Synthetic Biology Journal ›› 2022, Vol. 3 ›› Issue (1): 238-251.DOI: 10.12211/2096-8280.2021-082
Efficient capture and assembly of AT-rich genomic fragments using ExoCET-BAC strategy
JIANG Chanjuan1,2, CUI Tianqi1, SUN Hongluan1, JIAO Nianzhi2, FU Jun1, ZHANG Youming1, WANG Hailong1
- 1.State Key Laboratory of Microbial Technology,Institute of Microbial Technology,Shandong University,Qingdao 266237,Shandong,China
2.Institute of Marine Science and Technology,Shandong University,Qingdao 266237,Shandong,China
-
Received:2021-08-06Revised:2021-08-28Online:2022-03-14Published:2022-02-28 -
Contact:ZHANG Youming, WANG Hailong
ExoCET-BAC策略高效抓取和组装高AT含量基因组大片段
姜婵娟1,2, 崔天琦1, 孙洪娈1, 焦念志2, 符军1, 张友明1, 王海龙1
- 1.山东大学微生物技术研究院,微生物技术国家重点实验室,山东 青岛 266237
2.山东大学海洋研究院,山东 青岛 266237
-
通讯作者:张友明,王海龙 -
作者简介:姜婵娟 (1990—),女,博士研究生。研究方向为微生物基因编辑与药物合成生物学等。E-mail:chanjuanjiang@163.com张友明 (1964—),男,教授,博士生导师。研究方向为研究方向为基因组编辑与合成生物学等。E-mail:zhangyouming@sdu.edu.cn王海龙 (1984—),男,教授,博士生导师。研究方向为微生物基因编辑与药物合成生物学等。E-mail:wanghailong@sdu.edu.cn -
基金资助:山东省泰山学者项目(tsqn201812008);山东大学齐鲁青年学者项目
CLC Number:
Cite this article
JIANG Chanjuan, CUI Tianqi, SUN Hongluan, JIAO Nianzhi, FU Jun, ZHANG Youming, WANG Hailong. Efficient capture and assembly of AT-rich genomic fragments using ExoCET-BAC strategy[J]. Synthetic Biology Journal, 2022, 3(1): 238-251.
姜婵娟, 崔天琦, 孙洪娈, 焦念志, 符军, 张友明, 王海龙. ExoCET-BAC策略高效抓取和组装高AT含量基因组大片段[J]. 合成生物学, 2022, 3(1): 238-251.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2021-082
| Characteristics | Source | |
|---|---|---|
| Strains | ||
| E. coli GB2005 | E. coli DH10B derivates in which fhuA::IS2, ΔrecET, ΔybcC; endogenous recET and DLP12 phage ybcC genes are deleted, and fhuA is mutated to make it resistant to T1 phage | Laboratory stock |
| E. coli GB05-dir | GB05 derivates expressing full-length RecE/RecT under the arabinose-inducible PBAD promoter | Laboratory stock |
| Plasmids | ||
| pSC101-BAD-ETgA-tet | Full-length RecE/RecT inducible (PBAD promoter) expression plasmid contains a temperature-sensitive pSC101 origin and a tetracycline resistance gene. | Laboratory stock |
| pBR322-amp-ccdB-rpsL | A pBR322 plasmid with the ampicillin resistance gene, kanamycin resistance gene, and counterselection genes ccdB and rpsl. | Laboratory stock |
| pBeloBAC11-hygccdB | A bacterial artificial chromosome (BAC) with the chloramphenicol resistance gene, hygromycin resistance gene and ccdB gene | Laboratory stock |
Tab. 1 Strains and plasmids used in this study
| Characteristics | Source | |
|---|---|---|
| Strains | ||
| E. coli GB2005 | E. coli DH10B derivates in which fhuA::IS2, ΔrecET, ΔybcC; endogenous recET and DLP12 phage ybcC genes are deleted, and fhuA is mutated to make it resistant to T1 phage | Laboratory stock |
| E. coli GB05-dir | GB05 derivates expressing full-length RecE/RecT under the arabinose-inducible PBAD promoter | Laboratory stock |
| Plasmids | ||
| pSC101-BAD-ETgA-tet | Full-length RecE/RecT inducible (PBAD promoter) expression plasmid contains a temperature-sensitive pSC101 origin and a tetracycline resistance gene. | Laboratory stock |
| pBR322-amp-ccdB-rpsL | A pBR322 plasmid with the ampicillin resistance gene, kanamycin resistance gene, and counterselection genes ccdB and rpsl. | Laboratory stock |
| pBeloBAC11-hygccdB | A bacterial artificial chromosome (BAC) with the chloramphenicol resistance gene, hygromycin resistance gene and ccdB gene | Laboratory stock |
| Oligos | Sequence (5′→3′) | Purpose | Template |
|---|---|---|---|
| pBR322-5pieces-1 | TTTTAGTTTATTTTTAAGACTTTTTAATACTGGATCATTCGTTTAAACACAAATGGCAAGGGCTAATG | Amplification of the pBR322 vector for 5-piece DNA assembly | pBR322-amp-ccdB-rpsL |
| pBR322-2 | GGGAACTGTGGAATTCTTAAATTAAATACCTTGTCGAGGTGTTTAAACGGTGTGGTAGCTCGCGTATT | ||
| F1-1 | ACCTCGACAAGGTATTTAAT | Amplification of fragments for DNA assembly | MIT 9301 genomic DNA |
| F1-2 | CAACATTCTTTCTTGCGATT | ||
| F2-1 | CATACCAATATTTATTATCT | ||
| F2-2 | AGACCTAAATAAATTAAATT | ||
| F3-1 | ATTTTGATTTATTTGGTTTG | ||
| F3-2 | TAGCAAACAATACAAAAACG | ||
| F4-1 | CTTTATTTAGAGAATTTAAT | ||
| F4-2 | TCTGTAATTCTTGCTTTTGCTCTACC | ||
| pBR322-7pieces-1 | TTAAAAAAACAACAGGTAGAGCAAAAGCAAGAATTACAGAGTTTAAACACAAATGGCAAGGGCTAATG | Amplification of the pBR322 vector for 7-piece DNA assembly. Using together with pBR322-2. | pBR322-amp-ccdB-rpsL |
| F5-1 | TGTTGCTTGTGATGAGGCTG | Amplification of fragments for DNA assembly | MIT 9301 genomic DNA |
| F5-2 | AGATTTAAGAAGGATTTTTG | ||
| F6-1 | GAAATTTTAATTGCCGATTT | ||
| F6-2 | CACATCCTGCACAATGAAAA | ||
| pBR322-9pieces-1 | AATGAGAAGAGAAAAGGGAATTTTCATTGTGCAG GATGTGGTTTAAACACAAATGGCAAGGGCTAATG | Amplification of the pBR322 vector for 9-piece DNA assembly. Using together with pBR322-2. | pBR322-amp-ccdB-rpsL |
| F7-1 | ATAATGAGAAGAGAAAAGGGA | Amplification of fragments for DNA assembly | MIT 9301 genomic DNA |
| F7-2 | ATTTTGAAGGCCTGGAATTA | ||
| F8-1 | GCAAAACTACTTTGCTTAAT | ||
| F8-2 | TTTTGATTTAAGTAAAAGAT | ||
| pBR322-11pieces-1 | TAATTCTTTATCCAATGTTTATCTTTTACTTAAATCA AAAGTTTAAACACAAATGGCAAGGGCTAATG | Amplification of the pBR322 vector for 11-piece DNA assembly. Using together with pBR322-2. | pBR322-amp-ccdB-rpsL |
| F9-1 | AGTAATTCTTTATCCAATGT | Amplification of fragments for DNA assembly | MIT 9301 genomic DNA |
| F9-2 | TCAATCTTTACTAAAAAAAT | ||
| F10-1 | ATTCTTGAATAATTTTACTCT | ||
| F10-2 | GAATGATCCAGTATTAAAAAG | ||
| BAC-11pieces-1 | TTTTAGTTTATTTTTAAGACTTTTTAATACTGGATCATTCGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector for 11-piece DNA assembly. | pBeloBAC11-hygccdB |
| BAC-11pieces-2 | GGGAACTGTGGAATTCTTAAATTAAATACCTTGTCGAGGTGTTTAAACCGGGTACCGAGCTCGAATTCG | ||
| BAC-49kb-1 | CCTTAAAAGATTGATTATTTTTCAACCATTATTCAATTTTCCCATCATAGATGGCATTAGTTATTCTCCCAGGTTTAAACGCGGCCGCCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 49 kb fragment | |
| BAC-49kb-2 | GAATTCGAGCTCGGTACCCGGCGGCCGCGGTTTAAACATAGATTGTGTATTGGCGTGTTTAAAAAAAAAATGGAAGAATGAATTAATACAGATTTAGTAACTAATCTA | ||
| BAC-21kb-1 | TCAATGTCCAGAACTTAAAGAGGTATTACGCCTCATTGAAATAGGTCACTTTAGCAATGGGGATAAAGAATTGTTTAAACGCGGCCGCCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 21 kb fragment | |
| BAC-21kb-2 | GAATTCGAGCTCGGTACCCGGCGGCCGCGTTTAAACATTGCTAATCCCCTCCTTGCATCTTGCAAAGATACTTGTTGAGGAGTAGTGACAACTATAGCTCCAGAAATA | ||
| BAC-82kb-1 | TTTTACCTGGCATATCTGGCGGAATAAATACTGACATATTTTCTACAATTCCTAGTAAAGGTACTCCGAGTTGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 82 kb fragment | |
| BAC-82kb-2 | GAATTCGAGCTCGGTACCCGGTTTAAACAATCTTATTGAAGACATTAGTGAGGAAAAAAAATTACCTCCTAATATCGTTGAAGCAGCCTTGCGCGAAGCT | ||
| BAC-65kb-1 | ATTTACCAAATCCAAGGAATGAAGCAGTAGAAAATGATCTAATTGTTGATAATAAATGCTTTATAGAATTAGGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 65 kb fragment | |
| BAC-65kb-2 | GAATTCGAGCTCGGTACCCGGTTTAAACCTTTATTAAATAGCTTGACCGGACATGATCCATTCTTTGTTATGGCGGACTTTGAAGACTACCTAAACAAAC | ||
| BAC-22kb-1 | TATGGGATCAAAAAGAAATCCCAAGAATTGCTCAATTGGTATATTTGGAGTTAATTACGACGGGACATGTTCGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 22 kb fragment | |
| BAC-22kb -2 | GAATTCGAGCTCGGTACCCGGTTTAAACCCAACTACTCTTGATAATGGCCTATTAGAAGAAGTTGTTGAAGTTGCTAAAAAATACTCCAATAGATGTGAT | ||
| BAC-18kb-A1 | ATCAAATAAGGTATCTTTACTCAATTTGTAAAAAAGTTACTTACTATCCGATAATAGGATTAATCTGAGGCTGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 18 kb-A fragment | |
| BAC-18kb-A2 | GAATTCGAGCTCGGTACCCGGTTTAAACAATGATGAATGCCATAAGATTTATAAATATAAAATGTGCCAGGTTTGCCAAATAATGATTTGTTTGATTCAG | ||
| BAC-54kb-1 | CAAGGTAACAACTGCGAGAGTTAGAATAAGAACGTAAGCAAAAGCTTCCATAAGATTAAGTAATTAAGTTTAGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 54 kb fragment | |
| BAC-54kb-2 | GAATTCGAGCTCGGTACCCGGTTTAAACACACATTTACACTTAGTGTTTGAATTTCAAGTCTATTTCGGGTTGAATTAAATTGTTTTTTTATAGTAGTTA | ||
| BAC-25kb-1 | CACTGTAATTAGTGAAATAAACCCCTTCTGTCTCCTTTTTGATTTCTTCAGTCCATCTTTCTTGTGGAGCAAGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 25 kb fragment | |
| BAC-25kb-2 | GAATTCGAGCTCGGTACCCGGTTTAAACGGTCAACGAATTTCTAACGGTTCCGATTTCGACGAACAAACCGGTAAATTAAAAGAAGGGAACAAATCTTTA | ||
| BAC-20kb-1 | TCCCATAACATCAATTAAAAGCGCTATTTCTTATAAGAAATCTATTATTGAAGCGTTGCCAGAAAGTTCTAAGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 20 kb fragment | |
| BAC-20kb-2 | GAATTCGAGCTCGGTACCCGGTTTAAACAGAGGAGAACGAAAAAAAGGTAGTTCTCTTGTCACAGGATCTGAGGTGCAATCTCAGGCCAGTGGTGCAAGC | ||
| BAC-18kb-B1 | AAAATTTATATTAGAAACCATAGTCTCTTTAGTTTTACTTTTATTAGTTATTTTTAAATCAATAATTAATTCGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 18kb-B fragment | |
| BAC-18kb-B2 | GAATTCGAGCTCGGTACCCGGTTTAAACAGGGTTCTATAAAAGTTTTAATTAAATCAATAAGTGTTATTTGAAAATCCACCAAAAAATATAGAGAGCTTA | ||
| BAC-10kb-1 | TTGAGATGTTAGATATTGTAGTTAACAAAAAAAGAGAAGTGTTTAACGGTTTAAATAAACATAGGGACTATAGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 10 kb fragment | |
| BAC-10kb-2 | TAAATATTAATGATATTTCCTTTTATTCAATCCTTTACTATTTGAGCGGTGCAATATGCCCATCTTTGGTGTGTTTAAACCGGGTACCGAGCTCGAATTC | ||
| BAC-11kb-1 | ACGGAATGCAAGAGGATGGTGCTAGTAACGAAACATATACAGCATTAAACGGTTTCTATTCATTTGATAACGGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 11 kb fragment | |
| BAC-11kb-2 | GAATTCGAGCTCGGTACCCGGTTTAAACCGTTATCAAATGAATAATAACCATTAAATGCCATGTATGTATCGTTGTCGTTACCATCTTCGATGTTTGCGT | ||
| BAC-7kb-1 | TACAGGCATTCCGGCAGTTTTAAAATTACTGCCGGAAATGTTTCCTCCTGATTTCTCAAGAGCTGGAACTTGGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 7 kb fragment | |
| BAC-7kb-2 | GAAGTACGCTAATGTTTCGTCCACATTTGCGTCAGCAACATCGAGATCTCCGACTTCATAACCAGCACTGATGTTTAAACCGGGTACCGAGCTCGAATTC | ||
| BAC-12kb-1 | TAAAAGAATTGCAAAATGAGTCATATATAATTAAGAAAAATGATCAGGAAGGGGTTATAAAAAGTAATACTGGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 12 kb fragment | |
| BAC-12kb-2 | GAATTCGAGCTCGGTACCCGGTTTAAACCTTTAATAGACTCTTTGGCAATGAGTATCGCAGGAACTTCGATTTCTGTTTTTTTATCTTTACTTCTATGTT | ||
| BAC-67kb-1 | TCCTTGATTTAATGTCCAGTTGGGATAGTTATTTACCTAAAATGAAAAAATATAAAAAAGTTATAGGACATGGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 67 kb fragment | |
| BAC-67kb-2 | GAATTCGAGCTCGGTACCCGGTTTAAACGGATATCACTCTAAAGATATAAGGAATTTTGAAAATATCCCAAAAATCAATAAGTTGATGTTTGAGATAGCT | ||
| BAC-50kb-1 | GAGTTCAGACCCAGCAGATATTCCTCGATATAAAGCAGCCTAAATTACAAAAAGCTTGAAAACTATATAAATGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 50 kb fragment | |
| BAC-50kb-2 | GAATTCGAGCTCGGTACCCGGTTTAAACCTACCCAGGGAGAAGACATATCATGTAAATTAGTCTGAAATCCCATTTCTTCTAATTCATAGCCTATTGGAT |
Tab. 2 Oligonucleotide sequences used in this study
| Oligos | Sequence (5′→3′) | Purpose | Template |
|---|---|---|---|
| pBR322-5pieces-1 | TTTTAGTTTATTTTTAAGACTTTTTAATACTGGATCATTCGTTTAAACACAAATGGCAAGGGCTAATG | Amplification of the pBR322 vector for 5-piece DNA assembly | pBR322-amp-ccdB-rpsL |
| pBR322-2 | GGGAACTGTGGAATTCTTAAATTAAATACCTTGTCGAGGTGTTTAAACGGTGTGGTAGCTCGCGTATT | ||
| F1-1 | ACCTCGACAAGGTATTTAAT | Amplification of fragments for DNA assembly | MIT 9301 genomic DNA |
| F1-2 | CAACATTCTTTCTTGCGATT | ||
| F2-1 | CATACCAATATTTATTATCT | ||
| F2-2 | AGACCTAAATAAATTAAATT | ||
| F3-1 | ATTTTGATTTATTTGGTTTG | ||
| F3-2 | TAGCAAACAATACAAAAACG | ||
| F4-1 | CTTTATTTAGAGAATTTAAT | ||
| F4-2 | TCTGTAATTCTTGCTTTTGCTCTACC | ||
| pBR322-7pieces-1 | TTAAAAAAACAACAGGTAGAGCAAAAGCAAGAATTACAGAGTTTAAACACAAATGGCAAGGGCTAATG | Amplification of the pBR322 vector for 7-piece DNA assembly. Using together with pBR322-2. | pBR322-amp-ccdB-rpsL |
| F5-1 | TGTTGCTTGTGATGAGGCTG | Amplification of fragments for DNA assembly | MIT 9301 genomic DNA |
| F5-2 | AGATTTAAGAAGGATTTTTG | ||
| F6-1 | GAAATTTTAATTGCCGATTT | ||
| F6-2 | CACATCCTGCACAATGAAAA | ||
| pBR322-9pieces-1 | AATGAGAAGAGAAAAGGGAATTTTCATTGTGCAG GATGTGGTTTAAACACAAATGGCAAGGGCTAATG | Amplification of the pBR322 vector for 9-piece DNA assembly. Using together with pBR322-2. | pBR322-amp-ccdB-rpsL |
| F7-1 | ATAATGAGAAGAGAAAAGGGA | Amplification of fragments for DNA assembly | MIT 9301 genomic DNA |
| F7-2 | ATTTTGAAGGCCTGGAATTA | ||
| F8-1 | GCAAAACTACTTTGCTTAAT | ||
| F8-2 | TTTTGATTTAAGTAAAAGAT | ||
| pBR322-11pieces-1 | TAATTCTTTATCCAATGTTTATCTTTTACTTAAATCA AAAGTTTAAACACAAATGGCAAGGGCTAATG | Amplification of the pBR322 vector for 11-piece DNA assembly. Using together with pBR322-2. | pBR322-amp-ccdB-rpsL |
| F9-1 | AGTAATTCTTTATCCAATGT | Amplification of fragments for DNA assembly | MIT 9301 genomic DNA |
| F9-2 | TCAATCTTTACTAAAAAAAT | ||
| F10-1 | ATTCTTGAATAATTTTACTCT | ||
| F10-2 | GAATGATCCAGTATTAAAAAG | ||
| BAC-11pieces-1 | TTTTAGTTTATTTTTAAGACTTTTTAATACTGGATCATTCGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector for 11-piece DNA assembly. | pBeloBAC11-hygccdB |
| BAC-11pieces-2 | GGGAACTGTGGAATTCTTAAATTAAATACCTTGTCGAGGTGTTTAAACCGGGTACCGAGCTCGAATTCG | ||
| BAC-49kb-1 | CCTTAAAAGATTGATTATTTTTCAACCATTATTCAATTTTCCCATCATAGATGGCATTAGTTATTCTCCCAGGTTTAAACGCGGCCGCCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 49 kb fragment | |
| BAC-49kb-2 | GAATTCGAGCTCGGTACCCGGCGGCCGCGGTTTAAACATAGATTGTGTATTGGCGTGTTTAAAAAAAAAATGGAAGAATGAATTAATACAGATTTAGTAACTAATCTA | ||
| BAC-21kb-1 | TCAATGTCCAGAACTTAAAGAGGTATTACGCCTCATTGAAATAGGTCACTTTAGCAATGGGGATAAAGAATTGTTTAAACGCGGCCGCCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 21 kb fragment | |
| BAC-21kb-2 | GAATTCGAGCTCGGTACCCGGCGGCCGCGTTTAAACATTGCTAATCCCCTCCTTGCATCTTGCAAAGATACTTGTTGAGGAGTAGTGACAACTATAGCTCCAGAAATA | ||
| BAC-82kb-1 | TTTTACCTGGCATATCTGGCGGAATAAATACTGACATATTTTCTACAATTCCTAGTAAAGGTACTCCGAGTTGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 82 kb fragment | |
| BAC-82kb-2 | GAATTCGAGCTCGGTACCCGGTTTAAACAATCTTATTGAAGACATTAGTGAGGAAAAAAAATTACCTCCTAATATCGTTGAAGCAGCCTTGCGCGAAGCT | ||
| BAC-65kb-1 | ATTTACCAAATCCAAGGAATGAAGCAGTAGAAAATGATCTAATTGTTGATAATAAATGCTTTATAGAATTAGGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 65 kb fragment | |
| BAC-65kb-2 | GAATTCGAGCTCGGTACCCGGTTTAAACCTTTATTAAATAGCTTGACCGGACATGATCCATTCTTTGTTATGGCGGACTTTGAAGACTACCTAAACAAAC | ||
| BAC-22kb-1 | TATGGGATCAAAAAGAAATCCCAAGAATTGCTCAATTGGTATATTTGGAGTTAATTACGACGGGACATGTTCGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 22 kb fragment | |
| BAC-22kb -2 | GAATTCGAGCTCGGTACCCGGTTTAAACCCAACTACTCTTGATAATGGCCTATTAGAAGAAGTTGTTGAAGTTGCTAAAAAATACTCCAATAGATGTGAT | ||
| BAC-18kb-A1 | ATCAAATAAGGTATCTTTACTCAATTTGTAAAAAAGTTACTTACTATCCGATAATAGGATTAATCTGAGGCTGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 18 kb-A fragment | |
| BAC-18kb-A2 | GAATTCGAGCTCGGTACCCGGTTTAAACAATGATGAATGCCATAAGATTTATAAATATAAAATGTGCCAGGTTTGCCAAATAATGATTTGTTTGATTCAG | ||
| BAC-54kb-1 | CAAGGTAACAACTGCGAGAGTTAGAATAAGAACGTAAGCAAAAGCTTCCATAAGATTAAGTAATTAAGTTTAGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 54 kb fragment | |
| BAC-54kb-2 | GAATTCGAGCTCGGTACCCGGTTTAAACACACATTTACACTTAGTGTTTGAATTTCAAGTCTATTTCGGGTTGAATTAAATTGTTTTTTTATAGTAGTTA | ||
| BAC-25kb-1 | CACTGTAATTAGTGAAATAAACCCCTTCTGTCTCCTTTTTGATTTCTTCAGTCCATCTTTCTTGTGGAGCAAGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 25 kb fragment | |
| BAC-25kb-2 | GAATTCGAGCTCGGTACCCGGTTTAAACGGTCAACGAATTTCTAACGGTTCCGATTTCGACGAACAAACCGGTAAATTAAAAGAAGGGAACAAATCTTTA | ||
| BAC-20kb-1 | TCCCATAACATCAATTAAAAGCGCTATTTCTTATAAGAAATCTATTATTGAAGCGTTGCCAGAAAGTTCTAAGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 20 kb fragment | |
| BAC-20kb-2 | GAATTCGAGCTCGGTACCCGGTTTAAACAGAGGAGAACGAAAAAAAGGTAGTTCTCTTGTCACAGGATCTGAGGTGCAATCTCAGGCCAGTGGTGCAAGC | ||
| BAC-18kb-B1 | AAAATTTATATTAGAAACCATAGTCTCTTTAGTTTTACTTTTATTAGTTATTTTTAAATCAATAATTAATTCGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 18kb-B fragment | |
| BAC-18kb-B2 | GAATTCGAGCTCGGTACCCGGTTTAAACAGGGTTCTATAAAAGTTTTAATTAAATCAATAAGTGTTATTTGAAAATCCACCAAAAAATATAGAGAGCTTA | ||
| BAC-10kb-1 | TTGAGATGTTAGATATTGTAGTTAACAAAAAAAGAGAAGTGTTTAACGGTTTAAATAAACATAGGGACTATAGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 10 kb fragment | |
| BAC-10kb-2 | TAAATATTAATGATATTTCCTTTTATTCAATCCTTTACTATTTGAGCGGTGCAATATGCCCATCTTTGGTGTGTTTAAACCGGGTACCGAGCTCGAATTC | ||
| BAC-11kb-1 | ACGGAATGCAAGAGGATGGTGCTAGTAACGAAACATATACAGCATTAAACGGTTTCTATTCATTTGATAACGGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 11 kb fragment | |
| BAC-11kb-2 | GAATTCGAGCTCGGTACCCGGTTTAAACCGTTATCAAATGAATAATAACCATTAAATGCCATGTATGTATCGTTGTCGTTACCATCTTCGATGTTTGCGT | ||
| BAC-7kb-1 | TACAGGCATTCCGGCAGTTTTAAAATTACTGCCGGAAATGTTTCCTCCTGATTTCTCAAGAGCTGGAACTTGGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 7 kb fragment | |
| BAC-7kb-2 | GAAGTACGCTAATGTTTCGTCCACATTTGCGTCAGCAACATCGAGATCTCCGACTTCATAACCAGCACTGATGTTTAAACCGGGTACCGAGCTCGAATTC | ||
| BAC-12kb-1 | TAAAAGAATTGCAAAATGAGTCATATATAATTAAGAAAAATGATCAGGAAGGGGTTATAAAAAGTAATACTGGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 12 kb fragment | |
| BAC-12kb-2 | GAATTCGAGCTCGGTACCCGGTTTAAACCTTTAATAGACTCTTTGGCAATGAGTATCGCAGGAACTTCGATTTCTGTTTTTTTATCTTTACTTCTATGTT | ||
| BAC-67kb-1 | TCCTTGATTTAATGTCCAGTTGGGATAGTTATTTACCTAAAATGAAAAAATATAAAAAAGTTATAGGACATGGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 67 kb fragment | |
| BAC-67kb-2 | GAATTCGAGCTCGGTACCCGGTTTAAACGGATATCACTCTAAAGATATAAGGAATTTTGAAAATATCCCAAAAATCAATAAGTTGATGTTTGAGATAGCT | ||
| BAC-50kb-1 | GAGTTCAGACCCAGCAGATATTCCTCGATATAAAGCAGCCTAAATTACAAAAAGCTTGAAAACTATATAAATGTTTAAACCTCTAGAGTCGACCTGCAGG | Amplification of the BAC vector to clone the 50 kb fragment | |
| BAC-50kb-2 | GAATTCGAGCTCGGTACCCGGTTTAAACCTACCCAGGGAGAAGACATATCATGTAAATTAGTCTGAAATCCCATTTCTTCTAATTCATAGCCTATTGGAT |
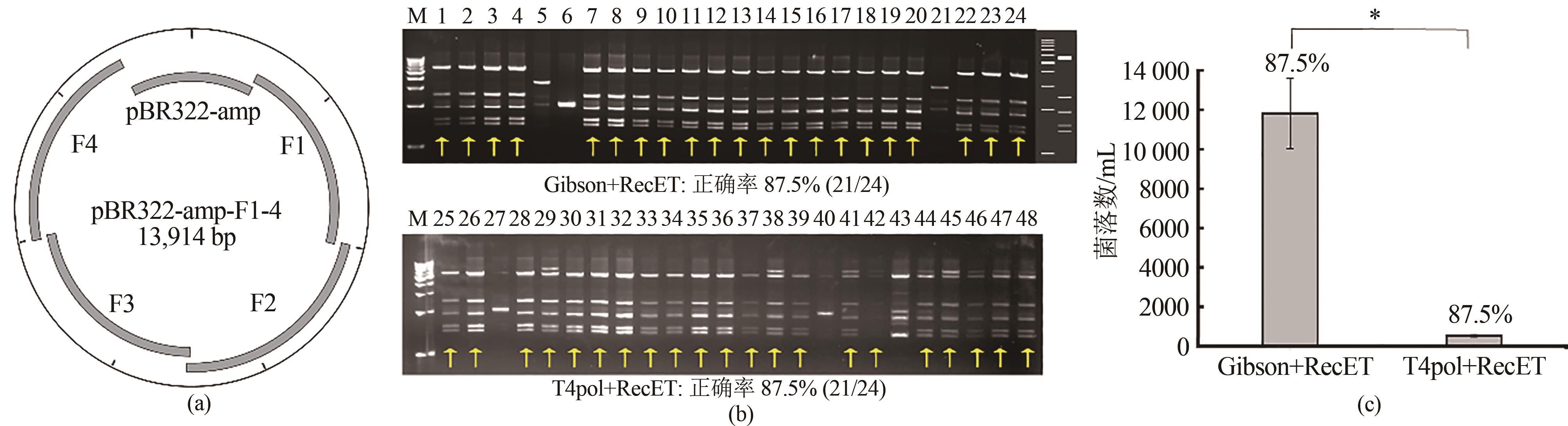
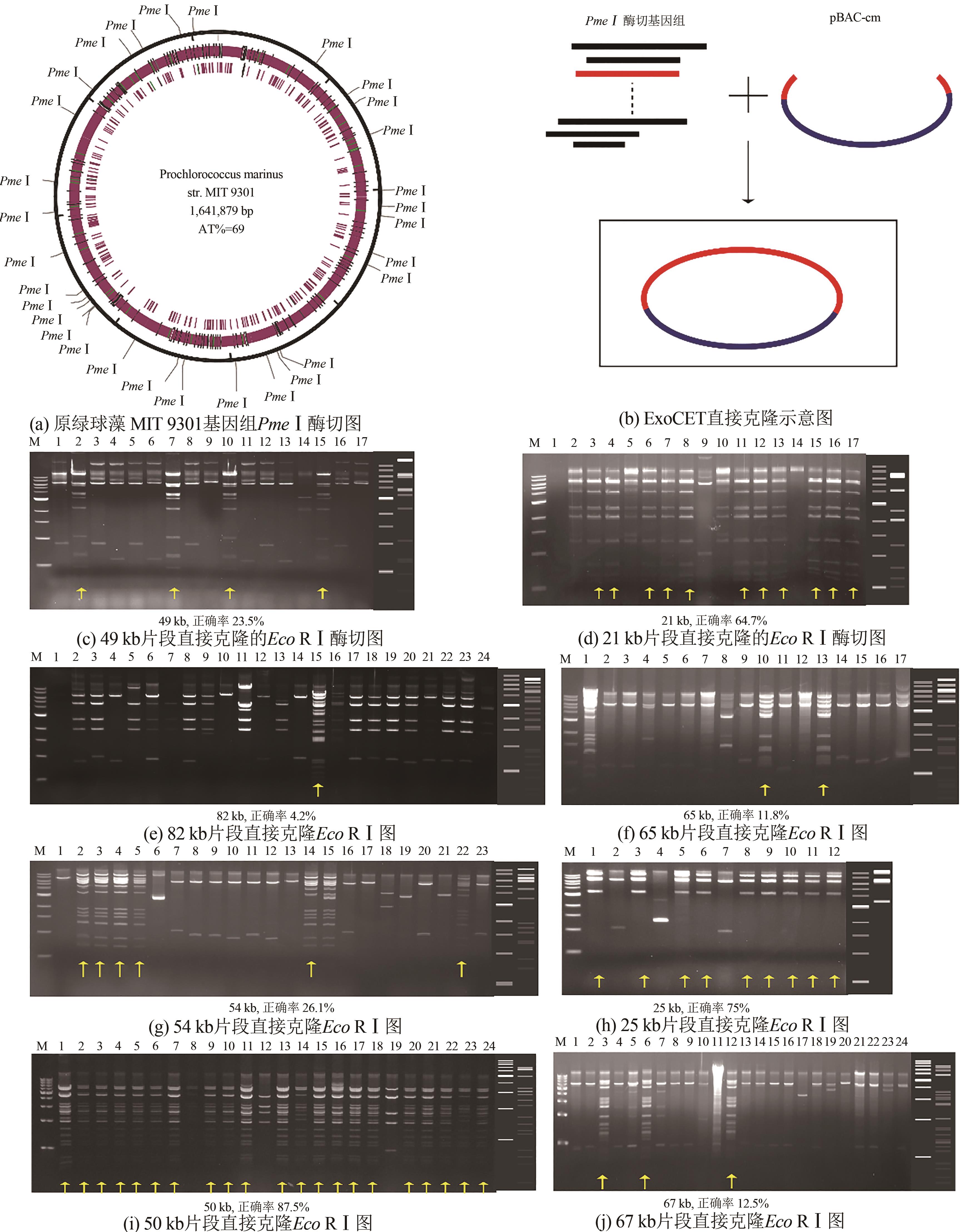
Fig. 1 Assembly of 5 DNA fragments using ExoCET(a) Schematic diagram for the assembly. (b) EcoRⅠ digestion map for assembled plamids. The right side of the gel imagines is the theoretical map for the restriction digestion. 1-24: Gibson+RecET, and 25-48: T4pol+RecET assembly. (c) Comparison of efficiencies for the assembly. Significance was analyzed by one-way ANOVA, and P < 0.05 was considered statistically significant (***P < 0.001, **P < 0.01, and *P < 0.05)
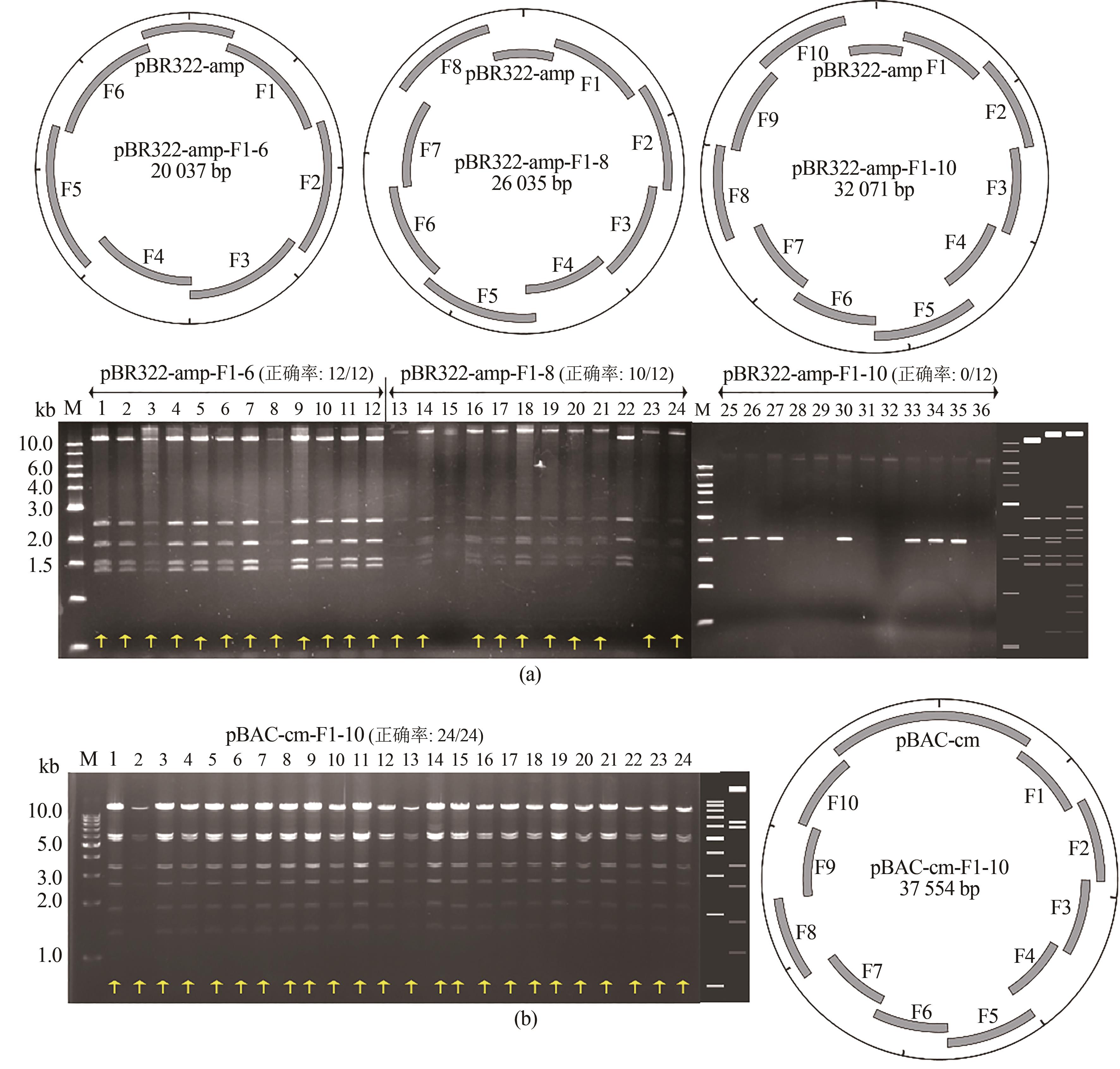
Fig. 2 Assembly of 7-11 fragments using ExoCET(a) Schematic diagrams and EcoRⅠ restriction analysis for the assembly using pBR322 vectors. 1-12: 7-piece assembly, 13-24: 9-piece assembly, and 25-36: 11-piece assembly. The right side of the gel imagine is the theoretical map for the restriction digestion of pBR322-amp-F1-6, pBR322-amp-F1-8 and pBR322-amp-F1-10 from left to right. (b) Schematic diagrams and EcoRⅤ restriction analysis for the 11-piece assembly using the BAC vector. The right side of the gel imagine is the theoretical map for the restriction digestion
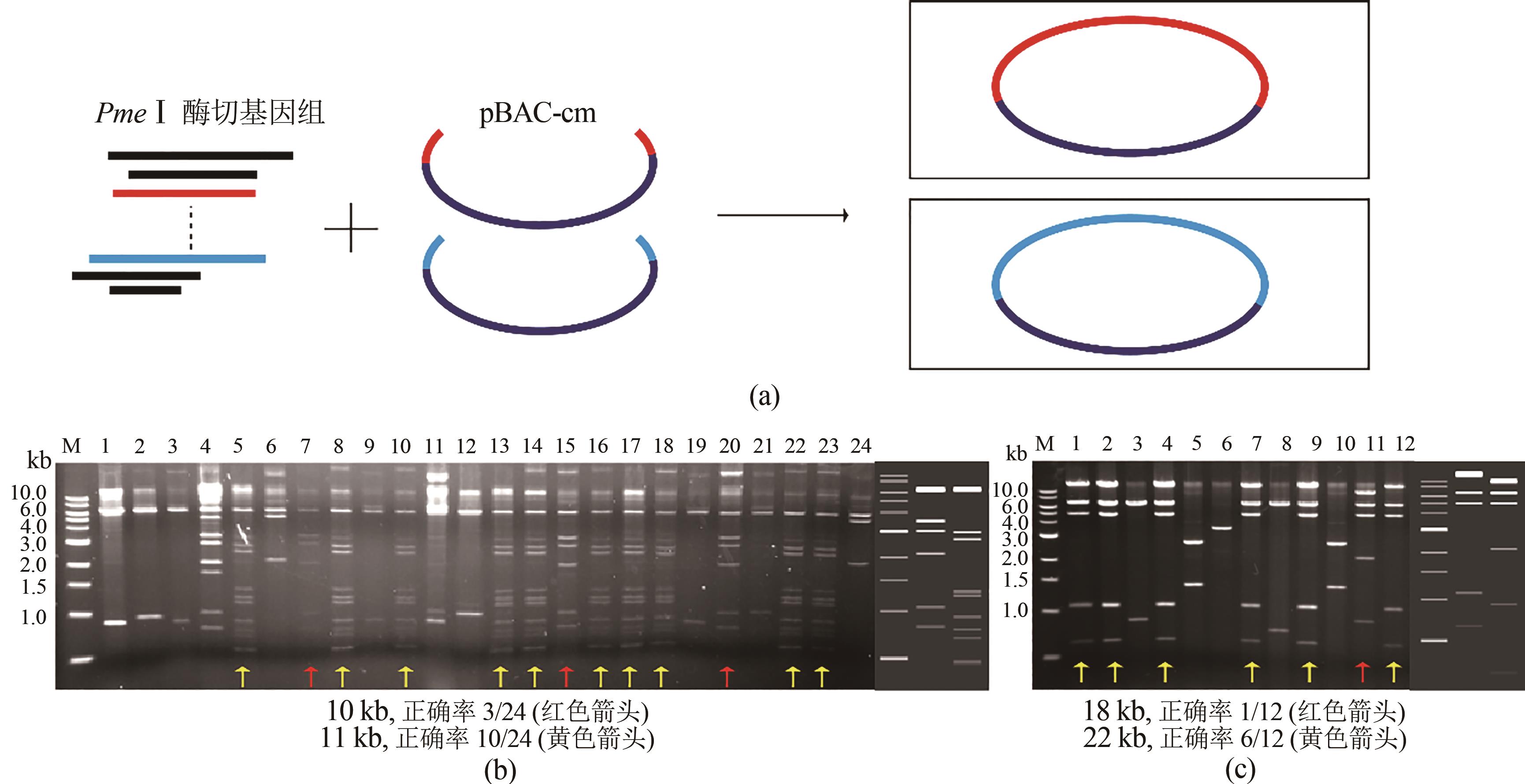
Fig. 4 ExoCET-BAC strategy for capturing two DNA fragments simultaneously(a) Schematic diagram for the strategy. (b) EcoRⅠrestriction map for the direct cloning of 10 kb and 11 kb fragments. The right side of the gel imagine is the theoretical restriction map for the direct cloning of 10 kb and 11 kb fragments. (c) EcoRⅠ restriction map for the direct cloning of 18 kb and 22 kb fragments. The right side of the gel imagine is the theoretical restriction map for the direct cloning of 18 kb and 22 kb fragments from left to right
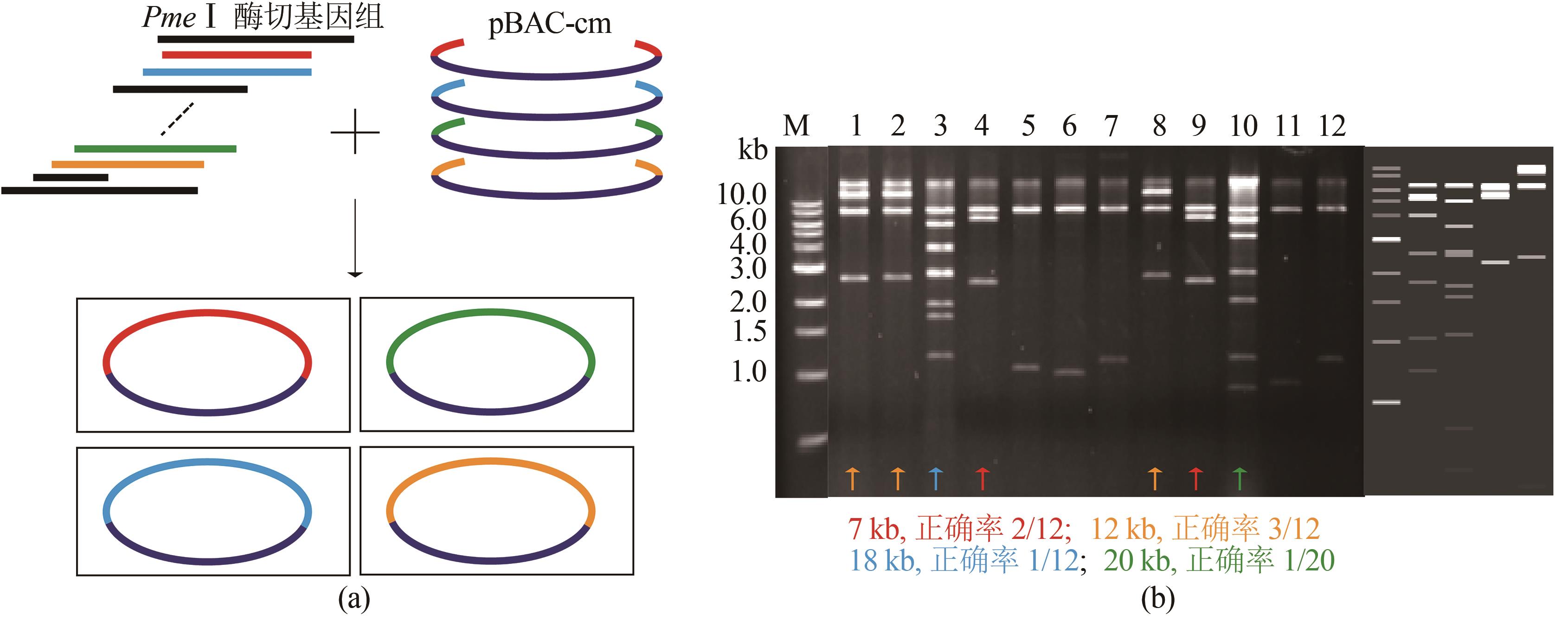
Fig. 5 ExoCET-BAC strategy for capturing four DNA fragments simultaneously(a) Schematic diagram for the strategy. (b) EcoRⅠrestriction map for the direct cloning of 7 kb, 12 kb, 18 kb and 20 kb fragments. The right side of the gel imagine is the theoretical restriction map for the direct cloning of 20 kb、18 kb、7 kb and 12 kb fragments from left to right
| 1 | YAMANAKA K, REYNOLDS K A, KERSTEN R D, et al. Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(5): 1957-1962. |
| 2 | LEE N C O, LARIONOV V, KOUPRINA N. Highly efficient CRISPR/Cas9-mediated TAR cloning of genes and chromosomal loci from complex genomes in yeast[J]. Nucleic Acids Research, 2015, 43(8): e55. |
| 3 | FU Jun, BIAN Xiaoying, HU Shenbiao, et al. Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting[J]. Nature Biotechnology, 2012, 30(5): 440-446. |
| 4 | JIANG Wenjun, ZHAO Xuejin, GABRIELI T, et al. Cas9-Assisted Targeting of CHromosome segments CATCH enables one-step targeted cloning of large gene clusters[J]. Nature Communications, 2015, 6: 8101. |
| 5 | JIANG Wenjun, ZHU Ting F. Targeted isolation and cloning of 100-kb microbial genomic sequences by Cas9-assisted targeting of chromosome segments[J]. Nature Protocols, 2016, 11(5): 960-975. |
| 6 | WANG Hailong, LI Zhen, JIA Ruonan, et al. ExoCET: exonuclease in vitro assembly combined with RecET recombination for highly efficient direct DNA cloning from complex genomes[J]. Nucleic Acids Research, 2018, 46(5): e28. |
| 7 | TAO Weixin, CHEN Li, ZHAO Chunhua, et al. In vitro packaging mediated one-step targeted cloning of natural product pathway[J]. ACS Synthetic Biology, 2019, 8(9): 1991-1997. |
| 8 | ENGHIAD B, HUANG Chunshuai, GUO Fang, et al. Cas12a-assisted precise targeted cloning using in vivo Cre-lox recombination[J]. Nature Communications, 2021, 12(1): 1171. |
| 9 | GREUNKE C, DUELL E R, D'AGOSTINO P M, et al. Direct Pathway Cloning (DiPaC) to unlock natural product biosynthetic potential[J]. Metabolic Engineering, 2018, 47: 334-345. |
| 10 | D'AGOSTINO P M, GULDER T A M. Direct pathway cloning combined with sequence- and ligation-independent cloning for fast biosynthetic gene cluster refactoring and heterologous expression[J]. ACS Synthetic Biology, 2018, 7(7): 1702-1708. |
| 11 | QUAN Jiayuan, TIAN Jingdong. Circular polymerase extension cloning for high-throughput cloning of complex and combinatorial DNA libraries[J]. Nature Protocols, 2011, 6(2): 242-251. |
| 12 | SHAO Zengyi, ZHAO Hua, Zhao Huimin. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways[J]. Nucleic Acids Research, 2009, 37(2): e16. |
| 13 | SHAO Zengyi, ZHAO Huimin. Manipulating natural product biosynthetic pathways via DNA assembler[J]. Current Protocols in Chemical Biology, 2014, 6(2): 65-100. |
| 14 | HIPPEL P H VON, JOHNSON N P, MARCUS A H. Fifty years of DNA “breathing”: reflections on old and new approaches[J]. Biopolymers, 2013, 99(12): 923-954. |
| 15 | GODISKA R, MEAD D, DHODDA V, et al. Linear plasmid vector for cloning of repetitive or unstable sequences in Escherichia coli [J]. Nucleic Acids Research, 2009, 38(6): e88. |
| 16 | HAO Tingting, XIE Zhoujie, WANG Min, et al. An anaerobic bacterium host system for heterologous expression of natural product biosynthetic gene clusters[J]. Nature Communications, 2019, 10: 3665. |
| 17 | ZHANG Youming, BUCHHOLZ F, MUYRERS J P P, et al. A new logic for DNA engineering using recombination in Escherichia coli [J]. Nature Genetics, 1998, 20(2): 123-128. |
| 18 | ZHANG Youming, MUYRERS J P, TESTA G, et al. DNA cloning by homologous recombination in Escherichia coli [J]. Nature Biotechnology, 2000, 18(12): 1314-1317. |
| 19 | WANG Hailong, LI Zhen, JIA Ruonan, et al. RecET direct cloning and Redαβ recombineering of biosynthetic gene clusters, large operons or single genes for heterologous expression[J]. Nature Protocols, 2016, 11(7): 1175-1190. |
| 20 | CHAI Yi, SHAN Shiping, WEISSMAN K J, et al. Heterologous expression and genetic engineering of the tubulysin biosynthetic gene cluster using red/ET recombineering and inactivation mutagenesis[J]. Chemistry & Biology, 2012, 19(3): 361-371. |
| 21 | SONG Chaoyi, LUAN Ji, CUI Qingwen, et al. Enhanced heterologous spinosad production from a 79-kb synthetic multi-operon assembly[J]. ACS Synthetic Biology, 2019, 8(1): 137-147. |
| 22 | FU Jun, WENZEL S C, PERLOVA O, et al. Efficient transfer of two large secondary metabolite pathway gene clusters into heterologous hosts by transposition[J]. Nucleic Acids Research, 2008, 36(17): e113. |
| 23 | ABBASI M N, FU Jun, BIAN Xiaoying, et al. Recombineering for genetic engineering of natural product biosynthetic pathways[J]. Trends in Biotechnology, 2020, 38(7): 715-728. |
| 24 | SONG Chaoyi, LUAN Ji, LI Ruijuan, et al. RedEx: a method for seamless DNA insertion and deletion in large multimodular polyketide synthase gene clusters[J]. Nucleic Acids Research, 2020, 48(22): e130. |
| 25 | WANG Hailong, BIAN Xiaoying, XIA Liqiu, et al. Improved seamless mutagenesis by recombineering using ccdB for counterselection[J]. Nucleic Acids Research, 2013, 42(5): e37. |
| 26 | BIAN Xiaoying, PLAZA A, YAN Fu, et al. Rational and efficient site-directed mutagenesis of adenylation domain alters relative yields of luminmide derivatives in vivo[J]. Biotechnology and Bioengineering, 2015, 112(7): 1343-1353. |
| 27 | NEWMAN D J, CRAGG G M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019[J]. Journal of Natural Products, 2020, 83(3): 770-803. |
| 28 | HUO Liujie, HUG J J, FU Chengzhang, et al. Heterologous expression of bacterial natural product biosynthetic pathways[J]. Natural Product Reports, 2019, 36(10): 1412-1436. |
| 29 | LI J S, DU Yongle, GU Di, et al. Discovery and biosynthesis of clostyrylpyrones from the obligate anaerobe clostridium roseum[J]. Organic Letters, 2020, 22(21): 8204-8209. |
| 30 | ZHANG Weimin, ZHAO Guanghou, LUO Zhouqing, et al. Engineering the ribosomal DNA in a megabase synthetic chromosome[J]. Science, 2017, 355(6329): eaaf3981. |
| 31 | XIE Zexiong, LI Bingzhi, MITCHELL L A, et al. “Perfect” designer chromosome V and behavior of a ring derivative[J]. Science, 2017, 355(6329): eaaf4704. |
| 32 | WU Yi, LI Bingzhi, ZHAO Meng, et al. Bug mapping and fitness testing of chemically synthesized chromosome X[J]. Science, 2017, 355(6329): eaaf4706. |
| 33 | SHEN Yue, WANG Yun, CHEN Tai, et al. Deep functional analysis of synII, a 770-kilobase synthetic yeast chromosome[J]. Science, 2017, 355(6329): eaaf4791. |
| 34 | GIBSON D G, GLASS J I, LARTIGUE C, et al. Creation of a bacterial cell controlled by a chemically synthesized genome[J]. Science, 2010, 329(5987): 52-56. |
| 35 | FREDENS J, WANG Kaihang, DE LA TORRE D, et al. Total synthesis of Escherichia coli with a recoded genome[J]. Nature, 2019, 569(7757): 514-518. |
| [1] | XI Mengyu, HU Yiling, GU Yucheng, GE Huiming. Genome mining-directed discovery for natural medicinal products [J]. Synthetic Biology Journal, 2024, 5(3): 447-473. |
| [2] | LEI Ru, TAO Hui, LIU Tiangang. Deep genome mining boosts the discovery of microbial terpenoids [J]. Synthetic Biology Journal, 2024, 5(3): 507-526. |
| [3] | SONG Yongxiang, ZHANG Xiufeng, LI Yanqin, XIAO Hua, YAN Yan. Resistance-gene directed discovery of bioactive natural products [J]. Synthetic Biology Journal, 2024, 5(3): 474-491. |
| [4] | CHEN Yongcan, SI Tong, ZHANG Jianzhi. Applications of automated synthetic biotechnology in DNA assembly and microbial chassis manipulation [J]. Synthetic Biology Journal, 2023, 4(5): 857-876. |
| [5] | ZHANG Fanzhong, XIANG Changjun, ZHANG Lihan. Advances and applications of evolutionary analysis and big-data guided bioinformatics in natural product research [J]. Synthetic Biology Journal, 2023, 4(4): 629-650. |
| [6] | CHEN Qingli, TONG Yigang. Merging the frontiers: synthetic biology for advanced bacteriophage design [J]. Synthetic Biology Journal, 2023, 4(2): 283-300. |
| [7] | TENG Xiaolong, SHI Shuobo. Optimization and development of CRISPR/Cas9 systems for genome editing [J]. Synthetic Biology Journal, 2023, 4(1): 67-85. |
| [8] | YANG Yi, MAO Yufeng, YANG Chunhe, WANG Meng, LIAO Xiaoping, MA Hongwu. Recent progress in computational tools for designing editing sequences used in microbial genetic manipulations [J]. Synthetic Biology Journal, 2023, 4(1): 30-46. |
| [9] | PAN Yingjia, XIA Siyang, DONG Chang, CAI Jin, LIAN Jiazhang. Mutator-driven continuous genome evolution of Saccharomyces cerevisiae [J]. Synthetic Biology Journal, 2023, 4(1): 225-240. |
| [10] | JIN Jiaoyu, ZHOU Jiahai. The mystery of Z-genome biosynthesis has been elucidated [J]. Synthetic Biology Journal, 2022, 3(1): 1-5. |
| [11] | Bo HE, Zongheng FU, Yi WU, Guangrong ZHAO. Research progress of synthetic mammalian genomics [J]. Synthetic Biology Journal, 2022, 3(1): 78-97. |
| [12] | Qingzhuo WANG, Ping SONG, He HUANG. Synthetic biotechnology drives the development of natural eukaryotic lipid cell factories [J]. Synthetic Biology Journal, 2021, 2(6): 920-941. |
| [13] | Qian YANG, Botao CHENG, Zhijun TANG, Wen LIU. Applications and prospects of genome mining in the discovery of natural products [J]. Synthetic Biology Journal, 2021, 2(5): 697-715. |
| [14] | Haibo ZHOU, Qiyao SHEN, Hanna CHEN, Zongjie WANG, Yuezhong LI, Youming ZHANG, Xiaoying BIAN. Genome mining for novel natural products in Sorangium cellulosum So0157-2 by heterologous expression [J]. Synthetic Biology Journal, 2021, 2(5): 837-849. |
| [15] | Xiaoluo HUANG, Junbiao DAI. DNA synthesis technology: foundation of DNA data storage [J]. Synthetic Biology Journal, 2021, 2(3): 335-353. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||