Synthetic Biology Journal ›› 2021, Vol. 2 ›› Issue (4): 509-527.DOI: 10.12211/2096-8280.2021-041
• Invited Review • Previous Articles Next Articles
In memory of Prof. Daniel I.C. Wang: Engineering Yarrowia lipolytica for the production of plant-based lipids: technical constraints and perspectives for a sustainable cellular agriculture economy
XU Peng1,2
- 1.Department of Chemical Engineering,Guangdong-Technion,Israel Institute of Technology,Shantou 515063,Guangdong,China
2.Department of Chemical,Biochemical and Environmental Engineering,University of Maryland,Baltimore County,Baltimore,MD 21250,USA
-
Received:2021-04-03Revised:2021-05-12Online:2021-09-10Published:2021-08-31
纪念王义翘教授:解脂耶氏酵母替代植物油脂的技术瓶颈及展望
徐鹏1,2
- 1.广东以色列理工学院化学工程系,广东 汕头 515063
2.马里兰大学巴尔的摩分校化学、生化与环境工程系,马里兰州 巴尔的摩 21250,美国
-
作者简介:徐鹏 (1980—),男,工学博士,于2020年获得“生物技术与生物工程王义翘教授奖” (Biotechnology & Bioengineering Daniel I.C. Wang Award)。研究方向主要涉及代谢工程、合成生物学、生化工程、生物分子绿色制造、智能控制和生化反应网络建模等。E-mail:peng.xu@gtiit.edu.cn
CLC Number:
Cite this article
XU Peng. In memory of Prof. Daniel I.C. Wang: Engineering Yarrowia lipolytica for the production of plant-based lipids: technical constraints and perspectives for a sustainable cellular agriculture economy[J]. Synthetic Biology Journal, 2021, 2(4): 509-527.
徐鹏. 纪念王义翘教授:解脂耶氏酵母替代植物油脂的技术瓶颈及展望[J]. 合成生物学, 2021, 2(4): 509-527.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2021-041
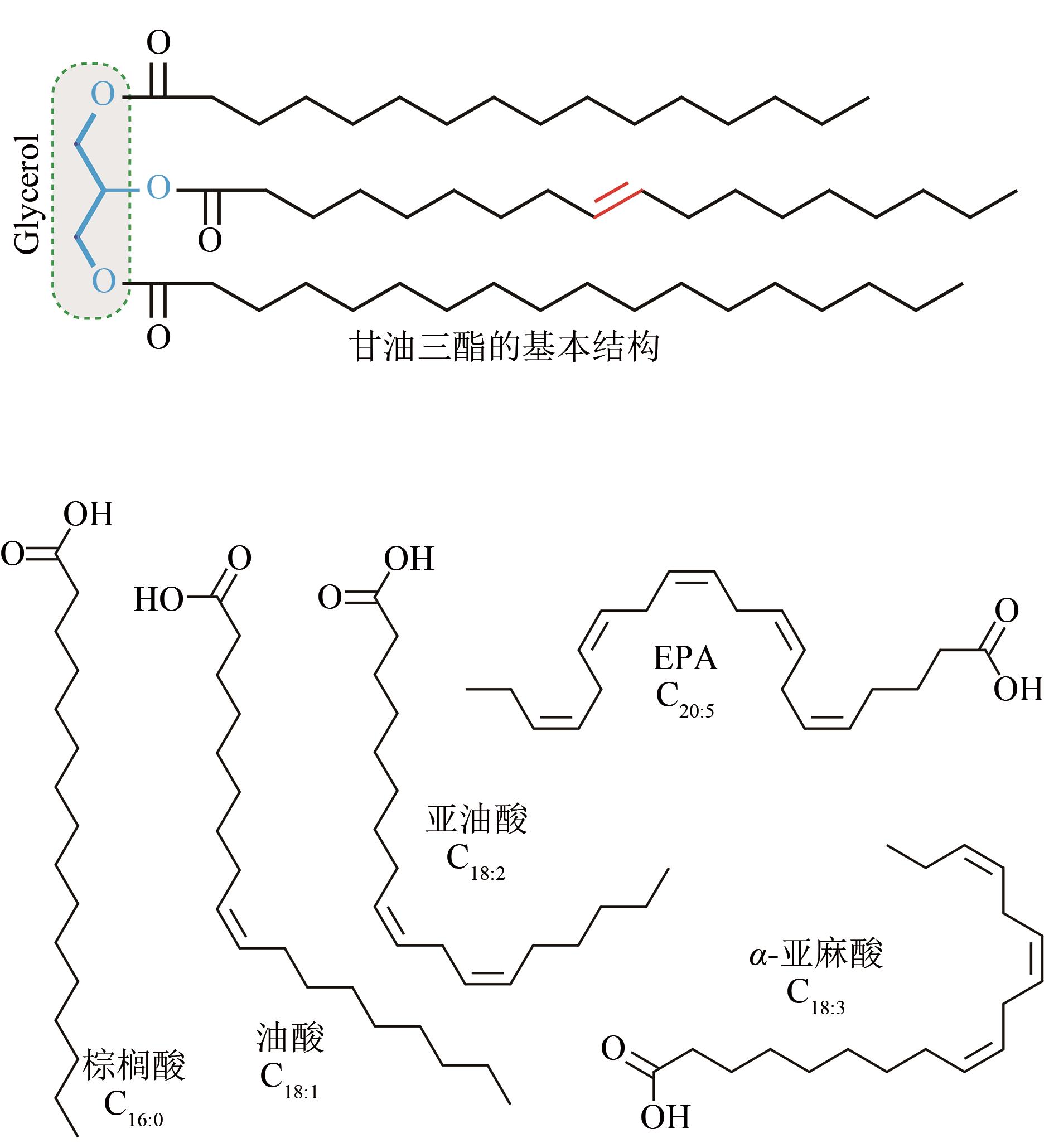
Fig. 1 The basic structure of triglyceride and the four important fatty acids: palmitic acid (C16∶0), oleic acid (C18∶1), linoleic acid (C18∶2), eicosapentaenoic acid (EPA, C20∶5) and α-linolenic acid (ALA, C18∶3).
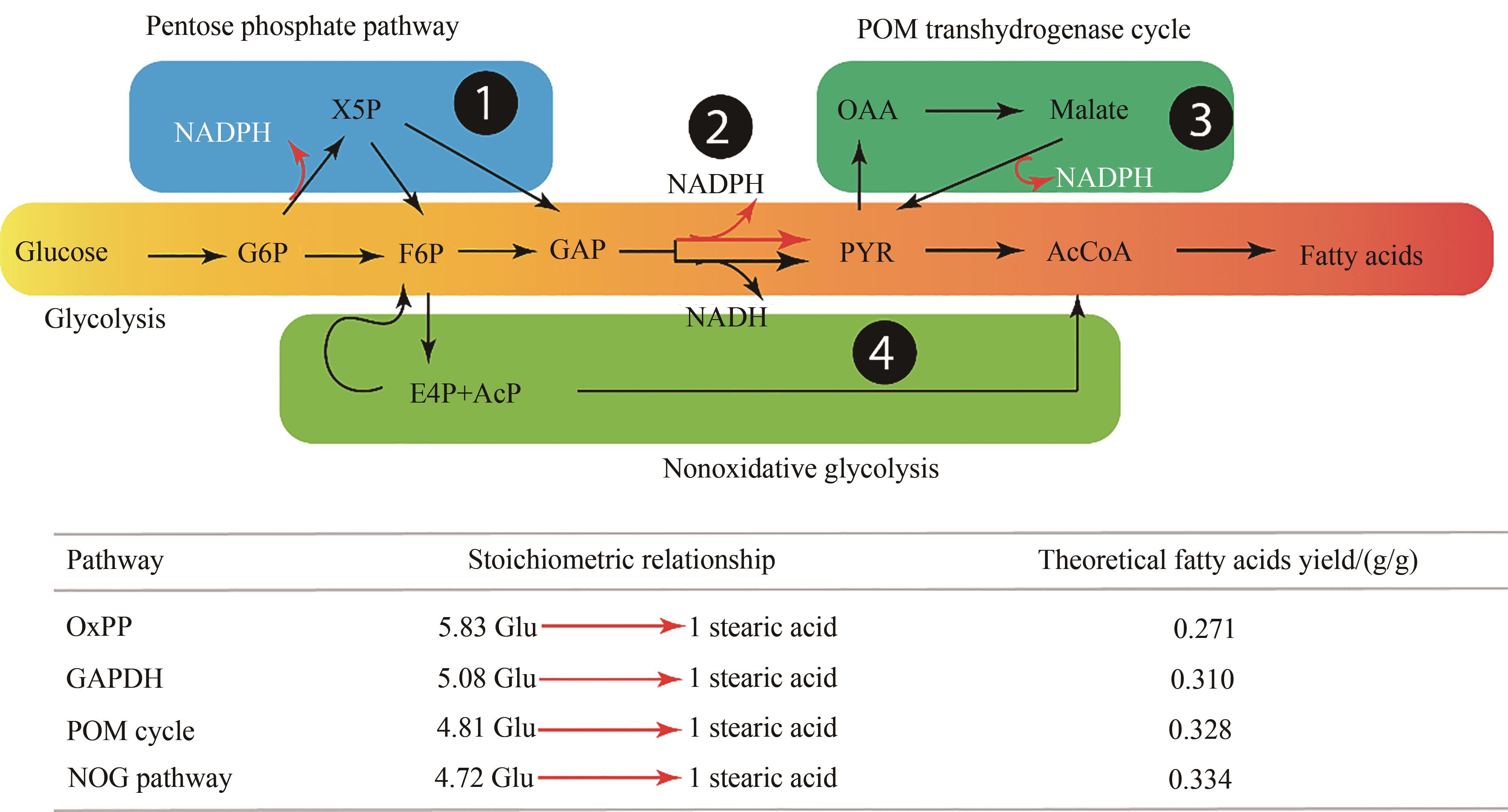
Fig. 4 Carbon conversion efficiency from NADPH for fatty acids synthesis pathways [56](OxPP—oxidative pentose phosphate pathway; GAPHD—NADPH-specific glyceraldehyde-3-phosphate dehydrogenase; POM cycle—pyruvate-oxaloacetate-malate transhydrogenase cycle; NOG—non-oxidative glycolytic pathway)
| 方程编号 Equation No. | 方程 Equations | 描述 Description |
|---|---|---|
| (1) | 细胞比生长速率 Specific growth rate | |
| (2) | 非油脂生物量积累 Oil-free cell growth | |
| (3) | 油脂积累 Lipid accumulation | |
| (4) | 底物消耗 Substrate consumption | |
| (5) | 总生物量 Total biomass | |
| (6) | 动态含油量 Oil content | |
| (7) | 动态过程得率 Process yield | |
| (8) | 总的生产强度 Overall productivity |
Tab. 1 Fermentation kinetics of the oil accumulation process in oleaginous yeast in batch culture
| 方程编号 Equation No. | 方程 Equations | 描述 Description |
|---|---|---|
| (1) | 细胞比生长速率 Specific growth rate | |
| (2) | 非油脂生物量积累 Oil-free cell growth | |
| (3) | 油脂积累 Lipid accumulation | |
| (4) | 底物消耗 Substrate consumption | |
| (5) | 总生物量 Total biomass | |
| (6) | 动态含油量 Oil content | |
| (7) | 动态过程得率 Process yield | |
| (8) | 总的生产强度 Overall productivity |
| 油料作物 Oil crop | 产量/[t/(hm2·a)] Production | 含油量/% Oil content | 最大产油得率/[t/(hm2·a)] Maximal oil yield |
|---|---|---|---|
| 黄豆 Soybean | 3.1 | 20 | 0.62 |
| 葵花籽 Sunflower seed | 1.7 | 50 | 0.85 |
| 油菜籽 Canola | 2.0 | 45 | 0.90 |
| 可可果 Cocoa bean | 0.8 | 50 | 0.40 |
| 棕榈果 Oil palm | 9.1 | 41 | 3.69 |
Tab. 2 The unit area output and maximal oil yield from major oil crops.
| 油料作物 Oil crop | 产量/[t/(hm2·a)] Production | 含油量/% Oil content | 最大产油得率/[t/(hm2·a)] Maximal oil yield |
|---|---|---|---|
| 黄豆 Soybean | 3.1 | 20 | 0.62 |
| 葵花籽 Sunflower seed | 1.7 | 50 | 0.85 |
| 油菜籽 Canola | 2.0 | 45 | 0.90 |
| 可可果 Cocoa bean | 0.8 | 50 | 0.40 |
| 棕榈果 Oil palm | 9.1 | 41 | 3.69 |
| 糖料作物 Sugar crop | 产量/[t/(hm2·a)] Production | 含糖量/% Sugar content | 最大产油得率①/[t/(hm2·a)] Maximal oil yield |
|---|---|---|---|
| 玉米 Corn | 10.7 (dry, 干重) | 74 (dry, 干重) | 2.15 |
| 红薯 Sweet potato | 25 (wet, 湿重) | 20 (wet, 湿重) | 1.36 |
| 甘蔗 Sugarcane | 80.9 (wet, 湿重) | 14 (wet, 湿重) | 3.07 |
Tab. 3 The maximal oil yield from Y. lipolytica converting sugar feedstock from corn, sweet potato or sugarcane.
| 糖料作物 Sugar crop | 产量/[t/(hm2·a)] Production | 含糖量/% Sugar content | 最大产油得率①/[t/(hm2·a)] Maximal oil yield |
|---|---|---|---|
| 玉米 Corn | 10.7 (dry, 干重) | 74 (dry, 干重) | 2.15 |
| 红薯 Sweet potato | 25 (wet, 湿重) | 20 (wet, 湿重) | 1.36 |
| 甘蔗 Sugarcane | 80.9 (wet, 湿重) | 14 (wet, 湿重) | 3.07 |
| 1 | AFEYAN N B, COONEY C L. Professor Daniel I C. Wang. a legacy of education, innovation, publication, and leadership [J]. Biotechnology and Bioengineering, 2020, 117(12): 3615-3627. |
| 2 | XUE Z, SHARPE P L, HONG S P, et al. Production of omega-3 eicosapentaenoic acid by metabolic engineering of Yarrowia lipolytica [J]. Nature Biotechnology, 2013, 31(8): 734-740. |
| 3 | SIMOPOULOS A P. The importance of the ratio of omega-6/omega-3 essential fatty acids [J]. Biomedicine & Pharmacotherapy, 2002, 56(8): 365-379. |
| 4 | VIJAY V, PIMM S L, JENKINS C N, et al. The impacts of oil palm on recent deforestation and biodiversity loss [J]. PLoS ONE, 2016, 11(7): e0159668. |
| 5 | CARLSON K M, HEILMAYR R, GIBBS H K, et al. Effect of oil palm sustainability certification on deforestation and fire in Indonesia [J]. Proceedings of the National Academy of Sciences, of the United States of America, 2018, 115(1): 121. |
| 6 | LEDESMA-AMARO R, NICAUD J M. Yarrowia lipolytica as a biotechnological chassis to produce usual and unusual fatty acids [J]. Progress in Lipid Research, 2016, 61: 40-50. |
| 7 | XU P, GU Q, WANG W, et al. Modular optimization of multi-gene pathways for fatty acids production in E. coli [J]. Nature Communications, 2013, 4: 1409. |
| 8 | HU P, CHAKRABORTY S, KUMAR A, et al. Integrated bioprocess for conversion of gaseous substrates to liquids [J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(14): 3773-3778. |
| 9 | XU J, LIU N, QIAO K, et al. Application of metabolic controls for the maximization of lipid production in semicontinuous fermentation [J]. Proceedings of the National Academy of Sciences of the United States of America, 2017, 114(27): E5308-E5316. |
| 10 | MA J, GU Y, MARSAFARI M, et al. Synthetic biology, systems biology, and metabolic engineering of Yarrowia lipolytica toward a sustainable biorefinery platform [J]. Journal of Industrial Microbiology and Biotechnology, 2020, 47(9/10): 845-862. |
| 11 | SCHWARTZ C M, HUSSAIN M S, BLENNER M, et al. Synthetic RNA polymerase III promoters facilitate high-efficiency CRISPR-Cas9-mediated genome editing in Yarrowia lipolytica [J]. ACS Synthetic Biology, 2016, 5(4): 356-359. |
| 12 | EDWARDS H, YANG Z, XU P. Characterization of Met25 as a color associated genetic marker in Yarrowia lipolytica [J]. Metabolic Engineering Communications, 2020, 11: e00147. |
| 13 | YANG Z, EDWARDS H, XU P. CRISPR-Cas12a/Cpf1-assisted precise, efficient and multiplexed genome-editing in Yarrowia lipolytica [J]. Metabolic Engineering Communications, 2020, 10: e00112. |
| 14 | YANG Z, BLENNER M. Genome editing systems across yeast species [J]. Current Opinion in Biotechnology, 2020, 66: 255-266. |
| 15 | YANG Z, XU P. Implementing CRISPR-Cas12a for efficient genome editing in Yarrowia lipolytica [M]// WHEELDON I, BLENNER M. Yarrowia lipolytica: methods and protocols. New York: Springer, 2021: 111-121. |
| 16 | CELIŃSKA E, LEDESMA-AMARO R, LARROUDE M, et al. Golden Gate Assembly system dedicated to complex pathway manipulation in Yarrowia lipolytica [J]. Microbial Biotechnology, 2017, 10(2): 450-455. |
| 17 | EGERMEIER M, SAUER M, MARX H. Golden Gate-based metabolic engineering strategy for wild-type strains of Yarrowia lipolytica [J]. FEMS Microbiology Letters, 2019, 366(4): fnz022. |
| 18 | LARROUDE M, PARK Y K, SOUDIER P, et al. A modular Golden Gate toolkit for Yarrowia lipolytica synthetic biology [J]. Microbial Biotechnology, 2019, 12(6): 1249-1259. |
| 19 | TONG Y, ZHOU J, ZHANG L, et al. A golden-gate based cloning toolkit to build violacein pathway libraries in Yarrowia lipolytica [J]. ACS Synthetic Biology, 2021, 10(1): 115-124. |
| 20 | WONG L, HOLDRIDGE B, ENGEL J, et al. Genetic tools for streamlined and accelerated pathway engineering in Yarrowia Lipolytica [M]// SANTOS C N S and AJIKUMAR P K. Microbial metabolic engineering: methods and protocols. New York: Springer, 2019, 155-177. |
| 21 | WONG L, ENGEL J, JIN E, et al. YaliBricks, a versatile genetic toolkit for streamlined and rapid pathway engineering in Yarrowia lipolytica [J]. Metabolic Engineering Communications, 2017, 5(): 68-77. |
| 22 | LIU H, MARSAFARI M, DENG L, et al. Understanding lipogenesis by dynamically profiling transcriptional activity of lipogenic promoters in Yarrowia lipolytica [J]. Applied Microbiology and Biotechnology, 2019, 103(7): 3167-3179. |
| 23 | GUO Z-P, BORSENBERGER V, CROUX C, et al. An artificial chromosome ylAC enables efficient assembly of multiple genes in Yarrowia lipolytica for biomanufacturing [J]. Communications Biology, 2020, 3(1): 199. |
| 24 | LV Y, EDWARDS H, ZHOU J, et al. Combining 26S rDNA and the Cre-loxP system for iterative gene integration and efficient marker curation in Yarrowia lipolytica [J]. ACS Synthetic Biology, 2019, 8(3): 568-576. |
| 25 | LIU J, WU X, YAO M, et al. Chassis engineering for microbial production of chemicals: from natural microbes to synthetic organisms [J]. Current Opinion in Biotechnology, 2020, 66: 105-112. |
| 26 | LIU Y, SU A, LI J, et al. Towards next-generation model microorganism chassis for biomanufacturing [J]. Applied Microbiology and Biotechnology, 2020, 104(21): 9095-9108. |
| 27 | PENDRILL F, PERSSON U M, GODAR J, et al. Agricultural and forestry trade drives large share of tropical deforestation emissions [J]. Global Environmental Change, 2019, 56: 1-10. |
| 28 | SCHROTH G, LÄDERACH P, MARTINEZ-VALLE A I, et al. Vulnerability to climate change of cocoa in West Africa: patterns, opportunities and limits to adaptation [J]. Science of the Total Environment, 2016, 556: 231-241. |
| 29 | MARKHAM K A, PALMER C M, CHWATKO M, et al. Rewiring Yarrowia lipolytica toward triacetic acid lactone for materials generation [J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(9): 2096. |
| 30 | LIU H, MARSAFARI M, WANG F, et al. Engineering acetyl-CoA metabolic shortcut for eco-friendly production of polyketides triacetic acid lactone in Yarrowia lipolytica [J]. Metabolic Engineering, 2019, 56: 60-68. |
| 31 | QIU X, XU P, ZHAO X, et al. Combining genetically-encoded biosensors with high throughput strain screening to maximize erythritol production in Yarrowia lipolytica [J]. Metabolic Engineering, 2020, 60: 66-76. |
| 32 | XU P, QIAO K, AHN W S, et al. Engineering Yarrowia lipolytica as a platform for synthesis of drop-in transportation fuels and oleochemicals [J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(39): 10848-10853. |
| 33 | SUN W, YANG Z, XU P. Engineering Yarrowia lipolytica for production of fatty alcohols with YaliBrick vectors[M]// WHEELDON I, BLENNER M.Yarrowia lipolytica: methods and protocols. New York: Springer, 2021: 159-173. |
| 34 | LI J, MA Y, LIU N,et al. Synthesis of high-titer alka(e)nes in Yarrowia lipolytica is enabled by a discovered mechanism [J]. Nature Communications, 2020, 11(1): 6198. |
| 35 | XUE Z, SHARPE P L, HONG S P, et al. Production of omega-3 eicosapentaenoic acid by metabolic engineering of Yarrowia lipolytica [J]. Nature Biotechnology, 2013, 31: 734-740. |
| 36 | LV Y, QIAN S, DU G, et al. Coupling feedback genetic circuits with growth phenotype for dynamic population control and intelligent bioproduction [J]. Metabolic Engineering, 2019, 54: 109-116. |
| 37 | LV Y, MARSAFARI M, KOFFAS M, et al. Optimizing oleaginous yeast cell factories for flavonoids and hydroxylated flavonoids biosynthesis[J]. ACS Synthetic Biology, 2019, 8(11): 2514-2523. |
| 38 | PALMER C M, MILLER K K, NGUYEN A, et al. Engineering 4-coumaroyl-CoA derived polyketide production in Yarrowia lipolytica through a β-oxidation mediated strategy [J]. Metabolic Engineering, 2020, 57: 174-181. |
| 39 | LIU H, WANG F, DENG L, et al. Genetic and bioprocess engineering to improve squalene production in Yarrowia lipolytica [J]. Bioresource Technology, 2020, 317: 123991. |
| 40 | HUANG Y Y, JIAN X X, LV Y B, et al. Enhanced squalene biosynthesis in Yarrowia lipolytica based on metabolically engineered acetyl-CoA metabolism [J]. Journal of Biotechnology, 2018, 281: 106-114. |
| 41 | TANG W-Y, WANG D-P, TIAN Y, et al. Metabolic engineering of Yarrowia lipolytica for improving squalene production [J]. Bioresource Technology, 2021, 323: 124652. |
| 42 | MARSAFARI M, XU P. Debottlenecking mevalonate pathway for antimalarial drug precursor amorphadiene biosynthesis in Yarrowia lipolytica [J]. Metabolic Engineering Communications, 2020, 10: e00121. |
| 43 | SÁEZ-SÁEZ J, WANG G, MARELLA E R, et al. Engineering the oleaginous yeast Yarrowia lipolytica for high-level resveratrol production [J]. Metabolic Engineering, 2020, 62: 51-61. |
| 44 | GU Y, MA J, ZHU Y, et al. Engineering Yarrowia lipolytica as a chassis for de novo synthesis of five aromatic-derived natural products and chemicals[J]. ACS Synthetic Biology, 2020, 9(8): 2096-2106. |
| 45 | GU Y, MA J, ZHU Y, et al. Refactoring ehrlich pathway for high-yield 2-phenylethanol production in Yarrowia lipolytica [J]. ACS Synthetic Biology, 2020, 9(3): 623-633. |
| 46 | SOARES G P A, SOUZA K S T, VILELA L F, et al. γ-Decalactone production by Yarrowia lipolytica and Lindnera saturnus in crude glycerol [J]. Preparative Biochemistry & Biotechnology, 2017, 47(6): 633-637. |
| 47 | BRAGA A, BELO I. Biotechnological production of γ-decalactone, a peach like aroma, by Yarrowia lipolytica [J]. World Journal of Microbiology and Biotechnology, 2016, 32(10): 169. |
| 48 | MARELLA E R, DAHLIN J, DAM M I, et al. A single-host fermentation process for the production of flavor lactones from non-hydroxylated fatty acids [J]. Metabolic Engineering, 2020, 61: 427-436. |
| 49 | MA J, GU Y, XU P. A roadmap to engineering antiviral natural products synthesis in microbes [J]. Current Opinion in Biotechnology, 2020, 66: 140-149. |
| 50 | MARSAFARI M, SAMIZADEH H, RABIEI B, et al. Biotechnological production of flavonoids: an update on plant metabolic engineering, microbial host selection, and genetically encoded biosensors [J]. Biotechnology Journal, 2020, 15(8): 1900432. |
| 51 | 汪庆卓, 黄和. 合成生物技术驱动天然的真核油脂细胞工厂开发[J]. 合成生物学, 2021, doi:10.12211/2096-8280.2020-090 . |
| WANG Q Z, HUANG H. Synthetic biotechnology drives the development of natural eukaryotic lipid cell factories [J]. Synthetic Biology Journal, 2021, doi: 10.12211/2096-8280.2020-090 . | |
| 52 | MUHAMMAD A, FENG X D, RASOOL A, et al. Production of plant natural products through engineered Yarrowia lipolytica [J]. Biotechnology Advances, 2020, 43: 107555. |
| 53 | WASYLENKO T M, AHN W S, STEPHANOPOULOS G. The oxidative pentose phosphate pathway is the primary source of NADPH for lipid overproduction from glucose in Yarrowia lipolytica [J]. Metabolic Engineering, 2015, 30: 27-39. |
| 54 | RATLEDGE C. The role of malic enzyme as the provider of NADPH in oleaginous microorganisms: a reappraisal and unsolved problems [J]. Biotechnology Letters, 2014, 36(8): 1557-1568. |
| 55 | WYNN J P, HAMID A B A, RATLEDGE C. The role of malic enzyme in the regulation of lipid accumulation in filamentous fungi [J]. Microbiology, 1999, 145(8): 1911-1917. |
| 56 | QIAO K, WASYLENKO T M, ZHOU K, et al. Lipid production in Yarrowia lipolytica is maximized by engineering cytosolic redox metabolism [J]. Nature Biotechnology, 2017, 35(2): 173-177. |
| 57 | JAYAKODY L N, JIN Y S. In-depth understanding of molecular mechanisms of aldehyde toxicity to engineer robust Saccharomyces cerevisiae [J]. Applied Microbiology and Biotechnology, 2021, 105(7): 2675-2692. |
| 58 | XU P, QIAO K, STEPHANOPOULOS G. Engineering oxidative stress defense pathways to build a robust lipid production platform in Yarrowia lipolytica [J]. Biotechnology and Bioengineering, 2017, 114(7): 1521-1530. |
| 59 | GU Y, XU P. Synthetic yeast brews neuroactive compounds [J]. Nature Chemical Biology, 2021, 17(1): 8-9. |
| 60 | SHUIB S, IBRAHIM I, MACKEEN M M, et al. First evidence for a multienzyme complex of lipid biosynthesis pathway enzymes in Cunninghamella bainieri [J]. Scientific Reports, 2018, 8(1): 3077. |
| 61 | RATLEDGE C. Regulation of lipid accumulation in oleaginous micro-organisms [J]. Biochemical Society Transactions, 2002, 30(6): 1047. |
| 62 | ZHOU Y J, BUIJS N A, ZHU Z, et al. Production of fatty acid-derived oleochemicals and biofuels by synthetic yeast cell factories [J]. Nature Communications, 2016, 7(1): 11709. |
| 63 | YU T, ZHOU Y J, WENNING L, et al. Metabolic engineering of Saccharomyces cerevisiae for production of very long chain fatty acid-derived chemicals [J]. Nature Communications, 2017, 8(1): 15587. |
| 64 | BELD J, LEE D J, BURKART M D. Fatty acid biosynthesis revisited: structure elucidation and metabolic engineering [J]. Molecular BioSystems, 2015, 11(1): 38-59. |
| 65 | ZHU Z, HU Y, TEIXEIRA P G, et al. Multidimensional engineering of Saccharomyces cerevisiae for efficient synthesis of medium-chain fatty acids [J]. Nature Catalysis, 2020, 3(1): 64-74. |
| 66 | GAJEWSKI J, PAVLOVIC R, FISCHER M, et al. Engineering fungal de novo fatty acid synthesis for short chain fatty acid production [J]. Nature Communications, 2017, 8(1): 14650. |
| 67 | SINGH K, GRAF B, LINDEN A, et al. Discovery of a regulatory subunit of the yeast fatty acid synthase [J]. Cell, 2020, 180(6): 1130-1143. |
| 68 | BLAZECK J, HILL A, LIU L, et al. Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production [J]. Nature Communications, 2014, 5: 3131. |
| 69 | ZHANG X K, NIE M Y, CHEN J, et al. Multicopy integrants of crt genes and co-expression of AMP deaminase improve lycopene production in Yarrowia lipolytica [J]. Journal of Biotechnology, 2019, 289: 46-54. |
| 70 | SILVERMAN A M, QIAO K, XU P, et al. Functional overexpression and characterization of lipogenesis-related genes in the oleaginous yeast Yarrowia lipolytica [J]. Applied Microbiology and Biotechnology, 2016, 100(8): 3781-3798. |
| 71 | CHANG L, TANG X, LU H, et al. Role of adenosine monophosphate deaminase during fatty acid accumulation in oleaginous fungus Mortierella alpina [J]. Journal of Agricultural and Food Chemistry, 2019, 67(34): 9551-9559. |
| 72 | SEIP J, JACKSON R, HE H, et al. Snf1 is a regulator of lipid accumulation in Yarrowia lipolytica [J]. Applied and Environmental Microbiology, 2013, 79(23): 7360-7370. |
| 73 | POMRANING K R, KIM Y M, NICORA C D, et al. Multi-omics analysis reveals regulators of the response to nitrogen limitation in Yarrowia lipolytica [J]. BMC Genomics, 2016, 17(1): 138. |
| 74 | KERKHOVEN E J, POMRANING K R, BAKER S E, et al. Regulation of amino-acid metabolism controls flux to lipid accumulation in Yarrowia lipolytica [J]. npj Systems Biology and Applications, 2016, 2: 16005. |
| 75 | BLAZECK J, HILL A, LIU L, et al. Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production [J]. Nature Communications, 2014, 5: 3131. |
| 76 | POMRANING K R, BREDEWEG E L, BAKER S E. Regulation of nitrogen metabolism by GATA zinc finger transcription factors in Yarrowia lipolytica [J]. mSphere, 2017, 2(1): e00038-17. |
| 77 | XU P. Analytical solution for a hybrid Logistic-Monod cell growth model in batch and continuous stirred tank reactor culture [J]. Biotechnology and Bioengineering, 2020, 117(3): 873-878. |
| 78 | ROEHR M, ZEHENTGRUBER O, KUBICEK C P. Kinetics of biomass formation and citric acid production by Aspergillus niger on pilot plant scale [J]. Biotechnology and Bioengineering, 1981, 23(11): 2433-2445. |
| 79 | CAI D, ZHU C, CHEN S. Microbial production of nattokinase: current progress, challenge and prospect [J]. World Journal of Microbiology and Biotechnology, 2017, 33(5): p. 84. |
| 80 | XU P, DING Z, QIAN Z, et al. Improved production of mycelial biomass and ganoderic acid by submerged culture of Ganoderma lucidum SB97 using complex media [J]. Enzyme and Microbial Technology, 2008, 42(4): 325-331. |
| 81 | SURENDHIRAN D, VIJAY M, SIVAPRAKASH B, et al. Kinetic modeling of microalgal growth and lipid synthesis for biodiesel production [J]. 3 Biotech, 2015, 5(5): 663-669. |
| 82 | PAPANIKOLAOU S, AGGELIS G. Modeling lipid accumulation and degradation in Yarrowia lipolytica cultivated on industrial fats [J]. Current Microbiology, 2003, 46(6): 0398-0402. |
| 83 | DULERMO R, GAMBOA-MELÉNDEZ H, DULERMO T, et al. The fatty acid transport protein Fat1p is involved in the export of fatty acids from lipid bodies in Yarrowia lipolytica [J]. FEMS Yeast Research, 2014, 14(6): 883-896. |
| 84 | RUMIN J, BONNEFOND H, SAINT-JEAN B, et al. The use of fluorescent Nile red and BODIPY for lipid measurement in microalgae [J]. Biotechnology for Biofuels, 2015, 8(1): 42. |
| 85 | FRIEDLANDER J, TSAKRAKLIDES V, KAMINENI A, et al. Engineering of a high lipid producing Yarrowia lipolytica strain [J]. Biotechnology for Biofuels, 2016, 9(1): 77. |
| 86 | MENG Y, YAO C, XUE S, et al. Application of Fourier transform infrared (FT-IR) spectroscopy in determination of microalgal compositions [J]. Bioresource Technology, 2014, 151: 347-354. |
| 87 | CHMIELARZ M, SAMPELS S, BLOMQVIST J, et al. FT-NIR: a tool for rapid intracellular lipid quantification in oleaginous yeasts [J]. Biotechnology for Biofuels, 2019, 12(1): 169. |
| 88 | NĚMCOVÁ A, GONOVÁ D, SAMEK O, et al. The use of raman spectroscopy to monitor metabolic changes in stressed Metschnikowia sp. yeasts [J]. Microorganisms, 2021, 9(2): 277. |
| 89 | BOWMAN E K, ALPER H S. Microdroplet-assisted screening of biomolecule production for metabolic engineering applications [J]. Trends in Biotechnology, 2020, 38(7): 701-714. |
| 90 | CAO W, CHENG S, YANG J, et al. Large-scale lipid analysis with C ̿ C location and sn-position isomer resolving power [J]. Nature Communications, 2020. 11(1): 375. |
| 91 | WANG M, WEI Y, JI B, et al. Advances in metabolic engineering of saccharomyces cerevisiae for cocoa butter equivalent production [J]. Frontiers in Bioengineering and Biotechnology, 2020, 8: 1194. |
| 92 | WEI Y, SIEWERS V, NIELSEN J. Cocoa butter-like lipid production ability of non-oleaginous and oleaginous yeasts under nitrogen-limited culture conditions [J]. Applied Microbiology and Biotechnology, 2017, 101(9): 3577-3585. |
| 93 | WEI Y, BERGENHOLM D, GOSSING M, et al. Expression of cocoa genes in Saccharomyces cerevisiae improves cocoa butter production [J]. Microbial Cell Factories, 2018, 17(1): 11. |
| 94 | XIONG D, ZHANG H Y, XIE Y F, et al. Conversion of mutton fat to cocoa butter equivalent by increasing the unsaturated fatty acids at the sn-2 position of triacylglycerol through fermentation by Yarrowia Lipolytica [J]. American Journal of Biochemistry and Biotechnology, 2015, 11(2): 57-65. |
| 95 | ZHAO L N, LI B, XIONG D, et al. Cocoa-butter-equivalent production from Yarrowia lipolytica by optimization of fermentation technology [J]. American Journal of Biochemistry and Biotechnology, 2016, 12(4): 196-205. |
| 96 | METZ J G, ROESSLER P, FACCIOTTI D, et al. Production of polyunsaturated fatty acids by polyketide synthases in both prokaryotes and eukaryotes [J]. Science, 2001, 293(5528): 290. |
| [1] | GUO Shuyuan, ZHANG Qiannan, Gulikezi· MAIMAITIREXIATI, YANG Yiqun, YU Tao. Advances in microbial production of liquid biofuels [J]. Synthetic Biology Journal, 2025, 6(1): 18-44. |
| [2] | SHAO Mingwei, SUN Simian, YANG Shimao, CHEN Guoqiang. Bioproduction based on extremophiles [J]. Synthetic Biology Journal, 2024, 5(6): 1419-1436. |
| [3] | ZHAO Liang, LI Zhenshuai, FU Liping, LYU Ming, WANG Shi’an, ZHANG Quan, LIU Licheng, LI Fuli, LIU Ziyong. Progress in biomanufacturing of lipids and single cell protein from one-carbon compounds [J]. Synthetic Biology Journal, 2024, 5(6): 1300-1318. |
| [4] | ZHU Fanghuan, CEN Xuecong, CHEN Zhen. Research progress of diols production by microbes [J]. Synthetic Biology Journal, 2024, 5(6): 1367-1385. |
| [5] | YU Wei, GAO Jiaoqi, ZHOU Yongjin. Bioconversion of one carbon feedstocks for producing organic acids [J]. Synthetic Biology Journal, 2024, 5(5): 1169-1188. |
| [6] | CHEN Xiwei, ZHANG Huaran, ZOU Yi. Biosynthesis and metabolic engineering of fungal non-ribosomal peptides [J]. Synthetic Biology Journal, 2024, 5(3): 571-592. |
| [7] | XIE Huang, ZHENG Yilei, SU Yiting, RUAN Jingyi, LI Yongquan. An overview on reconstructing the biosynthetic system of actinomycetes for polyketides production [J]. Synthetic Biology Journal, 2024, 5(3): 612-630. |
| [8] | ZHAO Jingyu, ZHANG Jian, QI Qingsheng, WANG Qian. Research progress in biosensors based on bacterial two-component systems [J]. Synthetic Biology Journal, 2024, 5(1): 38-52. |
| [9] | SUN Huili, CUI Jinyu, LUAN Guodong, LYU Xuefeng. Progress of cyanobacterial synthetic biotechnology for efficient light-driven carbon fixation and ethanol production [J]. Synthetic Biology Journal, 2023, 4(6): 1161-1177. |
| [10] | YAN Xiongying, WANG Zhen, LOU Jiyun, ZHANG Haoyu, HUANG Xingyu, WANG Xia, YANG Shihui. Progress in the construction of microbial cell factories for efficient biofuel production [J]. Synthetic Biology Journal, 2023, 4(6): 1082-1121. |
| [11] | WU Yujie, LIU Xinxin, LIU Jianhui, Yang Kaiguang, SUI Zhigang, ZHANG Lihua, ZHANG Yukui. Research progress of strain screening and quantitative analysis of key molecules based on high-throughput liquid chromatography and mass spectrometry [J]. Synthetic Biology Journal, 2023, 4(5): 1000-1019. |
| [12] | SUN Meili, WANG Kaifeng, LU Ran, JI Xiaojun. Rewiring and application of Yarrowia lipolytica chassis cell [J]. Synthetic Biology Journal, 2023, 4(4): 779-807. |
| [13] | CHENG Zhenzhen, ZHANG Jian, GAO Cong, LIU Liming, CHEN Xiulai. Progress in metabolic engineering of microorganisms for the utilization of formate [J]. Synthetic Biology Journal, 2023, 4(4): 756-778. |
| [14] | GAO Xianyun, NIU Lingxue, JIAN Ni, GUAN Ningzi. Applications of microbial synthetic biology in the diagnosis and treatment of diseases [J]. Synthetic Biology Journal, 2023, 4(2): 263-282. |
| [15] | LIU Jiayu, YANG Zhihan, YANG Lei, ZHU Liying, ZHU Zhengming, JIANG Ling. Advances in the development of Clostridium tyrobutyricum cell factories driven by synthetic biotechnology [J]. Synthetic Biology Journal, 2022, 3(6): 1174-1200. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||