1,正丁醇增加55.9%。乙醇3.0 g/L,增加233%;丙酮2.1 g/L,下降61.1% | 18.4 | [ |
| C. acetobutylicum ATCC 824 | △pta△buk +adhE1D485G | TargeTron连续中断pta和buk后,过表达点突变的adhE1 D485G (对NADH和NADPH均有较好亲和力),正丁醇增加60.2%;乙醇1.1 g/L,增加22.2%;丙酮1.5 g/L,降低72.2% | 18.9 | [ |
| C. beijerinckii NCIMB 8052 | △pta△buk | TargeTron连续中断pta和buk后意外发现可实现葡萄糖木糖同步发酵,消除了CCR效应,正丁醇增产36.4% | 12.64 | [ |
| C. beijerinckiiCC101 | +ctfAB+adhE2 | 过表达酸回用途径和正丁醇合成途径关键酶,正丁醇产量增加接近100% | 12 | [ |
| C. acetobutylicum ATCC 824 | △ldhA, △ctfAB, △ptb, △buk, +thl, hbd | 删除乳酸脱氢酶基因ldhA、ctfAB、ptb、buk,过表达thl和3-羟基丁酰辅酶A脱氢酶基因(hbd),增强主途径,连续发酵且辅以气提移除正丁醇抑制 | 550① | [ |
Tab. 3 Summary of n-butanol titer enhancement in Clostridium
| 出发菌株 | 基因型 | 增产策略及效果描述 | 正丁醇/g·L-1 | 文献 |
|---|---|---|---|---|
| C. acetobutylicum ATCC 824 | +pykA+pfkA | 过表达EMP途径的6-磷酸果糖激酶和丙酮酸激酶基因,增强碳源供应,正丁醇增加44.3% | 19.12 | [ |
| C. acetobutylicum ATCC 824 | +thl(R133G,H156N;G222V) | 过表达点突变的thl(3处点突变减少辅酶A对硫酯酶的反馈抑制),增强主途径代谢,乙醇和正丁醇产量分别增加46%和18%,丙酮几乎没有变化 | 12.4 | [ |
| C. diolis DSM15140 | +ter | 过表达不可逆的反式烯酰辅酶A还原酶(来自齿垢密螺旋体),正丁醇产量提升50.5% | 10.1 | [ |
| C. acetobutylicum ATCC 824 | △adc,+gshAB+adhE+ctfAB,+thl+hbd+crt+bcd | 正丁醇14.86g/L提升187%;乙醇产量3.25 g/L,提升278%;丙酮0.15 g/L,下降94.3% | 14.86 | [ |
| C. acetobutylicum ATCC 824 | △solR | 敲除产溶剂操纵子的负调控因子,正丁醇产量增加 | 17.8 | [ |
| C. acetobutylicum ATCC 824 | 野生型 | — | 11.71 | [ |
| C. acetobutylicum ATCC 824 | △solR | 敲除产溶剂操纵子的负调控因子,正丁醇产量小幅提升 | 14.6 | [ |
| C. acetobutylicum ATCC 824 | △solR+aad | 敲除产溶剂操纵子的负调控因子后过表达醇醛脱氢酶基因(aad) | 17.6 | [ |
| C. acetobutylicum ATCC 824 | 野生型 | — | 11.27 | [ |
| C. acetobutylicum ATCC 824 | +aad+thl | 过表达aad和thl增强主途径,但正丁醇产量和比例未见明显改变 | 11.34 | [ |
| C. acetobutylicum ATCC 824 | +aad | 过表达thl增强主途径,正丁醇产量和比例未见明显改变 | 11.86 | [ |
| C. acetobutylicum ATCC 824 | +aad-ctfB | 过表达aad及辅酶A转移酶基因ctfB的反义RNA。正丁醇产量小幅增加,但乙醇增加明显,导致正丁醇比例下降 | 13.19 | [ |
| C. acetobutylicum ATCC 824 | +aad D458G | 过表达点突变(485位天冬氨酸突变为甘氨酸)aad(对NADH和NADPH均有较好亲和力),正丁醇仅增加11.3%;乙醇大幅增加294% | 14.8 | [ |
| C. acetobutylicum ATCC 824 | +aad D458G +(Cb ald-Cl bdh) | 过表达aad D458G,以及拜氏梭菌来源的醛脱氢酶基因(ald),杨氏梭菌来源的正丁醇脱氢酶基因(bdh),试图增强主途径,正丁醇产量增加27.1%,乙醇和丙酮均未大幅增加 | 16.9 | [ |
| C. acetobutylicum ATCC 824 | △ack | TargeTron中断乙酸激酶基因(ack),试图阻断产乙酸途径,正丁醇产量提升22.9%,乙醇产量提升305% | 8.6 | [ |
| C. acetobutylicum ATCC 824 | △ack | TargeTron中断ack,试图阻断产乙酸途径,正丁醇产量几乎不变 | 11.3 | [ |
| C. acetobutylicum ATCC 824 | △pta | TargeTron中断乙酰磷酸转移酶基因pta,试图阻断产乙酸途径,溶剂产量大幅增加,其中正丁醇增加45.8% | 17.2 | [ |
| C. acetobutylicum ATCC 824 | △ptb | TargeTron中断丁酰磷酸转移酶基因ptb,试图阻断产丁酸途径,正丁醇减少32.8%;乙醇产量12.1 g/L,增加505%;丙酮5.3 g/L | 8 | [ |
| C. acetobutylicum ATCC 824 | △ptb | TargeTron中断丁ptb,试图阻断产丁酸途径,正丁醇增加17.8%,乙醇0.9 g/L,几乎不变 | 13.9 | [ |
| C. acetobutylicum ATCC 824 | △buk | TargeTron中断丁酸激酶基因buk,试图阻断产丁酸途径,正丁醇增加28.8%;乙醇1.9 g/L,增加111%;丙酮8 g/L,增加48.1% | 15.2 | [ |
| C. acetobutylicum ATCC 824 | △pta△buk | TargeTron连续中断pta和buk,正丁醇增加35.6% | 16 | [ |
| C. acetobutylicum ATCC 824 | △pta△buk +adhE1 | TargeTron连续中断pta和buk后,过表达醇醛脱氢酶基因adhE1,正丁醇增加55.9%。乙醇3.0 g/L,增加233%;丙酮2.1 g/L,下降61.1% | 18.4 | [ |
| C. acetobutylicum ATCC 824 | △pta△buk +adhE1D485G | TargeTron连续中断pta和buk后,过表达点突变的adhE1 D485G (对NADH和NADPH均有较好亲和力),正丁醇增加60.2%;乙醇1.1 g/L,增加22.2%;丙酮1.5 g/L,降低72.2% | 18.9 | [ |
| C. beijerinckii NCIMB 8052 | △pta△buk | TargeTron连续中断pta和buk后意外发现可实现葡萄糖木糖同步发酵,消除了CCR效应,正丁醇增产36.4% | 12.64 | [ |
| C. beijerinckiiCC101 | +ctfAB+adhE2 | 过表达酸回用途径和正丁醇合成途径关键酶,正丁醇产量增加接近100% | 12 | [ |
| C. acetobutylicum ATCC 824 | △ldhA, △ctfAB, △ptb, △buk, +thl, hbd | 删除乳酸脱氢酶基因ldhA、ctfAB、ptb、buk,过表达thl和3-羟基丁酰辅酶A脱氢酶基因(hbd),增强主途径,连续发酵且辅以气提移除正丁醇抑制 | 550① | [ |
| 菌株 | 基因型 | 比例提升策略和结果 | 比例 | 文献 |
|---|---|---|---|---|
| C. acetobutylicum ATCC 824 | △adc | TargeTron中断乙酰乙酸脱羧酶基因adc,丙酮产量下降,仅有0.12 g/L,丁醇产量降低 | — | [ |
| C. acetobutylicum ATCC 824 | △adc | TargeTron中断adc,丁醇减少46%,丙酮减少90%,乙醇减少约45% | 85.4% | [ |
| C. acetobutylicum EA2018 | △adc | TargeTron中断adc,丁醇14.1 g/L,比例由70%增加至82%;乙醇2.8 g/L,下降46.1%;丙酮0.1 g/L,下降90.8% | 82.9% | [ |
| C. acetobutylicum ATCC 824 | △pta△adc | TargeTron连续中断pta和adc,三者产量均下降 | 89.1% | [ |
| C. acetobutylicum ATCC 824 | △ctfA | TargeTron中断CoA转移酶基因ctfA,乙醇/丁醇产量降低,丙酮完全消失 | — | [ |
| C. acetobutylicum ATCC 824 | △ctfB | TargeTron中断CoA转移酶基因ctfA,乙醇/丁醇产量降低,丙酮完全消失 | — | [ |
| C. acetobutylicum ATCC 824 | △ctfB | TargeTron中断ctfB,丁醇5g/L,乙醇0.3 g/L,减少66.7%;丙酮消失 | 94.3% | [ |
| C. acetobutylicum ATCC 824 | △ctfA | TargeTron中断ctfA,丁醇产量降低47%,乙醇产量降低;丙酮消失 | 79.6% | [ |
| C. acetobutylicum ATCC 824 | △pta△ctfA | TargeTron连续中断pta和ctfA,三者均有所降低 | 86.9% | [ |
| C. acetobutylicum ATCC 824 | △pta△ctfB | TargeTron连续中断pta和ctfB,丁醇0.4 g/L,乙醇0.2 g/L,降低77.8%;丙酮消失 | 66.6% | [ |
| C. acetobutylicum ATCC 824 | △pta△buk | TargeTron连续中断pta和buk,丁醇16 g/L,增加35.6%,乙醇1.5 g/L;丙酮2.4 g/L | 80.4% | [ |
| C. acetobutylicum ATCC 824 | △pta△buk+adhE1 | TargeTron连续中断pta和buk,过表达adhE1,丁醇18.4 g/L,增加55.9%;乙醇3.0 g/L,增加233%;丙酮2.1 g/L,下降61.1% | 78.3% | [ |
| C. acetobutylicum ATCC 824 | △pta△buk+adhE1D485G | TargeTron连续中断pta和buk,过表达adhE1D485G,丁醇18.9 g/L,增加60.2%;乙醇1.1 g/L,增加22.2%;丙酮1.5 g/L,降低72.2% | 87.8% | [ |
Tab. 4 Summary of n-butanol ratio enhancement in Clostridium
| 菌株 | 基因型 | 比例提升策略和结果 | 比例 | 文献 |
|---|---|---|---|---|
| C. acetobutylicum ATCC 824 | △adc | TargeTron中断乙酰乙酸脱羧酶基因adc,丙酮产量下降,仅有0.12 g/L,丁醇产量降低 | — | [ |
| C. acetobutylicum ATCC 824 | △adc | TargeTron中断adc,丁醇减少46%,丙酮减少90%,乙醇减少约45% | 85.4% | [ |
| C. acetobutylicum EA2018 | △adc | TargeTron中断adc,丁醇14.1 g/L,比例由70%增加至82%;乙醇2.8 g/L,下降46.1%;丙酮0.1 g/L,下降90.8% | 82.9% | [ |
| C. acetobutylicum ATCC 824 | △pta△adc | TargeTron连续中断pta和adc,三者产量均下降 | 89.1% | [ |
| C. acetobutylicum ATCC 824 | △ctfA | TargeTron中断CoA转移酶基因ctfA,乙醇/丁醇产量降低,丙酮完全消失 | — | [ |
| C. acetobutylicum ATCC 824 | △ctfB | TargeTron中断CoA转移酶基因ctfA,乙醇/丁醇产量降低,丙酮完全消失 | — | [ |
| C. acetobutylicum ATCC 824 | △ctfB | TargeTron中断ctfB,丁醇5g/L,乙醇0.3 g/L,减少66.7%;丙酮消失 | 94.3% | [ |
| C. acetobutylicum ATCC 824 | △ctfA | TargeTron中断ctfA,丁醇产量降低47%,乙醇产量降低;丙酮消失 | 79.6% | [ |
| C. acetobutylicum ATCC 824 | △pta△ctfA | TargeTron连续中断pta和ctfA,三者均有所降低 | 86.9% | [ |
| C. acetobutylicum ATCC 824 | △pta△ctfB | TargeTron连续中断pta和ctfB,丁醇0.4 g/L,乙醇0.2 g/L,降低77.8%;丙酮消失 | 66.6% | [ |
| C. acetobutylicum ATCC 824 | △pta△buk | TargeTron连续中断pta和buk,丁醇16 g/L,增加35.6%,乙醇1.5 g/L;丙酮2.4 g/L | 80.4% | [ |
| C. acetobutylicum ATCC 824 | △pta△buk+adhE1 | TargeTron连续中断pta和buk,过表达adhE1,丁醇18.4 g/L,增加55.9%;乙醇3.0 g/L,增加233%;丙酮2.1 g/L,下降61.1% | 78.3% | [ |
| C. acetobutylicum ATCC 824 | △pta△buk+adhE1D485G | TargeTron连续中断pta和buk,过表达adhE1D485G,丁醇18.9 g/L,增加60.2%;乙醇1.1 g/L,增加22.2%;丙酮1.5 g/L,降低72.2% | 87.8% | [ |

Fig. 4 Reconstruction of n-butanol synthesis pathway in clostridia[Green lines represent carbohydrate metabolic pathway; the yellow lines represent the glycerol metabolic pathway; the lake blue represents the syngas metabolic pathway in gas-fermenting clostridia; the positive blue lines represent the synthesis pathway of n-butanol in Clostridium. dhaBCE—glycerol dehydratase; dhaT—1,3-propanediol dehydrogenase; gldA—glycerol-3-phosphate dehydrogenase; dhaKL—dihydroxyacetone phosphate kinase; mgsA—methylglyoxal synthase; yqhD—alcohol dehydrogenase; fucO—1,2-propanediol dehydrogenase; hk—hexokinase; gpi—fructose-6-phosphate isomerase; gapdh—glyceraldehyde-3-phosphate dehydrogenase; codh—carbon monoxide dehydrogenase; acs—acetyl-CoA synthase; fdh—formate dehydrogenase; fhs—formyl-tetrahydrofolate synthase; folD—methylenetetrahydrofolate cyclase/dehydrogenase; metF—methylenetetrahydrofolate reductase; metTr—methyltransferase; cat1—CoA transferase (Genes appeared in Figure 3 is not listed here)]
| 菌株 | 基因型 | 底物 | 正丁醇/g·L-1 | 文献 |
|---|---|---|---|---|
| C. cellulolyticum ATCC 35319 | +ato hbd crt bcd,adhE2 | 微晶纤维素 | 0.12 | [ |
| C. thermocellum DSM 1313 | △hpt△Clo1313△ldh+(Tt_thl-Tt_hbd, Tt_crt-St_ter-Ts_bad-Ts_bdh) b + Tt_thlM2-Tt_hbdM-St_terM | 微晶纤维素 | 0.357 | [ |
| C. cellulovoran DSM 743B | +adhE2 | 微晶纤维素 | 1.42 | [ |
| C. cellulovoran DSM 743B | +adhE2 | 预处理后玉米芯 | 3.36 | [ |
| C. cellulovoran DSM 743B | +adhE2 | 微晶纤维素 | 4.0 | [ |
| C. cellulovoran DSM 743B | Clocel*: +adhE1, ctfAB-adc | 碱处理玉米芯 | 3.47 | [ |
| C. cellulovoran DSM 743B | △xylR△araR+xylT+ter+cat1+adhE1 | 碱处理玉米芯 | 4.96 | [ |
| C. tyrobutyricum ATCC 25755 | △cat1+adhE2 | 葡萄糖 | 26.2 | [ |
| C. tyrobutyricum ATCC 25755 | △ack+adhE2 | 葡萄糖 | 10 | [ |
| C. tyrobutyricum ATCC 25755 | △ack-adhE2, ctfAB | 葡萄糖 | 12 | [ |
| C. tyrobutyricum ATCC 25755 | △ack-adhE2, xylT, xylA, xylB | 葡萄糖/木糖 | 12 | [ |
| C. ljungdahlii | +adhE, | 合成气 | 0.148 | [ |
| C. autoethanogenum | +thlA, hbd, crt, bcd, adhE, bdhA | 合成气 | 1.54 | [ |
| C. acetobutylicum ATCC 824 | M5 (△pSOL1) | 葡萄糖 | 0 | [ |
| C. acetobutylicum ATCC 824 | M5,+aad | 葡萄糖 | 6.23 | [ |
| C. acetobutylicum ATCC 824 | M5,+aad | 葡萄糖 | 10.23 | [ |
| C. acetobutylicum ATCC 824 | M5,+thl+aad | 葡萄糖 | 8 | [ |
| C. acetobutylicum ATCC 824 | M5,+aad△ack | 葡萄糖 | 6.82 | [ |
Tab. 5 Summary of n-butanol synthesis pathway reconstruction in Clostridium
| 菌株 | 基因型 | 底物 | 正丁醇/g·L-1 | 文献 |
|---|---|---|---|---|
| C. cellulolyticum ATCC 35319 | +ato hbd crt bcd,adhE2 | 微晶纤维素 | 0.12 | [ |
| C. thermocellum DSM 1313 | △hpt△Clo1313△ldh+(Tt_thl-Tt_hbd, Tt_crt-St_ter-Ts_bad-Ts_bdh) b + Tt_thlM2-Tt_hbdM-St_terM | 微晶纤维素 | 0.357 | [ |
| C. cellulovoran DSM 743B | +adhE2 | 微晶纤维素 | 1.42 | [ |
| C. cellulovoran DSM 743B | +adhE2 | 预处理后玉米芯 | 3.36 | [ |
| C. cellulovoran DSM 743B | +adhE2 | 微晶纤维素 | 4.0 | [ |
| C. cellulovoran DSM 743B | Clocel*: +adhE1, ctfAB-adc | 碱处理玉米芯 | 3.47 | [ |
| C. cellulovoran DSM 743B | △xylR△araR+xylT+ter+cat1+adhE1 | 碱处理玉米芯 | 4.96 | [ |
| C. tyrobutyricum ATCC 25755 | △cat1+adhE2 | 葡萄糖 | 26.2 | [ |
| C. tyrobutyricum ATCC 25755 | △ack+adhE2 | 葡萄糖 | 10 | [ |
| C. tyrobutyricum ATCC 25755 | △ack-adhE2, ctfAB | 葡萄糖 | 12 | [ |
| C. tyrobutyricum ATCC 25755 | △ack-adhE2, xylT, xylA, xylB | 葡萄糖/木糖 | 12 | [ |
| C. ljungdahlii | +adhE, | 合成气 | 0.148 | [ |
| C. autoethanogenum | +thlA, hbd, crt, bcd, adhE, bdhA | 合成气 | 1.54 | [ |
| C. acetobutylicum ATCC 824 | M5 (△pSOL1) | 葡萄糖 | 0 | [ |
| C. acetobutylicum ATCC 824 | M5,+aad | 葡萄糖 | 6.23 | [ |
| C. acetobutylicum ATCC 824 | M5,+aad | 葡萄糖 | 10.23 | [ |
| C. acetobutylicum ATCC 824 | M5,+thl+aad | 葡萄糖 | 8 | [ |
| C. acetobutylicum ATCC 824 | M5,+aad△ack | 葡萄糖 | 6.82 | [ |
| 出发菌株 | 基因型 | 策略及结果描述 | 正丁醇/g·L-1 | 文献 |
|---|---|---|---|---|
| C. acetobutylicumATCC 824 | +talA | 过表达PPP途径转醛醇酶基因(tal),木糖利用加速,但仍受葡萄糖抑制,发酵时间变短,丁醇产量增加 | 约8 | [ |
| C. acetobutylicumATCC 824 | +tal, tkl, rpe, rpi | 过表达PPP途径转醛醇酶基因(tal)、转酮醇酶基因(tkl)、核糖-5-磷酸异构酶基因(rpe)、核酮糖-5-磷酸3-差向异构酶基因(rpi),木糖利用增快,溶剂产量提升42% | 约5.5 | [ |
| C. acetobutylicum ATCC 824 | △glcG, +xylT, xylA, xylB | TargeTron中断PTS系统组成基因(glcG),过表达内源木糖转运蛋白基因(xylT)、木糖异构酶基因(xylA)和木酮糖激酶基因(xylB),基本消除CCR效应,溶剂产量增加24% | 9.11 | [ |
| C. acetobutylicumEA 2018 | △glcG, +xylT, xylA, xylB | 在EA2018宿主中,以TargeTron中断PTS系统组成基因(glcG),过表达丙酮丁醇梭菌ATCC824来源的木糖转运蛋白基因(xylT)、木糖异构酶基因(xylA)和木酮糖激酶基因(xylB),基本消除CCR效应,溶剂产量增加50% | 13.19 | [ |
| C. acetobutylicumATCC 824 | △ccpA | TargeTron中断多效调控子ccpA基因,突变株可同步利用葡萄糖和木糖,有机酸残留减少,丁醇得率增加 | 12.05 | [ |
| C. acetobutylicumATCC 824 | ccpA (V302N) | 对CcpA蛋白进行了定点突变(V302N),以削弱其与HPr-Ser46-P的结合能力,可有效缓解CcpA对木糖代谢的负调控作用,从而实现葡萄糖、木糖的同步发酵 | 8.61 | [ |
| C. acetobutylicum ATCC 824 | +xyloperon (cac1344-1349, mutated CRE) | 在过表达木糖代谢途径操纵子之前,突变基因上携带的的碳代谢抑制识别基序,在增强木糖代谢的同时,从而豁免碳代谢抑制效应,木糖利用增多、变快,但仍明显落后于葡萄糖 | — | [ |
| C. acetobutylicumATCC 824 | HPr (CRISPR/dCas9) | 使用CRISPR/dCas9转录下调Hpr激酶(CCR效应的感应酶)表达水平,木糖利用增多、变快,但仍明显落后于葡萄糖 | — | [ |
| C. beijerinckii NCIMB 8052 | △xylR, +xylT | TargeTron中断木糖代谢负调控子xylR基因,过表达内源木糖转运蛋白,基本消除CCR效应,溶剂增产35% | 11.27 | [ |
| C. beijerinckii NCIMB 8052 | △pta, △buk, △araR | TargeTron连续中断pta和buk后,继续中断araR,提升阿拉伯糖利用能力,消除了CCR效应,丁醇增产39.5% | 12.93 | [ |
| C. beijerinckii NCIMB 8052 | △xylR, △araR+xylT | TargeTron连续中断xylR和araR后,过表达xylT,同时提升木糖和阿拉伯糖利用能力,丁醇产量提升30.0% | 12.05 | [ |
| C. beijerinckii NCIMB 8052 | △pta△buk | TargeTron连续中断pta和buk后意外发现可实现葡萄糖木糖同步发酵,消除了CCR效应,丁醇增产36.4% | 12.64 | [ |
| C. tyrobutyricum | △ack, +adhE2, +xylT,xylA, xylB | 过表达丙酮丁醇梭菌来源的adhE2实现丁醇生产;删除ack基因减少乙酸副产物,过表达xylT以及xylAB增强木糖利用,葡萄糖木糖可同步利用,丁醇产量增加 | 12 | [ |
| C. cellulovorans | ΔxylR, ΔaraR, +xylT, adhE1 | 过表达adhE1实现丁醇生产;TargeTron中断木糖和阿拉伯糖代谢负调控基因xylR和araR,过表达来自拜氏梭菌的xylT,增强木糖利用,提升丁醇产量 | 2.92 | [ |
Tab. 6 Summary of pentose metabolism pathway engineering in n-butanol-producing Clostridium
| 出发菌株 | 基因型 | 策略及结果描述 | 正丁醇/g·L-1 | 文献 |
|---|---|---|---|---|
| C. acetobutylicumATCC 824 | +talA | 过表达PPP途径转醛醇酶基因(tal),木糖利用加速,但仍受葡萄糖抑制,发酵时间变短,丁醇产量增加 | 约8 | [ |
| C. acetobutylicumATCC 824 | +tal, tkl, rpe, rpi | 过表达PPP途径转醛醇酶基因(tal)、转酮醇酶基因(tkl)、核糖-5-磷酸异构酶基因(rpe)、核酮糖-5-磷酸3-差向异构酶基因(rpi),木糖利用增快,溶剂产量提升42% | 约5.5 | [ |
| C. acetobutylicum ATCC 824 | △glcG, +xylT, xylA, xylB | TargeTron中断PTS系统组成基因(glcG),过表达内源木糖转运蛋白基因(xylT)、木糖异构酶基因(xylA)和木酮糖激酶基因(xylB),基本消除CCR效应,溶剂产量增加24% | 9.11 | [ |
| C. acetobutylicumEA 2018 | △glcG, +xylT, xylA, xylB | 在EA2018宿主中,以TargeTron中断PTS系统组成基因(glcG),过表达丙酮丁醇梭菌ATCC824来源的木糖转运蛋白基因(xylT)、木糖异构酶基因(xylA)和木酮糖激酶基因(xylB),基本消除CCR效应,溶剂产量增加50% | 13.19 | [ |
| C. acetobutylicumATCC 824 | △ccpA | TargeTron中断多效调控子ccpA基因,突变株可同步利用葡萄糖和木糖,有机酸残留减少,丁醇得率增加 | 12.05 | [ |
| C. acetobutylicumATCC 824 | ccpA (V302N) | 对CcpA蛋白进行了定点突变(V302N),以削弱其与HPr-Ser46-P的结合能力,可有效缓解CcpA对木糖代谢的负调控作用,从而实现葡萄糖、木糖的同步发酵 | 8.61 | [ |
| C. acetobutylicum ATCC 824 | +xyloperon (cac1344-1349, mutated CRE) | 在过表达木糖代谢途径操纵子之前,突变基因上携带的的碳代谢抑制识别基序,在增强木糖代谢的同时,从而豁免碳代谢抑制效应,木糖利用增多、变快,但仍明显落后于葡萄糖 | — | [ |
| C. acetobutylicumATCC 824 | HPr (CRISPR/dCas9) | 使用CRISPR/dCas9转录下调Hpr激酶(CCR效应的感应酶)表达水平,木糖利用增多、变快,但仍明显落后于葡萄糖 | — | [ |
| C. beijerinckii NCIMB 8052 | △xylR, +xylT | TargeTron中断木糖代谢负调控子xylR基因,过表达内源木糖转运蛋白,基本消除CCR效应,溶剂增产35% | 11.27 | [ |
| C. beijerinckii NCIMB 8052 | △pta, △buk, △araR | TargeTron连续中断pta和buk后,继续中断araR,提升阿拉伯糖利用能力,消除了CCR效应,丁醇增产39.5% | 12.93 | [ |
| C. beijerinckii NCIMB 8052 | △xylR, △araR+xylT | TargeTron连续中断xylR和araR后,过表达xylT,同时提升木糖和阿拉伯糖利用能力,丁醇产量提升30.0% | 12.05 | [ |
| C. beijerinckii NCIMB 8052 | △pta△buk | TargeTron连续中断pta和buk后意外发现可实现葡萄糖木糖同步发酵,消除了CCR效应,丁醇增产36.4% | 12.64 | [ |
| C. tyrobutyricum | △ack, +adhE2, +xylT,xylA, xylB | 过表达丙酮丁醇梭菌来源的adhE2实现丁醇生产;删除ack基因减少乙酸副产物,过表达xylT以及xylAB增强木糖利用,葡萄糖木糖可同步利用,丁醇产量增加 | 12 | [ |
| C. cellulovorans | ΔxylR, ΔaraR, +xylT, adhE1 | 过表达adhE1实现丁醇生产;TargeTron中断木糖和阿拉伯糖代谢负调控基因xylR和araR,过表达来自拜氏梭菌的xylT,增强木糖利用,提升丁醇产量 | 2.92 | [ |
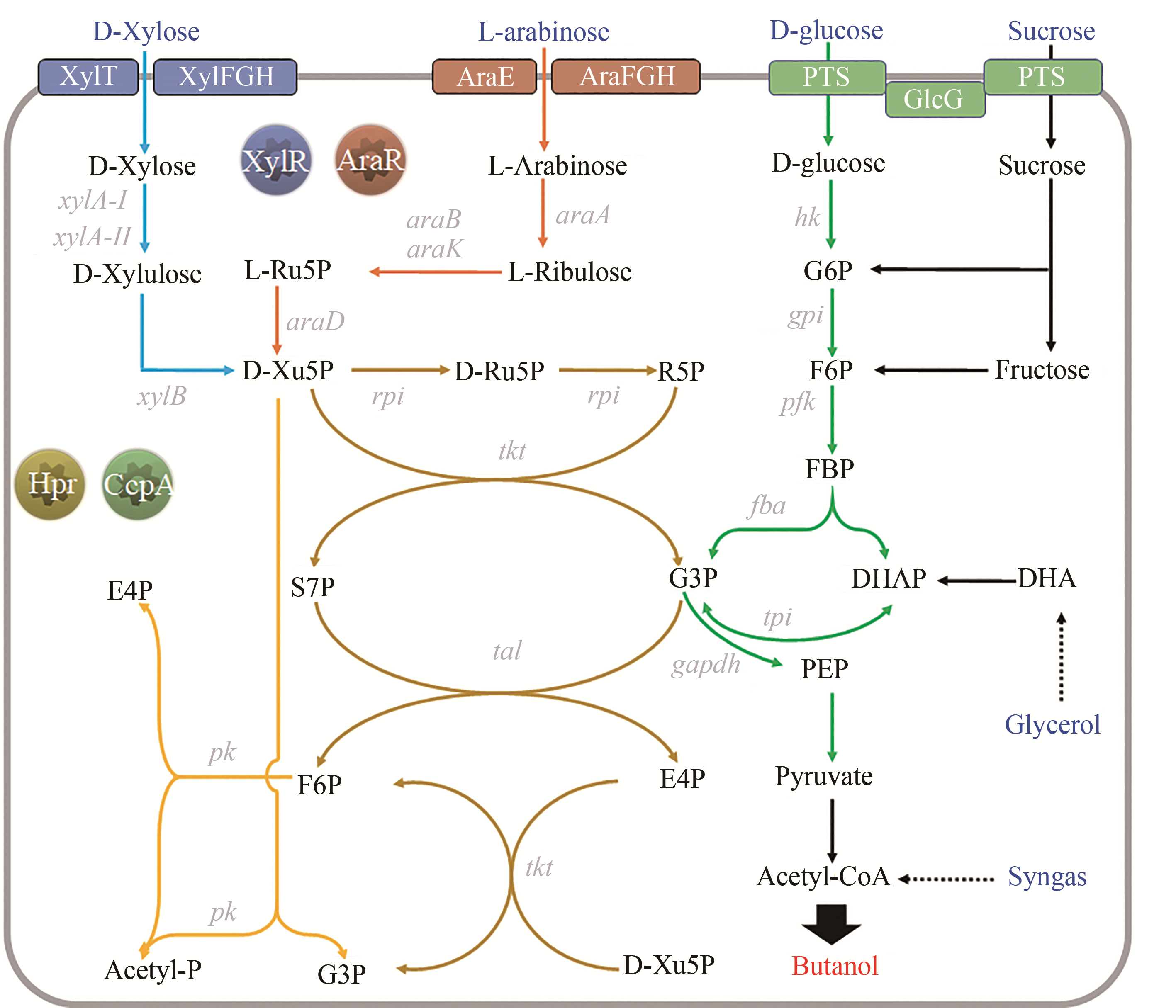
Fig. 5 Pentose metabolism pathway and involved regulatory proteins in n-butanol-producing clostridia[XylT/XylFGH—xylose-specific transporter; AraE/AraFGH—arabinose-specific transporter; PTS—phosphotransferase system; GlcG—enzyme II of PTS, closely related to the CCR; G6P—glucose-6-phosphate; F6P—fructose-6-phosphate; FBP—fructose-1,6-Diphosphate; E4P—erythrose-4-phosphate; S7P—sedum heptulose-7-phosphate; Ru5P—ribulose-5-phosphate; Xu5P—xylulose-5-phosphate; G3P—3-Glyceraldehyde phosphate; DHA—dihydroxyacetone; DHAP—dihydroxyacetone phosphate; PEP—phosphoenolpyruvate; xylA-I/II—xylose isomerase; xylB—xylulose kinase; araA—arabinose isomerase; araB/K—ribulose kinase; araD—ribulose-5-phosphate isomerase; rpi—5-phosphoribose isomerase; tal—transaldolase; tkt—transketolase; pk—phosphoketolase (Genes appeared in Fig. 2~4 are not listed here)]
| 1 | 谭天伟. 合成生物学—绿色生物制造产业发展趋势 [J]. 生物产业技术, 2015(6): 11-15. |
| TAN T W. Synthetic biology—development trend of green biological manufacturing industry [J]. Biotechnology&Business, 2015(6): 11-15. | |
| 2 | GU Y, JIANG Y, WU H, et al. Economical challenges to microbial producers of butanol: feedstock, butanol ratio and titer [J]. Biotechnology Journal, 2011, 6(11): 1348-1357. |
| 3 | WEN Z, LI Q, LIU J, et al. Consolidated bioprocessing for butanol production of cellulolytic Clostridia: development and optimization [J]. Microbial Biotechnology, 2020, 13(2): 410-422. |
| 4 | 戴宗杰, 董红军, 朱岩, 等. 生物丁醇代谢工程的研究进展 [J]. 生物加工过程, 2013, 11(2): 58-64. |
| DAI Z J, DONG H J, ZHU Y, et al. Metabolic engineering for biobutanol production: a review [J]. Chinese Journal of Bioprocess Engineering, 2013, 11(2): 58-64. | |
| 5 | 顾阳, 杨晟, 姜卫红. 产溶剂梭菌分子遗传操作技术研究进展 [J]. 生物工程学报, 2013, 29(8): 1133-1145. |
| GU Y, YANG S, JIANG W H. Development in molecular genetic manipulation of solventogenic clostridia [J]. Chinese Journal of Biotechnology, 2013, 29(8): 1133-1145. | |
| 6 | MOON H G, Y-S JANG, CHO C, et al. One hundred years of clostridial butanol fermentation [J]. FEMS Microbiology Letters, 2016, 363(3):fnw001. |
| 7 | NAWAB S, WANG N, MA X, et al. Genetic engineering of non-native hosts for 1-butanol production and its challenges: a review [J]. Microbial Cell Factories, 2020, 19(1):79. |
| 8 | REN C, WEN Z, XU Y, et al. Clostridia: a flexible microbial platform for the production of alcohols [J]. Current Opinion in Chemical Biology, 2016, 35:65-72. |
| 9 | CHEN C T, LIAO J C. Frontiers in microbial 1-butanol and isobutanol production [J]. FEMS Microbiology Letters, 2016, 363(5): fnw020. |
| 10 | ATSUMI S, CANN A F, CONNOR M R, et al. Metabolic engineering of Escherichia coli for 1-butanol production [J]. Metabolic Engineering, 2008, 10(6): 305-311. |
| 11 | STEEN E J, CHAN R, PRASAD N, et al. Metabolic engineering of Saccharomyces cerevisiae for the production of n-butanol [J]. Microbial Cell Factories, 2008, 7:36. |
| 12 | INUI M, SUDA M, KIMURA S, et al. Expression of Clostridium acetobutylicum butanol synthetic genes in Escherichia coli [J]. Applied Microbiology and Biotechnology, 2008, 77(6): 1305-1316. |
| 13 | NIELSEN D R, LEONARD E, S-H YOON, et al. Engineering alternative butanol production platforms in heterologous bacteria [J]. Metabolic Engineering, 2009, 11(4-5): 262-273. |
| 14 | BEREZINA O V, ZAKHAROVA N V, BRANDT A, et al. Reconstructing the clostridial n-butanol metabolic pathway in Lactobacillus brevis [J]. Applied Microbiology and Biotechnology, 2010, 87(2): 635-646. |
| 15 | YU A, ZHAO Y, PANG Y, et al. An oleaginous yeast platform for renewable 1-butanol synthesis based on a heterologous CoA-dependent pathway and an endogenous pathway [J]. Microbial Cell Factories, 2018, 17:166. |
| 16 | WANG M, HU L, FAN L, et al. Enhanced 1-butanol production in engineered Klebsiella pneumoniae by NADH regeneration [J]. Energy & Fuels, 2015, 29(3): 1823-1829. |
| 17 | WANG M M, FAN L H, TAN T W. 1-Butanol production from glycerol by engineered Klebsiella pneumoniae [J]. RSC Advances, 2014, 4(101): 57791-57798. |
| 18 | LAN E I, RO S Y, LIAO J C. Oxygen-tolerant coenzyme A-acylating aldehyde dehydrogenase facilitates efficient photosynthetic n-butanol biosynthesis in cyanobacteria [J]. Energy & Environmental Science, 2013, 6(9): 2672-2681. |
| 19 | LAN E I, LIAO J C. ATP drives direct photosynthetic production of 1-butanol in cyanobacteria [J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(16): 6018-6023. |
| 20 | SI T, LUO Y, XIAO H, et al. Utilizing an endogenous pathway for 1-butanol production in Saccharomyces cerevisiae [J]. Metabolic Engineering, 2014, 22:60-68. |
| 21 | LIAN J, SI T, NAIR N U, et al. Design and construction of acetyl-CoA overproducing Saccharomyces cerevisiae strains [J]. Metabolic Engineering, 2014, 24:139-149. |
| 22 | BRANDUARDI P, LONGO V, BERTERAME N M, et al. A novel pathway to produce butanol and isobutanol in Saccharomyces cerevisiae [J]. Biotechnology for Biofuels, 2013, 6(1):68. |
| 23 | SHEN C R, LAN E I, DEKISHIMA Y, et al. driving forces enable high-titer anaerobic 1-butanol synthesis in Escherichia coli [J]. Applied and Environmental Microbiology, 2011, 77(9): 2905-2915. |
| 24 | DELLOMONACO C, CLOMBURG J M, MILLER E N, et al. Engineered reversal of the beta-oxidation cycle for the synthesis of fuels and chemicals [J]. Nature, 2011, 476(7360): 355-U131. |
| 25 | PASZTOR A, KALLIO P, MALATINSZKY D, et al. A Synthetic O2-tolerant butanol pathway exploiting native fatty acid biosynthesis in Escherichia coli [J]. Biotechnology and Bioengineering, 2015, 112(1): 120-128. |
| 26 | SHEN C R, LIAO J C. Metabolic engineering of Escherichia coli for 1-butanol and 1-propanol production via the keto-acid pathways [J]. Metabolic Engineering, 2008, 10(6): 312-320. |
| 27 | SAINI M, CHEN M H, C-J CHIANG, et al. Potential production platform of n-butanol in Escherichia coli [J]. Metabolic Engineering, 2015, 27: 76-82. |
| 28 | OHTAKE T, PONTRELLI S, LAVINA W A, et al. Metabolomics-driven approach to solving a CoA imbalance for improved 1-butanol production in Escherichia coli [J]. Metabolic Engineering, 2017, 41: 135-143. |
| 29 | DONG H, ZHAO C, ZHANG T, et al. A systematically chromosomally engineered Escherichia coli efficiently produces butanol [J]. Metabolic Engineering, 2017, 44: 284-292. |
| 30 | SHI S, SI T, LIU Z, et al. Metabolic engineering of a synergistic pathway for n-butanol production in Saccharomyces cerevisiae [J]. Scientific Reports, 2016, 6(1):25675. |
| 31 | FATHIMA A M, CHUANG D, LAVINA W A, et al. Iterative cycle of widely targeted metabolic profiling for the improvement of 1-butanol titer and productivity in Synechococcus elongatus [J]. Biotechnology for Biofuels, 2018, 11(1):188. |
| 32 | LIU X, MIAO R, LINDBERG P, et al. Modular engineering for efficient photosynthetic biosynthesis of 1-butanol from CO2 in cyanobacteria [J]. Energy & Environmental Science, 2019, 12(9): 2765-2777. |
| 33 | BHANDIWAD A, SHAW A J, GUSS A, et al. Metabolic engineering of Thermoanaerobacterium saccharolyticum for n-butanol production [J]. Metabolic Engineering, 2014, 21:17-25. |
| 34 | ZHANG J, ZONG W, HONG W, et al. Exploiting endogenous CRISPR-Cas system for multiplex genome editing in Clostridium tyrobutyricum and engineer the strain for high-level butanol production [J]. Metabolic Engineering, 2018, 47: 49-59. |
| 35 | NOGUCHI T, TASHIRO Y, YOSHIDA T, et al. Efficient butanol production without carbon catabolite repression from mixed sugars with Clostridium saccharoperbutylacetonicum N1-4 [J]. Journal of Bioscience and Bioengineering, 2013, 116(6): 716-721. |
| 36 | SABRA W, GROEGER C, SHARMA P N, et al. Improved n-butanol production by a non-acetone producing Clostridium pasteurianum DSMZ 525 in mixed substrate fermentation [J]. Applied Microbiology and Biotechnology, 2014, 98(9): 4267-4276. |
| 37 | SILLERS R, AL-HINAI M A, PAPOUTSAKIS E T. Aldehyde-alcohol dehydrogenase and/or thiolase overexpression coupled with CoA transferase downregulation lead to higher alcohol titers and selectivity in Clostridium acetobutylicum fermentations [J]. Biotechnology and Bioengineering, 2009, 102(1): 38-49. |
| 38 | FORMANEK J, MACKIE R, BLASCHEK H P. Enhanced butanol production by Clostridium beijerinckii BA101 grown in semidefined P2 medium containing 6 percent maltodextrin or glucose [J]. Applied and Environmental Microbiology, 1997, 63(6): 2306-2310. |
| 39 | WEN Z, LEDESMA-AMARO R, LU M, et al. Metabolic engineering of Clostridium cellulovorans to improve butanol production by consolidated bioprocessing [J]. ACS Synthetic Biology, 2020, 9(2): 304-315. |
| 40 | KÖPKE M, HELD C, HUJER S, et al. Clostridium ljungdahlii represents a microbial production platform based on syngas [J]. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(29): 13087. |
| 41 | SHEN S, GU Y, CHAI C, et al. Enhanced alcohol titre and ratio in carbon monoxide-rich off-gas fermentation of Clostridium carboxidivorans through combination of trace metals optimization with variable-temperature cultivation [J]. Bioresource Technology, 2017, 239: 236-243. |
| 42 | KOEPKE M, LIEW F M. Production of butanol from carbon monoxide by a recombinant microorganism: EP patent application, 2012, WO 2012/053905 A1. |
| 43 | WEN Z, WU M, LIN Y, et al. Artificial symbiosis for acetone-butanol-ethanol (ABE) fermentation from alkali extracted deshelled corn cobs by co-culture of Clostridium beijerinckii and Clostridium cellulovorans [J]. Microbial cell factories, 2014, 13(1):92. |
| 44 | WEN Z, MINTON N P, ZHANG Y, et al. Enhanced solvent production by metabolic engineering of a twin-clostridial consortium [J]. Metabolic Engineering, 2017, 39:38-48. |
| 45 | Y-S JANG, LEE J Y, LEE J, et al. Enhanced butanol production obtained by reinforcing the direct butanol-forming route in Clostridium acetobutylicum [J]. mBio, 2012, 3(5): e00314-12. |
| 46 | BOND-WATTS B B, BELLEROSE R J, CHANG M C Y. Enzyme mechanism as a kinetic control element for designing synthetic biofuel pathways [J]. Nature Chemical Biology, 2011, 7(4): 222-227. |
| 47 | LAN E I, LIAO J C. Metabolic engineering of cyanobacteria for 1-butanol production from carbon dioxide [J]. Metabolic Engineering, 2011, 13(4): 353-363. |
| 48 | ATSUMI S, HANAI T, LIAO J C. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels [J]. Nature, 2008, 451(7174): 86-U13. |
| 49 | ATSUMI S, LIAO J C. Directed evolution of Methanococcus jannaschii citramalate synthase for biosynthesis of 1-propanol and 1-butanol by Escherichia coli [J]. Applied and Environmental Microbiology, 2008, 74(24): 7802-8. |
| 50 | WEN Z, LU M, LEDESMA-AMARO R, et al. TargeTron technology applicable in solventogenic clostridia: revisiting 12 years’ advances [J]. Biotechnology Journal, 2020, 15(1): e1900284. |
| 51 | YU M, ZHANG Y, TANG I C, et al. Metabolic engineering of Clostridium tyrobutyricum for n-butanol production [J]. Metabolic Engineering, 2011, 13(4): 373-382. |
| 52 | WU Y, YANG Y P, REN C, et al. Molecular modulation of pleiotropic regulator CcpA for glucose and xylose coutilization by solvent-producing Clostridium acetobutylicum [J]. Metabolic Engineering, 2015, 28: 169-179. |
| 53 | WIEGEL J, TANNER R, RAINEY F A. An introduction to the family Clostridiaceae [M]. New York: Springer, 2006. |
| 54 | MERMELSTEIN L D, WELKER N E, BENNETT G N, et al. Expression of cloned homologous fermentative genes in Clostridium acetobutylicum ATCC 824 [J]. Nature Biotechnology, 1992, 10(2): 190-195. |
| 55 | NÖLLING J, BRETON G, OMELCHENKO M V, et al. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum [J]. Journal of Bacteriology, 2001, 183(16): 4823. |
| 56 | KWON S W, PAARI K A, MALAVIYA A, et al. Synthetic biology tools for genome and transcriptome engineering of solventogenic Clostridium [J]. Frontiers in Bioengineering and Biotechnology, 2020, 8:282. |
| 57 | MCALLISTER K N, SORG J A. CRISPR genome editing systems in the genus Clostridium: a timely advancement [J]. Journal of Bacteriology, 2019, 201(16): e00219-19. |
| 58 | JOSEPH R C, KIM N M, SANDOVAL N R. Recent developments of the synthetic biology toolkit for Clostridium [J]. Frontiers in Microbiology, 2018, 9:154. |
| 59 | MINTON N P, EHSAAN M, HUMPHREYS C M, et al. A roadmap for gene system development in Clostridium [J]. Anaerobe, 2016, 41:104-112. |
| 60 | PYNE M E, BRUDER M, MOO-YOUNG M, et al. Technical guide for genetic advancement of underdeveloped and intractable Clostridium [J]. Biotechnology Advances, 2014, 32(3): 623-641. |
| 61 | WILKINSON S R, YOUNG M. Targeted integration of genes into the Clostridium acetobutylicum chromosome [J]. Microbiology, 1994, 140(1): 89-95. |
| 62 | DESAI R P, PAPOUTSAKIS E T. Antisense RNA strategies for metabolic engineering of Clostridium acetobutylicum [J]. Applied and Environmental Microbiology, 1999, 65(3): 936-945. |
| 63 | TUMMALA S B, WELKER N E, PAPOUTSAKIS E T. Design of antisense RNA constructs for downregulation of the acetone formation pathway of Clostridium acetobutylicum [J]. Journal of Bacteriology, 2003, 185(6): 1923-1934. |
| 64 | GUO H T, KARBERG M, LONG M, et al. Group Ⅱ introns designed to insert into therapeutically relevant DNA target sites in human cells [J]. Science, 2000, 289(5478): 452-457. |
| 65 | HEAP J T, PENNINGTON O J, CARTMAN S T, et al. The ClosTron: a universal gene knock-out system for the genus Clostridium [J]. Journal of Microbiological Methods, 2007, 70(3): 452-464. |
| 66 | SHAO L, HU S, YANG Y, et al. Targeted gene disruption by use of a group Ⅱ intron (targetron) vector in Clostridium acetobutylicum [J]. Cell Research, 2007, 17(11): 963-965. |
| 67 | HEAP J T, KUEHNE S A, EHSAAN M, et al. The ClosTron: Mutagenesis in Clostridium refined and streamlined [J]. Journal of Microbiological Methods, 2010, 80(1): 49-55. |
| 68 | MOHR G, HONG W, ZHANG J, et al. A targetron system for gene targeting in thermophiles and its application in Clostridium thermocellum [J]. Plos ONE, 2013, 8(7):e69032. |
| 69 | LIU Y-J, ZHANG J, CUI G-Z, et al. Current progress of targetron technology: development, improvement and application in metabolic engineering [J]. Biotechnology Journal, 2015, 10(6): 855-865. |
| 70 | ARGYROS D A, TRIPATHI S A, BARRETT T F, et al. High ethanol titers from cellulose by using metabolically engineered thermophilic, anaerobic microbes [J]. Applied and Environmental Microbiology, 2011, 77(23): 8288-8294. |
| 71 | AL-HINAI M A, FAST A G, PAPOUTSAKIS E T. Novel system for efficient isolation of Clostridium double-crossover allelic exchange mutants enabling markerless chromosomal gene deletions and DNA integration [J]. Applied and Environmental Microbiology, 2012, 78(22): 8112-8121. |
| 72 | CARTMAN S T, KELLY M L, HEEG D, et al. Precise manipulation of the Clostridium difficile chromosome reveals a lack of association between the tcdC genotype and toxin production [J]. Applied and Environmental Microbiology, 2012, 78(13): 4683-4690. |
| 73 | ZHANG N, SHAO L, JIANG Y, et al. I-SceI-mediated scarless gene modification via allelic exchange in Clostridium [J]. Journal of Microbiological Methods, 2015, 108: 49-60. |
| 74 | WANG Y, ZHANG Z T, SEO S O, et al. Markerless chromosomal gene deletion in Clostridium beijerinckii using CRISPR/Cas9 system [J]. Journal of Biotechnology, 2015, 200: 1-5. |
| 75 | LI Q, CHEN J, MINTON N P, et al. CRISPR-based genome editing and expression control systems in Clostridium acetobutylicum and Clostridium beijerinckii [J]. Biotechnology journal, 2016, 11(7): 961-972. |
| 76 | WANG Y, ZHANG Z-T, S-O SEO, et al. Bacterial genome editing with CRISPR-Cas9: deletion, integration, single nucleotide modification, and desirable “clean” mutant selection in Clostridium beijerinckii as an example [J]. ACS Synthetic Biology, 2016, 5(7): 721-732. |
| 77 | XU T, LI Y, SHI Z, et al. Efficient genome editing in Clostridium cellulolyticum via CRISPR-Cas9 nickase [J]. Applied and Environmental Microbiology, 2015, 81(13): 4423-4431. |
| 78 | HONG W, ZHANG J, CUI G, et al. Multiplexed CRISPR-Cpf1-mediated genome editing in Clostridium difficile toward the understanding of pathogenesis of C. difficile infection [J]. ACS Synthetic Biology, 2018, 7(6): 1588-1600. |
| 79 | CHUNG D. Harnessing heterologous and endogenous CRISPR-Cas machineries for efficient markerless genome editing in Clostridium [J]. Scientific Reports, 2016, 6: 25666 |
| 80 | LI Q, SEYS F M, MINTON N P, et al. CRISPR-Cas9(D10A) nickase-assisted base editing in the solvent producer Clostridium beijerinckii [J]. Biotechnology and Bioengineering, 2019, 116(6): 1475-1483. |
| 81 | BRUDER M R, PYNE M E, MOO-YOUNG M, et al. Extending CRISPR-Cas9 technology from genome editing to transcriptional engineering in the genus Clostridium [J]. Applied and Environmental Microbiology, 2016, 82(20): 6109-6119. |
| 82 | CHO C, LEE S Y. Efficient gene knockdown in Clostridium acetobutylicum by synthetic small regulatory RNAs [J]. Biotechnology and Bioengineering, 2017, 114(2): 374-383. |
| 83 | LANCKRIET A, TIMBERMONT L, HAPPONEN L J, et al. Generation of single-copy transposon insertions in Clostridium perfringens by electroporation of phage mu DNA transposition complexes [J]. Applied and Environmental Microbiology, 2009, 75(9): 2638-2642. |
| 84 | VIDAL J E, CHEN J, LI J, et al. Use of an EZ-Tn5-based random mutagenesis system to identify a novel toxin regulatory locus in Clostridium perfringens strain 13 [J]. PLoS One, 2009, 4(7): e6232. |
| 85 | CARTMAN S T, MINTON N P. A mariner-based transposon system for in vivo random mutagenesis of Clostridium difficile [J]. Applied and Environmental Microbiology, 2010, 76(4): 1103-1109. |
| 86 | ZHANG Y, XU S, CHAI C, et al. Development of an inducible transposon system for efficient random mutagenesis in Clostridium acetobutylicum [J]. FEMS Microbiology Letters, 2016, 363(8):fnw065. |
| 87 | STRECKER J, LADHA A. RNA-guided DNA insertion with CRISPR-associated transposases [J]. Science, 2019, 365(6448): 48-53. |
| 88 | HEAP J T, EHSAAN M, COOKSLEY C M, et al. Integration of DNA into bacterial chromosomes from plasmids without a counter-selection marker [J]. Nucleic Acids Research, 2012, 40(8): e59. |
| 89 | HUANG H, CHAI C, YANG S, et al. Phage serine integrase-mediated genome engineering for efficient expression of chemical biosynthetic pathway in gas-fermenting Clostridium ljungdahlii [J]. Metabolic Engineering, 2019, 52: 293-302. |
| 90 | DONG H, TAO W, GONG F, et al. A functional recT gene for recombineering of Clostridium [J]. Journal of Biotechnology, 2014, 173: 65-67. |
| 91 | JIANG Y, LIU J, JIANG W, et al. Current status and prospects of industrial bio-production of n-butanol in China [J]. Biotechnology Advances, 2015, 33(7): 1493-1501. |
| 92 | VENTURA J-R S, HU H, JAHNG D. Enhanced butanol production in Clostridium acetobutylicum ATCC 824 by double overexpression of 6-phosphofructokinase and pyruvate kinase genes [J]. Applied Microbiology and Biotechnology, 2013, 97(16): 7505-7516. |
| 93 | MANN M S, LUETKE-EVERSLOH T. Thiolase engineering for enhanced butanol production in Clostridium acetobutylicum [J]. Biotechnology and Bioengineering, 2013, 110(3): 887-897. |
| 94 | LI A, WEN Z, FANG D, et al. Developing Clostridium diolis as a biorefinery chassis by genetic manipulation [J]. Bioresource Technology, 2020, 305: 123066. |
| 95 | HOU X, PENG W, XIONG L, et al. Engineering Clostridium acetobutylicum for alcohol production [J]. Journal of Biotechnology, 2013, 166(1/2): 25-33. |
| 96 | NAIR R V, GREEN E M, WATSON D E, et al. Regulation of the sol locus genes for butanol and acetone formation in Clostridium acetobutylicum ATCC 824 by a putative transcriptional repressor [J]. Journal of Bacteriology, 1999, 181(1): 319-330. |
| 97 | HARRIS L M, BLANK L, DESAI R P, et al. Fermentation characterization and flux analysis of recombinant strains of Clostridium acetobutylicum with an inactivated solR gene [J]. Journal of Industrial Microbiology & Biotechnology, 2001, 27(5): 322-328. |
| 98 | BORMANN S, BAER Z C, SREEKUMAR S, et al. Engineering Clostridium acetobutylicum for production of kerosene and diesel blendstock precursors [J]. Metabolic Engineering, 2014, 25: 124-130. |
| 99 | COOKSLEY C M, ZHANG Y, WANG H, et al. Targeted mutagenesis of the Clostridium acetobutylicum acetone-butanol-ethanol fermentation pathway [J]. Metabolic Engineering, 2012, 14(6): 630-641. |
| 100 | LEHMANN D, RADOMSKI N, LUETKE-EVERSLOH T. New insights into the butyric acid metabolism of Clostridium acetobutylicum [J]. Applied Microbiology and Biotechnology, 2012, 96(5): 1325-1339. |
| 101 | LIU J, JIANG Y, CHEN J, et al. Metabolic engineering and adaptive evolution of Clostridium beijerinckii to increase solvent production from corn stover hydrolysate [J]. Journal of Agricultural and Food Chemistry, 2020, 68(30): 7916-7925. |
| 102 | LU C, YU L, VARGHESE S, et al. Enhanced robustness in acetone-butanol-ethanol fermentation with engineered Clostridium beijerinckii overexpressing adhE2 and ctfAB [J]. Bioresource Technology, 2017, 243: 1000-1008. |
| 103 | NGOC-PHUONG-THAO N, RAYNAUD C, MEYNIAL-SALLES I, et al. Reviving the Weizmann process for commercial n-butanol production [J]. Nature Communications, 2018, 9:3682. |
| 104 | GREEN E M, BOYNTON Z L, HARRIS L M, et al. Genetic manipulation of acid formation pathways by gene inactivation in Clostridium acetobutylicum ATCC 824 [J]. Microbiology, 1996, 142 (8): 2079-2086. |
| 105 | WANG Y, LI X, MILNE C B, et al. Development of a gene knockout system using mobile group II introns (Targetron) and genetic disruption of acid production pathways in Clostridium beijerinckii [J]. Applied and environmental microbiology, 2013, 79(19): 5853-63. |
| 106 | ZHAO Y, TOMAS C A, RUDOLPH F B, et al. Intracellular butyryl phosphate and acetyl phosphate concentrations in Clostridium acetobutylicum and their implications for solvent formation [J]. Applied & Environmental Microbiology, 2005, 71(1): 530-537. |
| 107 | MERMELSTEIN L D, PAPOUTSAKIS E T, PETERSEN D J, et al. Metabolic engineering of Clostridium acetobutylicum ATCC 824 for increased solvent production by enhancement of acetone formation enzyme activities using a synthetic acetone operon [J]. Biotechnology and Bioengineering, 1993, 42(9): 1053-1060. |
| 108 | JIANG Y, XU C, DONG F, et al. Disruption of the acetoacetate decarboxylase gene in solvent-producing Clostridium acetobutylicum increases the butanol ratio [J]. Metabolic Engineering, 2009, 11(4-5): 284-291. |
| 109 | LEHMANN D, HOENICKE D, EHRENREICH A, et al. Modifying the product pattern of Clostridium acetobutylicum [J]. Applied Microbiology and Biotechnology, 2012, 94(3): 743-754. |
| 110 | BAO T, FENG J, JIANG W, et al. Recent advances in n-butanol and butyrate production using engineered Clostridium tyrobutyricum [J]. World Journal of Microbiology & Biotechnology, 2020, 36(9): 138. |
| 111 | YU L, ZHAO J, XU M, et al. Metabolic engineering of Clostridium tyrobutyricum for n-butanol production: effects of CoA transferase [J]. Applied Microbiology and Biotechnology, 2015, 99(11): 4917-4930. |
| 112 | ZHANG J, YU L, XU M, et al. Metabolic engineering of Clostridium tyrobutyricum for n-butanol production from sugarcane juice [J]. Applied Microbiology and Biotechnology, 2017, 101(10): 4327-4337. |
| 113 | YU L, XU M, TANG I C, et al. Metabolic engineering of Clostridium tyrobutyricum for n-butanol production from maltose and soluble starch by overexpressing α-glucosidase [J]. Applied Microbiology and Biotechnology, 2015, 99(14): 6155-6165. |
| 114 | DU Y, JIANG W, YU M, et al. Metabolic process engineering of Clostridium tyrobutyricum delta ack-adhE2 for enhanced n-butanol production from glucose: effects of methyl viologen on NADH availability, flux distribution, and fermentation kinetics [J]. Biotechnology and Bioengineering, 2015, 112(4): 705-715. |
| 115 | WEN Z, LEDESMA-AMARO R, LIN J, et al. Improved n-butanol production from Clostridium cellulovorans by integrated metabolic and evolutionary engineering [J]. Applied and Environmental Microbiology, 2019, 85(7): e02560-18. |
| 116 | BAO T, ZHAO J, LI J, et al. n-Butanol and ethanol production from cellulose by Clostridium cellulovorans overexpressing heterologous aldehyde/alcohol dehydrogenases [J]. Bioresource Technology, 2019, 285: 121316. |
| 117 | GAIDA S M, LIEDTKE A, JENTGES A H W, et al. Metabolic engineering of Clostridium cellulolyticum for the production of n-butanol from crystalline cellulose [J]. Microbial Cell Factories, 2016, 15:6. |
| 118 | TIAN L, CONWAY P M, CERVENKA N D, et al. Metabolic engineering of Clostridium thermocellum for n-butanol production from cellulose [J]. Biotechnology for Biofuels, 2019, 12:186. |
| 119 | JIA D, JIANG W, GU Y. Research progresses in gas-fermenting clostridia [J]. Microbiology China, 2019, 46(2): 374-387. |
| 120 | ZHAO R, LIU Y, ZHANG H, et al. CRISPR-Cas12a-mediated gene deletion and regulation in Clostridium ljungdahlii and its application in carbon flux redirection in synthesis gas fermentation [J]. ACS Synthetic Biology, 2019, 8(10): 2270-2279. |
| 121 | LEMGRUBER R D S P, VALGEPEA K, TAPPEL R, et al. Systems-level engineering and characterisation of Clostridium autoethanogenum through heterologous production of poly-3-hydroxybutyrate (PHB) [J]. Metabolic Engineering, 2019, 53: 14-23. |
| 122 | VALGEPEA K, LOI K Q, BEHRENDORFF J B, et al. Arginine deiminase pathway provides ATP and boosts growth of the gas-fermenting acetogen Clostridium autoethanogenum [J]. Metabolic Engineering, 2017, 41: 202-211. |
| 123 | W-C KAO, LIN D-S, CHENG C-L, et al. Enhancing butanol production with Clostridium pasteurianum CH4 using sequential glucose–glycerol addition and simultaneous dual-substrate cultivation strategies [J]. Bioresource Technology, 2013, 135: 324-330. |
| 124 | PYNE M E, SOKOLENKO S, LIU X, et al. Disruption of the reductive 1,3-propanediol pathway triggers production of 1,2-propanediol for sustained glycerol fermentation by Clostridium pasteurianum [J]. Applied and Environmental Microbiology, 2016, 82(17): 5375-5388. |
| 125 | SCHWARZ K M, GROSSE-HONEBRINK A, DERECKA K, et al. Towards improved butanol production through targeted genetic modification of Clostridium pasteurianum [J]. Metabolic Engineering, 2017, 40: 124-137. |
| 126 | CORNILLOT E, NAIR R V, PAPOUTSAKIS E T, et al. The genes for butanol and acetone formation in Clostridium acetobutylicum ATCC 824 reside on a large plasmid whose loss leads to degeneration of the strain [J]. Journal of Bacteriology, 1997, 179(17): 5442-5447. |
| 127 | NAIR R V, PAPOUTSAKIS E T. Expression of plasmid-encoded aad in Clostridium acetobutylicum M5 restores vigorous butanol production [J]. Journal of Bacteriology, 1994, 176(18): 5843-5846. |
| 128 | SILLERS R, CHOW A, TRACY B, et al. Metabolic engineering of the non-sporulating, non-solventogenic Clostridium acetobutylicum strain M5 to produce butanol without acetone demonstrate the robustness of the acid-formation pathways and the importance of the electron balance [J]. Metabolic Engineering, 2008, 10(6): 321-332. |
| 129 | LEE J Y, Y-S JANG, LEE J, et al. Metabolic engineering of Clostridium acetobutylicum M5 for highly selective butanol production [J]. Biotechnology Journal, 2009, 4(10): 1432-1440. |
| 130 | OU J, XU N, ERNST P, et al. Process engineering of cellulosic n-butanol production from corn-based biomass using Clostridium cellulovorans [J]. Process Biochemistry, 2017, 62: 144-150. |
| 131 | YU L, XU M, TANG I C, et al. Metabolic engineering of Clostridium tyrobutyricum for n-butanol production through co-utilization of glucose and xylose [J]. Biotechnology and Bioengineering, 2015, 112(10): 2134-2141. |
| 132 | GU Y, JIANG Y, YANG S, et al. Utilization of economical substrate-derived carbohydrates by solventogenic clostridia: pathway dissection, regulation and engineering [J]. Current Opinion in Biotechnology, 2014, 29: 124-131. |
| 133 | GU Y, LI J, ZHANG L, et al. Improvement of xylose utilization in Clostridium acetobutylicum via expression of the talA gene encoding transaldolase from Escherichia coli [J]. Journal of Biotechnology, 2009, 143(4): 284-287. |
| 134 | JIN L, ZHANG H, CHEN L, et al. Combined overexpression of genes involved in pentose phosphate pathway enables enhanced D-xylose utilization by Clostridium acetobutylicum [J]. Journal of Biotechnology, 2014, 173: 7-9. |
| 135 | XIAO H, GU Y, NING Y, et al. Confirmation and elimination of xylose metabolism bottlenecks in glucose phosphoenolpyruvate-dependent phosphotransferase system-deficient Clostridium acetobutylicum for simultaneous utilization of glucose, xylose, and arabinose [J]. Applied and Environmental Microbiology, 2011, 77(22): 7886-7895. |
| 136 | LI Z, XIAO H, JIANG W, et al. Improvement of solvent production from xylose mother liquor by engineering the xylose metabolic pathway in Clostridium acetobutylicum EA 2018 [J]. Applied Biochemistry and Biotechnology, 2013, 171(3): 555-568. |
| 137 | REN C, GU Y, HU S, et al. Identification and inactivation of pleiotropic regulator CcpA to eliminate glucose repression of xylose utilization in Clostridium acetobutylicum [J]. Metabolic Engineering, 2010, 12(5): 446-454. |
| 138 | BRUDER M, MOO-YOUNG M, CHUNG D A, et al. Elimination of carbon catabolite repression in Clostridium acetobutylicum-a journey toward simultaneous use of xylose and glucose [J]. Applied Microbiology and Biotechnology, 2015, 99(18): 7579-7588. |
| 139 | XIAO H, LI Z L, JIANG Y, et al. Metabolic engineering of D-xylose pathway in Clostridium beijerinckii to optimize solvent production from xylose mother liquid [J]. Metabolic Engineering, 2012, 14(5): 569-578. |
| 140 | GU Y, DING Y, REN C, et al. Reconstruction of xylose utilization pathway and regulons in Firmicutes [J]. BMC Genomics, 2010, 11:255. |
| 141 | LIU L, ZHANG L, TANG W, et al. Phosphoketolase pathway for xylose catabolism in Clostridium acetobutylicum Revealed by C-13 metabolic flux analysis [J]. Journal of Bacteriology, 2012, 194(19): 5413-5422. |
| 142 | SERVINSKY M D, GERMANE K L, LIU S, et al. Arabinose is metabolized via a phosphoketolase pathway in Clostridium acetobutylicum ATCC 824 [J]. Journal of Industrial Microbiology & Biotechnology, 2012, 39(12): 1859-1867. |
| 143 | ZHANG L, LEYN S A, GU Y, et al. Ribulokinase and transcriptional regulation of arabinose metabolism in Clostridium acetobutylicum [J]. Journal of Bacteriology, 2012, 194(5): 1055-1064. |
| 144 | SUN Z, CHEN Y, YANG C, et al. A novel three-component system-based regulatory model for D-xylose sensing and transport in Clostridium beijerinckii [J]. Molecular Microbiology, 2015, 95(4): 576-589. |
| 145 | LI J X, WANG C Y, YANG G H, et al. Molecular mechanism of environmental D-xylose perception by a XylFII-LytS complex in bacteria [J]. Proceedings of the National Academy of Sciences of the United States of America, 2017, 114(31): 8235-8240. |
| 146 | NESSLER S, FIEULAINE S, PONCET S, et al. HPr kinase/phosphorylase, the sensor enzyme of catabolite repression in Gram-positive bacteria: structural aspects of the enzyme and the complex with its protein substrate [J]. Journal of Bacteriology, 2003, 185(14): 4003-4010. |
| 147 | LORCA G L, CHUNG Y J, BARABOTE R D, et al. Catabolite repression and activation in Bacillus subtilis: dependency on CcpA, HPr, and HprK [J]. Journal of Bacteriology, 2005, 187(22): 7826-7839. |
| 148 | HU S, ZHENG H, GU Y, et al. Comparative genomic and transcriptomic analysis revealed genetic characteristics related to solvent formation and xylose utilization in Clostridium acetobutylicum EA 2018 [J]. BMC Genomics, 2011, 12:93. |
| 149 | PATAKOVA P, KOLEK J, SEDLAR K, et al. Comparative analysis of high butanol tolerance and production in clostridia [J]. Biotechnology Advances, 2018, 36(3): 721-738. |
| 150 | PEABODY G L, KAO K C. Recent progress in biobutanol tolerance in microbial systems with an emphasis on Clostridium [J]. FEMS Microbiology Letters, 2016, 363(5):fnw017. |
| 151 | BARAL N R, SHAH A. Microbial inhibitors: formation and effects on acetone-butanol-ethanol fermentation of lignocellulosic biomass [J]. Applied Microbiology and Biotechnology, 2014, 98(22): 9151-9172. |
| 152 | IBRAHEEM O, NDIMBA B K. Molecular adaptation mechanisms employed by ethanologenic bacteria in response to lignocellulose-derived inhibitory compounds [J]. International Journal of Biological Sciences, 2013, 9(6): 598-612. |
| 153 | WILLSON B J, KOVACS K, WILDING-STEELE T, et al. Production of a functional cell wall-anchored minicellulosome by recombinant Clostridium acetobutylicum ATCC 824 [J]. Biotechnol Biofuels, 2016, 9:109. |
| 154 | WEN Z, LEDESMA-AMARO R, LU M, et al. Combined evolutionary engineering and genetic manipulation improve low pH tolerance and butanol production in a synthetic microbial Clostridium community [J]. Biotechnology and Bioengineering, 2020, 117(7): 2008-2022. |
| [1] | GUO Shuyuan, ZHANG Qiannan, Gulikezi· MAIMAITIREXIATI, YANG Yiqun, YU Tao. Advances in microbial production of liquid biofuels [J]. Synthetic Biology Journal, 2025, 6(1): 18-44. |
| [2] | Xue BAI, Nan LIANG, Xinqi LI, Zhipeng MO, Shuhuan TONG, Meiqi YUE, Xiaojing JIA, Kang REN, Xiaojie XI, Wei CHAO. Research on the equipment, process, and commercialization progress of syngas fermentation [J]. Synthetic Biology Journal, 2025, (): 1-22. |
| [3] | Ping ZHANG, Weijiao ZHANG, Ruirui XU, Jianghua LI, Jian CHEN, Zhen KANG. Research advances on Mycosporine-like amino acids biosynthesis [J]. Synthetic Biology Journal, 2024, (): 1-14. |
| [4] | Chuangen TANG, Jing WANG, Shuo ZHANG, Haoning ZHANG, Zhen KANG. Recent advances in synthesis and mining strategies of functional peptides [J]. Synthetic Biology Journal, 2024, (): 1-18. |
| [5] | SUN Huili, CUI Jinyu, LUAN Guodong, LYU Xuefeng. Progress of cyanobacterial synthetic biotechnology for efficient light-driven carbon fixation and ethanol production [J]. Synthetic Biology Journal, 2023, 4(6): 1161-1177. |
| [6] | BAI Zhonghu, REN He, NIE Jianqi, SUN Yang. The recent progresses and applications of in-parallel fermentation technology [J]. Synthetic Biology Journal, 2023, 4(5): 904-915. |
| [7] | TAO Fei, SUN Tao, WANG Yu, WEI Ting, NI Jun, XU Ping. Challenges and opportunities in the research of Synechococcus chassis under the context of carbon peak and neutrality [J]. Synthetic Biology Journal, 2022, 3(5): 932-952. |
| [8] | CUI Jinyu, ZHANG Aidi, LUAN Guodong, LYU Xuefeng. Engineering microalgae for photosynthetic biosynthesis: progress and prospect [J]. Synthetic Biology Journal, 2022, 3(5): 884-900. |
| [9] | CAO Chenkai, LI Jialong, ZHANG Kechun. Progress in artificial metabolic pathways for biosynthesis of organic alcohols & acids [J]. Synthetic Biology Journal, 2021, 2(6): 902-919. |
| [10] | ZHANG Xiaolong, WANG Chenyun, LIU Yanfeng, LI Jianghua, LIU Long, DU Guocheng. Research progress of constructing efficient biomanufacturing system based on synthetic biotechnology [J]. Synthetic Biology Journal, 2021, 2(6): 863-875. |
| [11] | HOU Zhengjie, SUN Huizhong, BAI Song, CHEN Xinyue, CAO Chunyang, CHENG Jingsheng. Research progress of cyclic lipopeptide biosynthesis [J]. Synthetic Biology Journal, 2021, 2(4): 577-597. |
| [12] | ZHANG Hui, YUAN Yaomeng, ZHANG Chong, YANG Song, XING Xinhui. Research progresses and future prospects of synthetic methylotrophic cell factory for methanol assimilation [J]. Synthetic Biology Journal, 2021, 2(2): 222-233. |
| [13] | Jiaoqi GAO, Yongjin ZHOU. Advances in methanol bio-transformation [J]. Synthetic Biology Journal, 2020, 1(2): 158-173. |
| [14] | Sen XIAO, Litao HU, Zhicheng SHI, Fayin WANG, Siting YU, Guocheng DU, Jian CHEN, Zhen KANG. Research advances in biosynthesis of hyaluronic acid with controllable molecular weights [J]. Synthetic Biology Journal, 2024, (): 1-16. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||