Synthetic Biology Journal ›› 2022, Vol. 3 ›› Issue (1): 116-137.DOI: 10.12211/2096-8280.2021-079
• Invited Review • Previous Articles Next Articles
Developing C1-based metabolic network in methylotrophy for biotransformation
GUO Shuyuan1,2, WU Lianghuan1,2, LIU Xiangjian1,2, WANG Bo1,2, YU Tao1,2
- 1.Center for Synthetic Biochemistry,Shenzhen Institute of Synthetic Biology,Shenzhen Institutes of Advanced Technology,Chinese Academy of Sciences,Shenzhen 518055,Guangdong,China
2.CAS Key Laboratory of Quantitative Engineering Biology,Shenzhen Institute of Synthetic Biology,Shenzhen Institutes of Advanced Technology,Chinese Academy of Sciences,Shenzhen 518055,Guangdong,China
-
Received:2021-07-23Revised:2021-10-21Online:2022-03-14Published:2022-02-28 -
Contact:YU Tao
微生物中一碳代谢网络构建的进展与挑战
郭姝媛1,2, 吴良焕1,2, 刘香健1,2, 王博1,2, 于涛1,2
- 1.中国科学院深圳先进技术研究院,深圳合成生物学创新研究院,合成生物化学研究中心,广东 深圳 518055
2.中国科学院深圳先进技术研究院,深圳合成生物学创新研究院,中国科学院定量工程生物学重点实验室,广东 深圳 518055
-
通讯作者:于涛 -
作者简介:郭姝媛 (1991—),女,博士,博士后。主要从事基于酿酒酵母的甲醇生物转化及产物合成研究。E-mail:sy.guo@siat.ac.cn于涛 (1986—),男,博士,研究员。主要从事酿酒酵母的合成生物学研究。E-mail:tao.yu@siat.ac.cn -
基金资助:中国博士后科学基金(2020M682973);国家重点研发计划(2020YFA0907800);国家自然科学基金(32071416);广东省基础与应用基础研究基金(2020A1515110927);深圳合成生物学创新研究院项目(JCHZ20200003)
CLC Number:
Cite this article
GUO Shuyuan, WU Lianghuan, LIU Xiangjian, WANG Bo, YU Tao. Developing C1-based metabolic network in methylotrophy for biotransformation[J]. Synthetic Biology Journal, 2022, 3(1): 116-137.
郭姝媛, 吴良焕, 刘香健, 王博, 于涛. 微生物中一碳代谢网络构建的进展与挑战[J]. 合成生物学, 2022, 3(1): 116-137.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2021-079
| 宿主 | 发酵培养基 | 产物(产量) | 备注 | 参考 文献 |
|---|---|---|---|---|
| 巴斯德毕赤酵母 | 甲醇,基础盐培养基 | 6-甲基水杨酸(2.2 g/L) | ①表达磷酸泛酰巯基乙胺基转移酶(Aspergillus nidulans)、6-甲基水杨酸合成酶(Aspergillus terrus);②5 L生物反应器 | [ |
| 甲醇,葡萄糖,基础盐培养基 | 香柏酮(208 mg/L) | ①表达前二烯加氧酶(Hyoscyamus muticus)、细胞色素P450还原酶(Arabidopsis thaliana)、缬烯合酶(Callitropsis nootkatensis)、乙醇脱氢酶(Pichia pastoris)、部分羟基戊二酰辅酶A还原酶(Saccharomyces cerevisiae);②补料分批 | [ | |
| 葡萄糖(40 g/L),草酸(0.2 g/L),酵母提取物(7.5 g/L),蛋白胨(10 g/L),基础盐培养基 | 透明质酸 (2.5 MDa;0.8~1.7 g/L) | ①表达透明质酸合成酶2(Xenopus laevis)、UDP-葡萄糖脱氢酶(Xenopus laevis)、UDP-葡萄糖焦磷酸化酶(P. pastoris)、UDP-N-乙酰氨基葡萄糖焦磷酸化酶(P. pastoris)、磷酸葡萄糖异构酶(P. pastoris);②1 L生物反应器 | [ | |
| 甲醇(1%),组氨酸(0.05%),生物素(0.01%) | 富马酸(0.76 g/L),苹果酸(42.28 g/L),琥珀酸(9.42 g/L) | ①表达丙酮酸羧化酶(Pichia pastoris)、苹果酸脱氢酶1(Pichia pastoris)、富马酸酶(Pichia pastoris);②0.6 L生物反应器 | [ | |
| 酵母提取物(1%),蛋白胨(2%),葡萄糖(2%) | 番茄红素(1.141 μg/g细胞干重),β-胡萝卜素(339 μg/g细胞干重) | ①表达香叶酰二磷酸合成酶(Erwinia uredovora)、茄红素合成酶(Erwinia uredovora)、茄红素去饱和酶(Erwinia uredovora)、番茄红素环化酶(Ficus carica);②摇瓶培养 | [ | |
| 甲醇(0.5%),甘油,基础盐培养基 | 单那可林 J(593.9 mg/L),洛伐他汀(250.8 mg/L) | ①途径分割策略;②5 L生物反应器 | [ | |
| 甲醇(0.5%),甘油,基础盐培养基 | 洛伐他汀(419 mg/L) | ①表达洛伐他汀合成模块B-C-A-F-D-G(Aspergillus terreus)、细胞色素P450还原酶(Aspergillus terreus)、磷酸泛酰巯基乙胺基转移酶(Aspergillus nidulans)、他汀外排蛋白(Aspergillus terreus);②5 L生物反应器 | [ | |
| 汉逊酵母 | 甲醇(0.2%),胆碱(0.2%),葡萄糖(0.25%) | 青霉素(1 mg/L) | ①表达非核糖体肽合成酶(Penicillium chrysogenum)、异青霉素N合成酶(Penicillium chrysogenum)、异青霉素素N酰基转移酶(Penicillium chrysogenum)、苯乙酰CoA连接酶(Penicillium chrysogenum);②恒化器 | [ |
| 甲醇芽孢杆菌 | 甲醇,基础盐培养基 | L-谷氨酸(58 g/L) | ① 50 ℃培养;②补料分批 | [ |
| 甲醇(150 mmol/L),酵母提取物(0.25 g/L),基础盐培养基 | L-赖氨酸(11 g/L) | ①表达天冬氨酸激酶(Bacillus methanolicus);② 3 L生物反应器 | [ | |
| 甲醇(150 mmol/L),基础盐培养基 | L-赖氨酸(65 g/L), L-谷氨酸(28 g/L) | ①表达丙酮酸羧化酶(Bacillus methanolicus)、高丝氨酸脱氢酶(Bacillus methanolicus);②3 L生物反应器 | [ | |
| 甲醇(200 mmol/L),酵母提取物(0.025%),基础盐培养基 | L-赖氨酸(777 mg/L) | ①表达赖氨酸外排基因(Corynebacterium glutamicum);② 500 mL摇瓶发酵 | [ | |
| 甲醇(200 mmol/L),酵母提取物(0.025%),基础盐培养基 | 尸胺(11.3 g/L) | ①表达赖氨酸脱羧酶(Escherichia coli);② 3 L生物反应器 | [ | |
| 甲醇(200 mmol/L),诱导剂,基础盐培养基 | 尸胺(10.2 g/L) | ①诱导的启动子系统和θ-and rolling circle-复制系统;② 3 L生物反应器 | [ | |
| 甲醇(200 mmol/L),5'-磷酸吡哆醛(20 μmol/L),基础盐培养基 | γ-氨基丁酸(9 g/L) | ①表达谷氨酸脱羧酶(Sulfobacillus thermosulfidooxidans);② 3 L生物反应器 | [ | |
| 扭脱甲基杆菌 AM1 | 甲醇(0.5%),琥珀酸(0.5%),基础盐培养基 | 聚羟基烷酸酯 (PHA,35.1%±4.36%) | ①表达PHA合成酶(Aeromonas caviae);② Co2+缺陷条件;③摇瓶培养 | [ |
| 甲醇(125 mmol/L),琥珀酸(20 mmol/L),乙胺(20 mmol/L),基础盐培养基 | 1-丁醇(15.2 mg/L) | ①表达烯酰辅酶A还原酶(Treponema denticola)、乙醇脱氢酶(Clostridium acetobutylicum)、巴豆酸酶(Methylobacterium extorquens AM1);②摇瓶培养 | [ | |
| 甲醇(240 mmol/L),基础盐培养基 | 衣康酸(31.6 mg/L± 5.5 mg/L) | ①表达乌头酸脱羧酶(Aspergillus terreus);② 2 L生物反应器 | [ | |
| 甲醇(125 mmol/L),基础盐培养基 | 1-丁醇(25.5 mg/L) | ①适应性进化筛选突变株耐受丁醇达到0.5%;②摇瓶培养 | [ | |
| 甲醇(1%),基础盐培养基 | 甲羟戊酸(2.67 g/L) | ①生物感应器辅助;②产率0.55 mol乙酰CoA/mol甲醇;③ 5 L-生物反应器 | [ | |
| 甲醇(3 g/L),基础盐培养基 | 2-羟基异丁酸(2.1 g/L) | ①表达(R)-3-羟基丁基辅酶A特异性辅酶B12依赖性诱变酶(Bacillus massiliosenegalensis JC6);②产率0.11 g/g甲醇,生物反应器 | [ | |
| 甲醇(125 mmol/L),琥珀酸(15 mmol/L),基础盐培养基 | 3-羟基丙酸(69.8 mg/L) | ①表达丙二酰辅酶A还原酶(Chlorofexus aurantiacus);②摇瓶培养 | [ | |
| 甲醇(0.5%),琥珀酸(15 mmol/L),基础盐培养基 | 3-羟基丙酸(0.857 g/L) | ①ΔhprA,表达己糖磷酸合成酶(Bacillus methanolicus)、磷酸己糖异构酶(Bacillus methanolicus)、磷酸果糖激酶(Bacillus methanolicus)、6-磷酸葡萄糖脱氢酶(Bacillus methanolicus)、丙二酰辅酶A还原酶(Chlorofexus aurantiacus);② 2.5 L生物反应器 | [ | |
| 甲醇(125 mmol/L),琥珀酸(15 mmol/L),基础盐培养基 | 异丁醇(19 mg/L) | ①ΔldhA,表达2-酮异戊酸脱羧酶(Lactococcus lactis)、醇脱氢酶(Lactococcus lactis)、乙酰乳酸合酶(Bacillus subtilis);②摇瓶培养 | [ | |
| 甲醇(123 mmol/L),基础盐培养基 | 甲基富马酸,(2S)-甲基琥珀酸(0.65 g/L) | ①ΔphaC,表达硫酯酶;②摇瓶培养;产率0.17 g/g甲醇;③钴缺陷条件 | [ | |
| 甲醇(123 mmol/L),基础盐培养基 | α-蛇麻烯(1.65 g/L) | ①表达α-蛇麻烯合成酶(Zingiber zerumbet)、法尼焦磷酸合成酶(Saccharomyces cerevisiae)、异源甲羟戊酸途径;② 2.4 L生物反应器 | [ | |
| 甲醇(1%),基础盐培养基 | 甲羟戊酸(2.22 g/L) | ①表达HMG-CoA 合成酶(Enterococcus faecalistiters)、HMG-CoA还原酶(Enterococcus faecalistiters)、乙酰乙酰CoA 硫解酶(Ralstonia eutropha);②产率28.4 mg/g甲醇,5 L生物反应器 | [ | |
| 甲醇,琥珀酸盐,基础盐 培养基 | 丁二烯前体(0.6 mg/L) | ①表达羟乙烷基噻唑激酶(Escherichia coli)、磷酸异戊烯基激酶(Methanothermobacter thermautotrophicus)、乙醇脱氢酶(Clostridium acetobutylicum);②摇瓶培养 | [ | |
| 扭脱甲基杆菌 ATCC 55366 | 甲醇,基础盐培养基 | 绿色荧光蛋白(4 g/L) | ①表达不同的启动子;产率80 mg/g甲醇;② 20 L生物反应器 | [ |
| 甲醇(1%),异丙基苯甲酸(20 mg/L),基础盐培养基 | 功能性聚羟基烷酸酯(PHA,35.1%±4.36%) | ①表达聚羟基脂肪酸酯合成酶2(Pseudomonas fluorescens GK13);②含有碳碳双键的功能性PHA;3.摇瓶培养 | [ | |
| 甲基杆菌 MB200 | 甲醇(1.2%),基础盐培养基 | 乙醛酸(14.38 mg/mL) | ①表达羟基丙酮酸还原酶(M. extorquens);②摇瓶培养 | [ |
| 甲醇(1.2%),基础盐培养基 | L-丝氨酸(11.4 g/L) | ①表达丝氨酸羟甲基转移酶(SHMT,M. extorquens);②静息发酵 | [ | |
| 甲醇(1.2%),基础盐培养基 | L-丝氨酸(6.6 g/L) | ①表达甲醇脱氢酶(Methylobacterium sp. MB200);②静息发酵 | [ | |
| 甲醇(1.2%),基础盐培养基 | L-丝氨酸(9.1 g/L) | Δgnd(6-磷酸葡萄糖脱氢酶) | [ | |
| 甲基杆菌MB 126 | 甲醇(125 mmol/L),基础盐培养基 | (R)-3-羟基丁酸(2.8 g/L) | ① Δhbd(3-羟基丁酸降解酶)、Δlip(硫辛酸合成酶)② 2 L生物反应器 | [ |
| Protomonas extorquens NR-1 | 甲醇(1%),基础盐培养基 | L-丝氨酸(54.5 g/L) | ①静息发酵;② 2 L生物反应器,产率0.183 mol丝氨酸/mol甲醇 | [ |
| 甲基菌属糖原ATCC突变体 | 甲醇(0.5%),蛋白胨(3 g/L),基础盐培养基 | L-谷氨酸(38.3 g/L),L-苏氨酸(11 g/L),L-赖氨酸(5.6 g/L) | ①NTG突变获取不同突变体,RV3、AL119、DHL122;② 5 L生物反应器 | [ |
| 嗜甲基菌属 Methylotrophus | 甲醇(500 mmol/L),硫酸铵 | L-赖氨酸(112 mmol) | ①表达ED 途径;② 1 L生物反应器 | [ |
| 甲烷球菌属 Maripaludis | 甲酸,乙酸(10 mmol/L),丙酮酸(20 mmol/L),丙氨酸(1 mmol/L),CO2/H2 | 香叶醇 | ①表达香叶醇合成酶(Ocimum basilicum);② 产率4.6 mg/g甲酸 | [ |
Tab. 1 Chemicals produced by native methylotrophs
| 宿主 | 发酵培养基 | 产物(产量) | 备注 | 参考 文献 |
|---|---|---|---|---|
| 巴斯德毕赤酵母 | 甲醇,基础盐培养基 | 6-甲基水杨酸(2.2 g/L) | ①表达磷酸泛酰巯基乙胺基转移酶(Aspergillus nidulans)、6-甲基水杨酸合成酶(Aspergillus terrus);②5 L生物反应器 | [ |
| 甲醇,葡萄糖,基础盐培养基 | 香柏酮(208 mg/L) | ①表达前二烯加氧酶(Hyoscyamus muticus)、细胞色素P450还原酶(Arabidopsis thaliana)、缬烯合酶(Callitropsis nootkatensis)、乙醇脱氢酶(Pichia pastoris)、部分羟基戊二酰辅酶A还原酶(Saccharomyces cerevisiae);②补料分批 | [ | |
| 葡萄糖(40 g/L),草酸(0.2 g/L),酵母提取物(7.5 g/L),蛋白胨(10 g/L),基础盐培养基 | 透明质酸 (2.5 MDa;0.8~1.7 g/L) | ①表达透明质酸合成酶2(Xenopus laevis)、UDP-葡萄糖脱氢酶(Xenopus laevis)、UDP-葡萄糖焦磷酸化酶(P. pastoris)、UDP-N-乙酰氨基葡萄糖焦磷酸化酶(P. pastoris)、磷酸葡萄糖异构酶(P. pastoris);②1 L生物反应器 | [ | |
| 甲醇(1%),组氨酸(0.05%),生物素(0.01%) | 富马酸(0.76 g/L),苹果酸(42.28 g/L),琥珀酸(9.42 g/L) | ①表达丙酮酸羧化酶(Pichia pastoris)、苹果酸脱氢酶1(Pichia pastoris)、富马酸酶(Pichia pastoris);②0.6 L生物反应器 | [ | |
| 酵母提取物(1%),蛋白胨(2%),葡萄糖(2%) | 番茄红素(1.141 μg/g细胞干重),β-胡萝卜素(339 μg/g细胞干重) | ①表达香叶酰二磷酸合成酶(Erwinia uredovora)、茄红素合成酶(Erwinia uredovora)、茄红素去饱和酶(Erwinia uredovora)、番茄红素环化酶(Ficus carica);②摇瓶培养 | [ | |
| 甲醇(0.5%),甘油,基础盐培养基 | 单那可林 J(593.9 mg/L),洛伐他汀(250.8 mg/L) | ①途径分割策略;②5 L生物反应器 | [ | |
| 甲醇(0.5%),甘油,基础盐培养基 | 洛伐他汀(419 mg/L) | ①表达洛伐他汀合成模块B-C-A-F-D-G(Aspergillus terreus)、细胞色素P450还原酶(Aspergillus terreus)、磷酸泛酰巯基乙胺基转移酶(Aspergillus nidulans)、他汀外排蛋白(Aspergillus terreus);②5 L生物反应器 | [ | |
| 汉逊酵母 | 甲醇(0.2%),胆碱(0.2%),葡萄糖(0.25%) | 青霉素(1 mg/L) | ①表达非核糖体肽合成酶(Penicillium chrysogenum)、异青霉素N合成酶(Penicillium chrysogenum)、异青霉素素N酰基转移酶(Penicillium chrysogenum)、苯乙酰CoA连接酶(Penicillium chrysogenum);②恒化器 | [ |
| 甲醇芽孢杆菌 | 甲醇,基础盐培养基 | L-谷氨酸(58 g/L) | ① 50 ℃培养;②补料分批 | [ |
| 甲醇(150 mmol/L),酵母提取物(0.25 g/L),基础盐培养基 | L-赖氨酸(11 g/L) | ①表达天冬氨酸激酶(Bacillus methanolicus);② 3 L生物反应器 | [ | |
| 甲醇(150 mmol/L),基础盐培养基 | L-赖氨酸(65 g/L), L-谷氨酸(28 g/L) | ①表达丙酮酸羧化酶(Bacillus methanolicus)、高丝氨酸脱氢酶(Bacillus methanolicus);②3 L生物反应器 | [ | |
| 甲醇(200 mmol/L),酵母提取物(0.025%),基础盐培养基 | L-赖氨酸(777 mg/L) | ①表达赖氨酸外排基因(Corynebacterium glutamicum);② 500 mL摇瓶发酵 | [ | |
| 甲醇(200 mmol/L),酵母提取物(0.025%),基础盐培养基 | 尸胺(11.3 g/L) | ①表达赖氨酸脱羧酶(Escherichia coli);② 3 L生物反应器 | [ | |
| 甲醇(200 mmol/L),诱导剂,基础盐培养基 | 尸胺(10.2 g/L) | ①诱导的启动子系统和θ-and rolling circle-复制系统;② 3 L生物反应器 | [ | |
| 甲醇(200 mmol/L),5'-磷酸吡哆醛(20 μmol/L),基础盐培养基 | γ-氨基丁酸(9 g/L) | ①表达谷氨酸脱羧酶(Sulfobacillus thermosulfidooxidans);② 3 L生物反应器 | [ | |
| 扭脱甲基杆菌 AM1 | 甲醇(0.5%),琥珀酸(0.5%),基础盐培养基 | 聚羟基烷酸酯 (PHA,35.1%±4.36%) | ①表达PHA合成酶(Aeromonas caviae);② Co2+缺陷条件;③摇瓶培养 | [ |
| 甲醇(125 mmol/L),琥珀酸(20 mmol/L),乙胺(20 mmol/L),基础盐培养基 | 1-丁醇(15.2 mg/L) | ①表达烯酰辅酶A还原酶(Treponema denticola)、乙醇脱氢酶(Clostridium acetobutylicum)、巴豆酸酶(Methylobacterium extorquens AM1);②摇瓶培养 | [ | |
| 甲醇(240 mmol/L),基础盐培养基 | 衣康酸(31.6 mg/L± 5.5 mg/L) | ①表达乌头酸脱羧酶(Aspergillus terreus);② 2 L生物反应器 | [ | |
| 甲醇(125 mmol/L),基础盐培养基 | 1-丁醇(25.5 mg/L) | ①适应性进化筛选突变株耐受丁醇达到0.5%;②摇瓶培养 | [ | |
| 甲醇(1%),基础盐培养基 | 甲羟戊酸(2.67 g/L) | ①生物感应器辅助;②产率0.55 mol乙酰CoA/mol甲醇;③ 5 L-生物反应器 | [ | |
| 甲醇(3 g/L),基础盐培养基 | 2-羟基异丁酸(2.1 g/L) | ①表达(R)-3-羟基丁基辅酶A特异性辅酶B12依赖性诱变酶(Bacillus massiliosenegalensis JC6);②产率0.11 g/g甲醇,生物反应器 | [ | |
| 甲醇(125 mmol/L),琥珀酸(15 mmol/L),基础盐培养基 | 3-羟基丙酸(69.8 mg/L) | ①表达丙二酰辅酶A还原酶(Chlorofexus aurantiacus);②摇瓶培养 | [ | |
| 甲醇(0.5%),琥珀酸(15 mmol/L),基础盐培养基 | 3-羟基丙酸(0.857 g/L) | ①ΔhprA,表达己糖磷酸合成酶(Bacillus methanolicus)、磷酸己糖异构酶(Bacillus methanolicus)、磷酸果糖激酶(Bacillus methanolicus)、6-磷酸葡萄糖脱氢酶(Bacillus methanolicus)、丙二酰辅酶A还原酶(Chlorofexus aurantiacus);② 2.5 L生物反应器 | [ | |
| 甲醇(125 mmol/L),琥珀酸(15 mmol/L),基础盐培养基 | 异丁醇(19 mg/L) | ①ΔldhA,表达2-酮异戊酸脱羧酶(Lactococcus lactis)、醇脱氢酶(Lactococcus lactis)、乙酰乳酸合酶(Bacillus subtilis);②摇瓶培养 | [ | |
| 甲醇(123 mmol/L),基础盐培养基 | 甲基富马酸,(2S)-甲基琥珀酸(0.65 g/L) | ①ΔphaC,表达硫酯酶;②摇瓶培养;产率0.17 g/g甲醇;③钴缺陷条件 | [ | |
| 甲醇(123 mmol/L),基础盐培养基 | α-蛇麻烯(1.65 g/L) | ①表达α-蛇麻烯合成酶(Zingiber zerumbet)、法尼焦磷酸合成酶(Saccharomyces cerevisiae)、异源甲羟戊酸途径;② 2.4 L生物反应器 | [ | |
| 甲醇(1%),基础盐培养基 | 甲羟戊酸(2.22 g/L) | ①表达HMG-CoA 合成酶(Enterococcus faecalistiters)、HMG-CoA还原酶(Enterococcus faecalistiters)、乙酰乙酰CoA 硫解酶(Ralstonia eutropha);②产率28.4 mg/g甲醇,5 L生物反应器 | [ | |
| 甲醇,琥珀酸盐,基础盐 培养基 | 丁二烯前体(0.6 mg/L) | ①表达羟乙烷基噻唑激酶(Escherichia coli)、磷酸异戊烯基激酶(Methanothermobacter thermautotrophicus)、乙醇脱氢酶(Clostridium acetobutylicum);②摇瓶培养 | [ | |
| 扭脱甲基杆菌 ATCC 55366 | 甲醇,基础盐培养基 | 绿色荧光蛋白(4 g/L) | ①表达不同的启动子;产率80 mg/g甲醇;② 20 L生物反应器 | [ |
| 甲醇(1%),异丙基苯甲酸(20 mg/L),基础盐培养基 | 功能性聚羟基烷酸酯(PHA,35.1%±4.36%) | ①表达聚羟基脂肪酸酯合成酶2(Pseudomonas fluorescens GK13);②含有碳碳双键的功能性PHA;3.摇瓶培养 | [ | |
| 甲基杆菌 MB200 | 甲醇(1.2%),基础盐培养基 | 乙醛酸(14.38 mg/mL) | ①表达羟基丙酮酸还原酶(M. extorquens);②摇瓶培养 | [ |
| 甲醇(1.2%),基础盐培养基 | L-丝氨酸(11.4 g/L) | ①表达丝氨酸羟甲基转移酶(SHMT,M. extorquens);②静息发酵 | [ | |
| 甲醇(1.2%),基础盐培养基 | L-丝氨酸(6.6 g/L) | ①表达甲醇脱氢酶(Methylobacterium sp. MB200);②静息发酵 | [ | |
| 甲醇(1.2%),基础盐培养基 | L-丝氨酸(9.1 g/L) | Δgnd(6-磷酸葡萄糖脱氢酶) | [ | |
| 甲基杆菌MB 126 | 甲醇(125 mmol/L),基础盐培养基 | (R)-3-羟基丁酸(2.8 g/L) | ① Δhbd(3-羟基丁酸降解酶)、Δlip(硫辛酸合成酶)② 2 L生物反应器 | [ |
| Protomonas extorquens NR-1 | 甲醇(1%),基础盐培养基 | L-丝氨酸(54.5 g/L) | ①静息发酵;② 2 L生物反应器,产率0.183 mol丝氨酸/mol甲醇 | [ |
| 甲基菌属糖原ATCC突变体 | 甲醇(0.5%),蛋白胨(3 g/L),基础盐培养基 | L-谷氨酸(38.3 g/L),L-苏氨酸(11 g/L),L-赖氨酸(5.6 g/L) | ①NTG突变获取不同突变体,RV3、AL119、DHL122;② 5 L生物反应器 | [ |
| 嗜甲基菌属 Methylotrophus | 甲醇(500 mmol/L),硫酸铵 | L-赖氨酸(112 mmol) | ①表达ED 途径;② 1 L生物反应器 | [ |
| 甲烷球菌属 Maripaludis | 甲酸,乙酸(10 mmol/L),丙酮酸(20 mmol/L),丙氨酸(1 mmol/L),CO2/H2 | 香叶醇 | ①表达香叶醇合成酶(Ocimum basilicum);② 产率4.6 mg/g甲酸 | [ |
| 宿主 | 主要 途径 | C1底物 | 共培养 底物 | 产品 (产量) | 特点 | 主要 策略 | 参考文献 |
|---|---|---|---|---|---|---|---|
| 谷氨酸棒状 杆菌 | RuMP | 甲醇 | 核糖(20/100 mmol/L);葡萄糖(50 mmol/L) | 尸胺(1.5 g/L) | 首次将甲醇转化为非天然产物-尸胺 | ①表达RuMP相关酶;②Δald(乙醛脱氢酶),Δfadh(甲醛脱氢酶);③表达赖氨酸脱羧酶(E. coli) | [ |
| RuMP | 甲醇 (15 g/L) | 木糖(4 g/L) | L-谷氨酸 (230 mg/L) | ①甲醇是主要的碳源,甲醇和木糖的利用率达到7.04∶1;②进化菌株甲醇耐受浓度为15 g/L | ①表达RuMP相关酶;②ΔAdhE(甲醛脱氢酶)、Δald(乙醛脱氢酶)、ΔrpiB(核糖磷酸异构酶);③表达木酮糖激酶、木酮糖异构酶;④过表达PPP途径、ED途径相关酶 | [ | |
| RuMP | 甲醇 | 木糖(4 g/L) | L-谷氨酸 (90 mg/L) | ①甲醇和木糖共利用可以明显提高细胞增长;②甲醇木糖利用率为3.83∶1;③胞内细胞13C甲醇标记度达到63% | ①表达RuMP相关酶;②ΔAdhE(甲醛脱氢酶)、Δald(乙醛脱氢酶);③表达木酮糖激酶、木酮糖异构酶;④ΔrpiB(核糖磷酸异构酶) | [ | |
| 大肠 杆菌 | RuMP | 甲醇 (60 mmol/L) | 葡萄糖、木糖、蛋白胨的混合物(1 g/L);酵母提取物(1 g/L) | 橘皮素(3.5 mg/L) | 首次报道E. coli利用甲醇合成黄酮类物质 | ①表达RuMP相关酶;②ΔfrmA(甲醛脱氢酶);③表达香豆酰CoA连接酶、查尔酮合成酶 | [ |
| RuMP | 甲醇 (6.4 g/L) | 葡萄糖(30~100 g/L)、酵母提取物(1 g/L); | 琥珀酸(68.75 g/L) | ①厌氧发酵加入甲醇提高琥珀酸产率0.98 g/g± 0.11 g/g甲醇;②甲醇氧化形成的NADH有助于琥珀酸形成 | ①表达RuMP相关酶;②5 L生物反应器 | [ | |
| MSC | 甲醇 (200 mmol/L); 甲酸 (20 mmol/L);CO2 | 木糖(30 mmol/L) | 乙醇(36 mmol/L) | ①首次报道基于丝氨酸循环构建合成甲基营养菌;②甲醇加入可以增加乙醇产量达到62%,甲醇利用率为0.7 mmol/(L·h)(以OD600计),木糖与甲醇消耗率为1∶0.7 | ①表达丝氨酸途径相关酶;②表达丙氨酸-乙醛酸转氨酶(Agt)、丝氨酸脱氢酶(Sdh)、苹果酸硫激酶(Mtk)、苹果酰CoA裂解酶(Mcl)等;③表达乙醛脱氢酶(Pdup)、乙醇脱氢酶(AdhB);④ΔaceB ΔglcB ΔgcvP Δgcl ΔfrdB ΔldhA | [ | |
| RuMP | 甲醇 | 葡萄糖(30 g/L),酵母提取物(10 g/L) | 丙酮(45.0 mmol/L ± 8.7 mmol/L) | ①两种策略共同提高甲醇利用率;②E. coli利用甲醇合成丙酮 | ①Δpgi(6-磷酸葡萄糖异构酶)、ΔfrmA(甲醛脱氢酶);②表达RuMP途径相关酶;③表达磷酸核糖异构酶(rpe)、转酮酶(tkt);④表达二磷酸果糖醛缩酶(fba)、景天庚糖双磷酸酶(glpX)、磷酸果糖激酶(pfk) | [ | |
| RuMP | 甲醇 (3.2 g/L), 甲醛 (0.15 g/L) | 葡萄糖(8~9 g/L),硫酸素焦磷酸盐(0.46 g/L) | 1,3-丙二醇 (508.3 mg/L ± 9.1 mg/L) | ①首次实现利用甲醇和丙酮酸合成1,3-丙二醇;②缩短途径,并有效提高1,3-丙二醇产量 | ①表达甲醇脱氢酶;② ΔfrmA(甲醛脱氢酶);③表达羟丁酸醛缩酶、酮酸脱羧酶、1,3-丁二酸氧化还原酶 | [ | |
| HACL | 甲醛 | 乙醇酸(1.2 g/L) | ①首次报道HACL途径,可以转化甲酸和甲醛;②首次利用甲醛合成乙醇酸和羟基异丁酸 | 表达羟酰CoA裂解酶(RuHACLG390N,Rhodospirillales bacterium)、酰基CoA还原酶(LmACR,Listeria monocytogenes) | [ | ||
| RuMP | 甲醇 (250 mmol/L) | 葡萄糖(10 g/L),酵母提取物(2 g/L) | 丙酮(58 mmol/L) | ①显著提升甲醇向丙酮的转化;②构建了甲醇依赖的菌株底盘 | ①Δpgi(6-磷酸葡萄糖异构酶)、Δedd(磷酸葡萄糖酸脱氢酶)、ΔrpiAB(核糖磷酸异构酶)、ΔfrmA(甲醛脱氢酶);②表达RuMP相关酶 | [ | |
| RuMP | 甲醇(50~ 100 mmol/L) | 葡萄糖(50 mmol/L),酵母提取物(1 g/L) | L-赖氨酸 (0.1 g/L) | 甲醇氧化所得的NADH转变为NADPH用于赖氨酸生产 | ①表达RuMP相关酶、ΔfrmA(甲醛脱氢酶);②表达Nudix水解酶、NADH激酶(POS5,Saccharomyces cerevisiae);③表达赖氨酸合成途径相关酶(lysC, dapA, dapB) | [ | |
| RuMP | 甲醇 | 木糖,核糖 | 乙醇(4.6 g/L); 1-丁醇(2 g/L) | ①构建甲醇依赖型木糖菌株;②甲醇与木糖利用率为1∶1 | ①表达RuMP相关酶;②ΔAdhE(甲醛脱氢酶)、Δald(乙醛脱氢酶)、ΔrpiAB(核糖磷酸异构酶);③表达腺苷酸环化酶 | [ |
Tab.2 Chemicals produced by synthetic methylotrophs
| 宿主 | 主要 途径 | C1底物 | 共培养 底物 | 产品 (产量) | 特点 | 主要 策略 | 参考文献 |
|---|---|---|---|---|---|---|---|
| 谷氨酸棒状 杆菌 | RuMP | 甲醇 | 核糖(20/100 mmol/L);葡萄糖(50 mmol/L) | 尸胺(1.5 g/L) | 首次将甲醇转化为非天然产物-尸胺 | ①表达RuMP相关酶;②Δald(乙醛脱氢酶),Δfadh(甲醛脱氢酶);③表达赖氨酸脱羧酶(E. coli) | [ |
| RuMP | 甲醇 (15 g/L) | 木糖(4 g/L) | L-谷氨酸 (230 mg/L) | ①甲醇是主要的碳源,甲醇和木糖的利用率达到7.04∶1;②进化菌株甲醇耐受浓度为15 g/L | ①表达RuMP相关酶;②ΔAdhE(甲醛脱氢酶)、Δald(乙醛脱氢酶)、ΔrpiB(核糖磷酸异构酶);③表达木酮糖激酶、木酮糖异构酶;④过表达PPP途径、ED途径相关酶 | [ | |
| RuMP | 甲醇 | 木糖(4 g/L) | L-谷氨酸 (90 mg/L) | ①甲醇和木糖共利用可以明显提高细胞增长;②甲醇木糖利用率为3.83∶1;③胞内细胞13C甲醇标记度达到63% | ①表达RuMP相关酶;②ΔAdhE(甲醛脱氢酶)、Δald(乙醛脱氢酶);③表达木酮糖激酶、木酮糖异构酶;④ΔrpiB(核糖磷酸异构酶) | [ | |
| 大肠 杆菌 | RuMP | 甲醇 (60 mmol/L) | 葡萄糖、木糖、蛋白胨的混合物(1 g/L);酵母提取物(1 g/L) | 橘皮素(3.5 mg/L) | 首次报道E. coli利用甲醇合成黄酮类物质 | ①表达RuMP相关酶;②ΔfrmA(甲醛脱氢酶);③表达香豆酰CoA连接酶、查尔酮合成酶 | [ |
| RuMP | 甲醇 (6.4 g/L) | 葡萄糖(30~100 g/L)、酵母提取物(1 g/L); | 琥珀酸(68.75 g/L) | ①厌氧发酵加入甲醇提高琥珀酸产率0.98 g/g± 0.11 g/g甲醇;②甲醇氧化形成的NADH有助于琥珀酸形成 | ①表达RuMP相关酶;②5 L生物反应器 | [ | |
| MSC | 甲醇 (200 mmol/L); 甲酸 (20 mmol/L);CO2 | 木糖(30 mmol/L) | 乙醇(36 mmol/L) | ①首次报道基于丝氨酸循环构建合成甲基营养菌;②甲醇加入可以增加乙醇产量达到62%,甲醇利用率为0.7 mmol/(L·h)(以OD600计),木糖与甲醇消耗率为1∶0.7 | ①表达丝氨酸途径相关酶;②表达丙氨酸-乙醛酸转氨酶(Agt)、丝氨酸脱氢酶(Sdh)、苹果酸硫激酶(Mtk)、苹果酰CoA裂解酶(Mcl)等;③表达乙醛脱氢酶(Pdup)、乙醇脱氢酶(AdhB);④ΔaceB ΔglcB ΔgcvP Δgcl ΔfrdB ΔldhA | [ | |
| RuMP | 甲醇 | 葡萄糖(30 g/L),酵母提取物(10 g/L) | 丙酮(45.0 mmol/L ± 8.7 mmol/L) | ①两种策略共同提高甲醇利用率;②E. coli利用甲醇合成丙酮 | ①Δpgi(6-磷酸葡萄糖异构酶)、ΔfrmA(甲醛脱氢酶);②表达RuMP途径相关酶;③表达磷酸核糖异构酶(rpe)、转酮酶(tkt);④表达二磷酸果糖醛缩酶(fba)、景天庚糖双磷酸酶(glpX)、磷酸果糖激酶(pfk) | [ | |
| RuMP | 甲醇 (3.2 g/L), 甲醛 (0.15 g/L) | 葡萄糖(8~9 g/L),硫酸素焦磷酸盐(0.46 g/L) | 1,3-丙二醇 (508.3 mg/L ± 9.1 mg/L) | ①首次实现利用甲醇和丙酮酸合成1,3-丙二醇;②缩短途径,并有效提高1,3-丙二醇产量 | ①表达甲醇脱氢酶;② ΔfrmA(甲醛脱氢酶);③表达羟丁酸醛缩酶、酮酸脱羧酶、1,3-丁二酸氧化还原酶 | [ | |
| HACL | 甲醛 | 乙醇酸(1.2 g/L) | ①首次报道HACL途径,可以转化甲酸和甲醛;②首次利用甲醛合成乙醇酸和羟基异丁酸 | 表达羟酰CoA裂解酶(RuHACLG390N,Rhodospirillales bacterium)、酰基CoA还原酶(LmACR,Listeria monocytogenes) | [ | ||
| RuMP | 甲醇 (250 mmol/L) | 葡萄糖(10 g/L),酵母提取物(2 g/L) | 丙酮(58 mmol/L) | ①显著提升甲醇向丙酮的转化;②构建了甲醇依赖的菌株底盘 | ①Δpgi(6-磷酸葡萄糖异构酶)、Δedd(磷酸葡萄糖酸脱氢酶)、ΔrpiAB(核糖磷酸异构酶)、ΔfrmA(甲醛脱氢酶);②表达RuMP相关酶 | [ | |
| RuMP | 甲醇(50~ 100 mmol/L) | 葡萄糖(50 mmol/L),酵母提取物(1 g/L) | L-赖氨酸 (0.1 g/L) | 甲醇氧化所得的NADH转变为NADPH用于赖氨酸生产 | ①表达RuMP相关酶、ΔfrmA(甲醛脱氢酶);②表达Nudix水解酶、NADH激酶(POS5,Saccharomyces cerevisiae);③表达赖氨酸合成途径相关酶(lysC, dapA, dapB) | [ | |
| RuMP | 甲醇 | 木糖,核糖 | 乙醇(4.6 g/L); 1-丁醇(2 g/L) | ①构建甲醇依赖型木糖菌株;②甲醇与木糖利用率为1∶1 | ①表达RuMP相关酶;②ΔAdhE(甲醛脱氢酶)、Δald(乙醛脱氢酶)、ΔrpiAB(核糖磷酸异构酶);③表达腺苷酸环化酶 | [ |
| 1 | CLOMBURG J M, CRUMBLEY A M, GONZALEZ R. Industrial biomanufacturing: The future of chemical production[J]. Science, 2017, 355(6320): aag0804. |
| 2 | 谭天伟, 陈必强, 张会丽, 等. 加快推进绿色生物制造 助力实现“碳中和”[J]. 化工进展, 2021, 40(3): 1137-1141. |
| TAN T W, CHEN B Q, ZHANG H L, et al. Accelerate promotion of green bio-manufacturing to help achieve “carbon neutrality”[J]. Chemical Industry and Engineering Progress, 2021, 40(3): 1137-1141. | |
| 3 | SINGH A K, KISHORE G M, PAKRASI H B. Emerging platforms for co-utilization of one-carbon substrates by photosynthetic organisms[J]. Current Opinion in Biotechnology, 2018, 53: 201-208. |
| 4 | DÜRRE P, EIKMANNS B J. C1-carbon sources for chemical and fuel production by microbial gas fermentation[J]. Current Opinion in Biotechnology, 2015, 35: 63-72. |
| 5 | KOPKE M, HELD C, HUJER S, et al. Clostridium ljungdahlii represents a microbial production platform based on syngas[J]. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(29): 13087-13092. |
| 6 | SCHEFFEN M, MARCHAL D G, BENEYTON T, et al. A new-to-nature carboxylation module to improve natural and synthetic CO2 fixation[J]. Nature Catalysis, 2021, 4(2): 105-115. |
| 7 | COTTON C A, EDLICH-MUTH C, BAR-EVEN A. Reinforcing carbon fixation: CO2 reduction replacing and supporting carboxylation[J]. Current Opinion in Biotechnology, 2018, 49: 49-56. |
| 8 | YISHAI O, LINDNER S N, GONZALEZ DE LA CRUZ J, et al. The formate bio-economy[J]. Current Opinion in Chemical Biology, 2016, 35: 1-9. |
| 9 | WANG Y, FAN L W, TUYISHIME P, et al. Synthetic methylotrophy: A practical solution for methanol-based biomanufacturing[J]. Trends in Biotechnology, 2020, 38(6): 650-666. |
| 10 | WU Y S, JIANG Z, LU X, et al. Domino electroreduction of CO2 to methanol on a molecular catalyst[J]. Nature, 2019, 575(7784): 639-642. |
| 11 | CHISTOSERDOVA L, LIDSTROM M E. Aerobic methylotrophic prokaryotes[M]//ROSENBERG E.The prokaryotes[M]. German: Springer Reference, 2013: 267. |
| 12 | 高教琪, 周雍进. 甲醇生物转化的机遇与挑战[J]. 合成生物学, 2020, 1(2): 158-173. |
| GAO J Q, ZHOU Y J. Advances in methanol bio-transformation[J]. Synthetic Biology Journal, 2020, 1(2): 158-173. | |
| 13 | CAMP H J M O DEN, ISLAM T, STOTT M B, et al. Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia [J]. Environmental Microbiology Reports, 2009, 1(5): 293-306. |
| 14 | ETTWIG K F, BUTLER M K, LE PASLIER D, et al. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria[J]. Nature, 2010, 464(7288): 543-548. |
| 15 | GAO L M, CAI M H, SHEN W, et al. Engineered fungal polyketide biosynthesis in Pichia pastoris: a potential excellent host for polyketide production[J]. Microbial Cell Factories, 2013, 12: 77. |
| 16 | WRIESSNEGGER T, AUGUSTIN P, ENGLEDER M, et al. Production of the sesquiterpenoid (+)-nootkatone by metabolic engineering of Pichia pastoris [J]. Metabolic Engineering, 2014, 24: 18-29. |
| 17 | JEONG E, SHIM W Y, KIM J H. Metabolic engineering of Pichia pastoris for production of hyaluronic acid with high molecular weight[J]. Journal of Biotechnology, 2014, 185: 28-36. |
| 18 | ZHANG T, GE C Y, DENG L, et al. C4-dicarboxylic acid production by overexpressing the reductive TCA pathway[J]. FEMS Microbiology Letters, 2015, 362(9): fnv052. |
| 19 | ARAYA-GARAY J M, FEIJOO-SIOTA L, ROSA-DOS-SANTOS F, et al. Construction of new Pichia pastoris X-33 strains for production of lycopene and β-carotene[J]. Applied Microbiology and Biotechnology, 2012, 93(6): 2483-2492. |
| 20 | LIU Y Q, TU X H, XU Q, et al. Engineered monoculture and co-culture of methylotrophic yeast for de novo production of monacolin J and lovastatin from methanol[J]. Metabolic Engineering, 2018, 45: 189-199. |
| 21 | LIU Y Q, BAI C X, XU Q, et al. Improved methanol-derived lovastatin production through enhancement of the biosynthetic pathway and intracellular lovastatin efflux in methylotrophic yeast[J]. Bioresources and Bioprocessing, 2018, 5: 22. |
| 22 | GIDIJALA L, KIEL J A K W, DOUMA R D, et al. An engineered yeast efficiently secreting penicillin[J]. PLoS One, 2009, 4(12): e8317. |
| 23 | BRAUTASET T, JAKOBSEN Ø M, JOSEFSEN K D, et al. Bacillus methanolicus: a candidate for industrial production of amino acids from methanol at 50 ℃[J]. Applied Microbiology and Biotechnology, 2007, 74(1): 22-34. |
| 24 | JAKOBSEN O M, BRAUTASET T, DEGNES K F, et al. Overexpression of wild-type aspartokinase increases L-lysine production in the thermotolerant methylotrophic bacterium Bacillus methanolicus [J]. Applied and Environmental Microbiology, 2009, 75(3): 652-661. |
| 25 | BRAUTASET T, JAKOBSEN Ø M, DEGNES K F, et al. Bacillus methanolicus pyruvate carboxylase and homoserine dehydrogenase I and II and their roles for L-lysine production from methanol at 50 ℃[J]. Applied Microbiology and Biotechnology, 2010, 87(3): 951-964. |
| 26 | NÆRDAL I, NETZER R, IRLA M, et al. L-lysine production by Bacillus methanolicus: genome-based mutational analysis and L-lysine secretion engineering[J]. Journal of Biotechnology, 2017, 244: 25-33. |
| 27 | NAERDAL I, PFEIFENSCHNEIDER J, BRAUTASET T, et al. Methanol-based cadaverine production by genetically engineered Bacillus methanolicus strains[J]. Microbial Biotechnology, 2015, 8(2): 342-350. |
| 28 | IRLA M, HEGGESET T M B, NÆRDAL I, et al. Genome-based genetic tool development for Bacillus methanolicus: θ-and rolling circle-replicating plasmids for inducible gene expression and application to methanol-based cadaverine production[J]. Frontiers in Microbiology, 2016, 7: 1481. |
| 29 | IRLA M, NÆRDAL I, BRAUTASET T, et al. Methanol-based γ-aminobutyric acid (GABA) production by genetically engineered Bacillus methanolicus strains[J]. Industrial Crops and Products, 2017, 106: 12-20. |
| 30 | ORITA I, NISHIKAWA K, NAKAMURA S, et al. Biosynthesis of polyhydroxyalkanoate copolymers from methanol by Methylobacterium extorquens AM1 and the engineered strains under cobalt-deficient conditions[J]. Applied Microbiology and Biotechnology, 2014, 98(8): 3715-3725. |
| 31 | HU B, LIDSTROM M E. Metabolic engineering of Methylobacterium extorquens AM1 for 1-butanol production[J]. Biotechnology for Biofuels, 2014, 7(1): 156. |
| 32 | LIM C K, VILLADA J C, CHALIFOUR A, et al. Designing and engineering Methylorubrum extorquens AM1 for itaconic acid production[J]. Frontiers in Microbiology, 2019, 10: 1027. |
| 33 | HU B, YANG Y M, BECK D A C, et al. Comprehensive molecular characterization of Methylobacterium extorquens AM1 adapted for 1-butanol tolerance[J]. Biotechnology for Biofuels, 2016, 9: 84. |
| 34 | LIANG W F, CUI L Y, CUI J Y, et al. Biosensor-assisted transcriptional regulator engineering for Methylobacterium extorquens AM1 to improve mevalonate synthesis by increasing the acetyl-CoA supply[J]. Metabolic Engineering, 2017, 39: 159-168. |
| 35 | ROHDE M T, TISCHER S, HARMS H, et al. Production of 2-hydroxyisobutyric acid from methanol by Methylobacterium extorquens AM1 expressing (R)-3-hydroxybutyryl coenzyme A-isomerizing enzymes[J]. Applied and Environmental Microbiology, 2017, 83(3). DOI: 10.1128/aem.02622-16 . |
| 36 | YANG Y M, CHEN W J, YANG J, et al. Production of 3-hydroxypropionic acid in engineered Methylobacterium extorquens AM1 and its reassimilation through a reductive route[J]. Microbial Cell Factories, 2017, 16(1): 179. |
| 37 | YUAN X J, CHEN W J, MA Z X, et al. Rewiring the native methanol assimilation metabolism by incorporating the heterologous ribulose monophosphate cycle into Methylorubrum extorquens [J]. Metabolic Engineering, 2021, 64: 95-110. |
| 38 | MA Z X, ZHANG M, ZHANG C T, et al. Metabolomic analysis improves bioconversion of methanol to isobutanol in Methylorubrum extorquens AM1[J]. Biotechnology Journal, 2021, 16(6): e2000413. |
| 39 | SONNTAG F, MÜLLER J E N, KIEFER P, et al. High-level production of ethylmalonyl-CoA pathway-derived dicarboxylic acids by Methylobacterium extorquens under cobalt-deficient conditions and by polyhydroxybutyrate negative strains[J]. Applied Microbiology and Biotechnology, 2015, 99(8): 3407-3419. |
| 40 | SONNTAG F, KRONER C, LUBUTA P, et al. Engineering Methylobacterium extorquens for de novo synthesis of the sesquiterpenoid α-humulene from methanol[J]. Metabolic Engineering, 2015, 32: 82-94. |
| 41 | ZHU W L, CUI J Y, CUI L Y, et al. Bioconversion of methanol to value-added mevalonate by engineered Methylobacterium extorquens AM1 containing an optimized mevalonate pathway[J]. Applied Microbiology and Biotechnology, 2016, 100(5): 2171-2182. |
| 42 | YANG J, ZHANG C T, YUAN X J, et al. Metabolic engineering of Methylobacterium extorquens AM1 for the production of butadiene precursor[J]. Microbial Cell Factories, 2018, 17(1): 194. |
| 43 | BÉLANGER L, FIGUEIRA M M, BOURQUE D, et al. Production of heterologous protein by Methylobacterium extorquens in high cell density fermentation[J]. FEMS Microbiology Letters, 2004, 231(2): 197-204. |
| 44 | HÖFER P, CHOI Y J, OSBORNE M J, et al. Production of functionalized polyhydroxyalkanoates by genetically modified Methylobacterium extorquens strains[J]. Microbial Cell Factories, 2010, 9: 70. |
| 45 | SHEN P H, WU B. Over-expression of a hydroxypyruvate reductase in Methylobacterium sp. MB200 enhances glyoxylate accumulation[J]. Journal of Industrial Microbiology and Biotechnology, 2007, 34(10): 657. |
| 46 | SHEN P H, CHAO H J, JIANG C J, et al. Enhancing production of L-serine by increasing the glyA gene expression in Methylobacterium sp. MB200[J]. Applied Biochemistry and Biotechnology, 2010, 160(3): 740-750. |
| 47 | CHAO H, WU B, SHEN P. Overexpression of the methanol dehydrogenase gene mxaF in Methylobacterium sp. MB200 enhances L-serine production[J]. Letters in Applied Microbiology, 2015, 61(4): 390-396. |
| 48 | LI X, WU B, ZHOU K, et al. Deletion of gene gnd encoding 6-phosphogluconate dehydrogenase promotes L-serine biosynthesis in a genetically engineered strain of Methylobacterium sp. MB200[J]. Biotechnology Letters, 2019, 41(1): 69-77. |
| 49 | HÖLSCHER T, BREUER U, ADRIAN L, et al. Production of the chiral compound (R)-3-hydroxybutyrate by a genetically engineered methylotrophic bacterium[J]. Applied and Environmental Microbiology, 2010, 76(16): 5585-5591. |
| 50 | HAGISHITA T, YOSHIDA T, IZUMI Y, et al. Efficient L-serine production from methanol and glycine by resting cells of Methylobacterium sp. strain MN43 [J]. Bioscience, Biotechnology, and Biochemistry, 1996, 60(10): 1604-1607. |
| 51 | MOTOYAMA H, ANAZAWA H, KATSUMATA R, et al. Amino acid production from methanol by Methylobacillus glycogenes mutants: isolation of L-glutamic acid hyper-producing mutants from M. glycogenes strains, and derivation of L-threonine and L-lysine-producing mutants from them[J]. Bioscience, Biotechnology, and Biochemistry, 1993, 57(1): 82-87. |
| 52 | ISHIKAWA K, GUNJI Y, YASUEDA H, et al. Improvement of L-lysine production by Methylophilus methylotrophus from methanol via the entner-doudoroff pathway, originating in Escherichia coli [J]. Bioscience, Biotechnology, and Biochemistry, 2008, 72(10): 2535-2542. |
| 53 | LYU Z, JAIN R, SMITH P, et al. Engineering the autotroph Methanococcus maripaludis for geraniol production[J]. ACS Synthetic Biology, 2016, 5(7): 577-581. |
| 54 | KALYUZHNAYA M G, PURI A W, LIDSTROM M E. Metabolic engineering in methanotrophic bacteria[J]. Metabolic Engineering, 2015, 29: 142-152. |
| 55 | NIELSEN A K, GERDES K, MURRELL J C. Copper-dependent reciprocal transcriptional regulation of methane monooxygenase genes in Methylococcus capsulatus and Methylosinus trichosporium [J]. Molecular Microbiology, 1997, 25(2): 399-409. |
| 56 | WHITAKER W B, SANDOVAL N R, BENNETT R K, et al. Synthetic methylotrophy: engineering the production of biofuels and chemicals based on the biology of aerobic methanol utilization[J]. Current Opinion in Biotechnology, 2015, 33: 165-175. |
| 57 | HARTNER F S, GLIEDER A. Regulation of methanol utilisation pathway genes in yeasts[J]. Microbial Cell Factories, 2006, 5: 39. |
| 58 | PARK H, LEE H, RO Y T, et al. Identification and functional characterization of a gene for the methanol: N,N'-dimethyl-4-nitrosoaniline oxidoreductase from Mycobacterium sp. strain JC1 (DSM 3803)[J]. Microbiology, 2010, 156(Pt 2): 463-471. |
| 59 | BYSTRYKH L V, GOVORUKHINA N I, DIJKHUIZEN L, et al. Tetrazolium-dye-linked alcohol dehydrogenase of the methylotrophic actinomycete Amycolatopsis methanolica is a three-component complex[J]. European Journal of Biochemistry, 1997, 247(1): 280-287. |
| 60 | NAGY I, VERHEIJEN S, DE SCHRIJVER A, et al. Characterization of the Rhodococcus sp. NI86/21 gene encoding alcohol: N,N'-dimethyl-4-nitrosoaniline oxidoreductase inducible by atrazine and thiocarbamate herbicides[J]. Archives of Microbiology, 1995, 163(6): 439-446. |
| 61 | ZHU T C, ZHAO T X, BANKEFA O E, et al. Engineering unnatural methylotrophic cell factories for methanol-based biomanufacturing: challenges and opportunities[J]. Biotechnology Advances, 2020, 39: 107467. |
| 62 | KROG A, HEGGESET T M B, MÜLLER J E N, et al. Methylotrophic Bacillus methanolicus encodes two chromosomal and one plasmid born NAD+ dependent methanol dehydrogenase paralogs with different catalytic and biochemical properties[J]. PLoS One, 2013, 8(3): e59188. |
| 63 | MÜLLER J E N, MEYER F, LITSANOV B, et al. Engineering Escherichia coli for methanol conversion[J]. Metabolic Engineering, 2015, 28: 190-201. |
| 64 | WANG X, WANG X L, LU X L, et al. Methanol fermentation increases the production of NAD(P)H-dependent chemicals in synthetic methylotrophic Escherichia coli [J]. Biotechnology for Biofuels, 2019, 12: 17. |
| 65 | WU T Y, CHEN C T, LIU J T J, et al. Characterization and evolution of an activator-independent methanol dehydrogenase from Cupriavidus necator N-1[J]. Applied Microbiology and Biotechnology, 2016, 100(11): 4969-4983. |
| 66 | WHITAKER W B, JONES J A, BENNETT R K, et al. Engineering the biological conversion of methanol to specialty chemicals in Escherichia coli [J]. Metabolic Engineering, 2017, 39: 49-59. |
| 67 | ROTH T B, WOOLSTON B M, STEPHANOPOULOS G, et al. Phage-assisted evolution of Bacillus methanolicus methanol dehydrogenase 2[J]. ACS Synthetic Biology, 2019, 8(4): 796-806. |
| 68 | TSURU D, ODA N, MATSUO Y, et al. Glutathione-independent formaldehyde dehydrogenase from Pseudomons putida: survey of functional groups with special regard for cysteine residues[J]. Bioscience, Biotechnology, and Biochemistry, 1997, 61(8): 1354-1357. |
| 69 | GUTHEIL W G, HOLMQUIST B, VALLEE B L. Purification, characterization, and partial sequence of the glutathione-dependent formaldehyde dehydrogenase from Escherichia coli: a class III alcohol dehydrogenase[J]. Biochemistry, 1992, 31(2): 475-481. |
| 70 | VORHOLT J A. Cofactor-dependent pathways of formaldehyde oxidation in methylotrophic bacteria[J]. Archives of Microbiology, 2002, 178(4): 239-249. |
| 71 | HATRONGJIT R, PACKDIBAMRUNG K. A novel NADP+-dependent formate dehydrogenase from Burkholderia stabilis 15516: Screening, purification and characterization[J]. Enzyme and Microbial Technology, 2010, 46(7): 557-561. |
| 72 | MÜLLER J E N, HEGGESET T M B, WENDISCH V F, et al. Methylotrophy in the thermophilic Bacillus methanolicus, basic insights and application for commodity production from methanol[J]. Applied Microbiology and Biotechnology, 2015, 99(2): 535-551. |
| 73 | RUßMAYER H, BUCHETICS M, GRUBER C, et al. Systems-level organization of yeast methylotrophic lifestyle[J]. BMC Biology, 2015, 13: 80. |
| 74 | BENNETT R K, GONZALEZ J E, WHITAKER W B, et al. Expression of heterologous non-oxidative pentose phosphate pathway from Bacillus methanolicus and phosphoglucose isomerase deletion improves methanol assimilation and metabolite production by a synthetic Escherichia coli methylotroph[J]. Metabolic Engineering, 2018, 45: 75-85. |
| 75 | LEßMEIER L, PFEIFENSCHNEIDER J, CARNICER M, et al. Production of carbon-13-labeled cadaverine by engineered Corynebacterium glutamicum using carbon-13-labeled methanol as co-substrate[J]. Applied Microbiology and Biotechnology, 2015, 99(23): 10163-10176. |
| 76 | WANG Y, FAN L W, TUYISHIME P, et al. Adaptive laboratory evolution enhances methanol tolerance and conversion in engineered Corynebacterium glutamicum [J]. Communications Biology, 2020, 3(1): 217. |
| 77 | TUYISHIME P, WANG Y, FAN L W, et al. Engineering Corynebacterium glutamicum for methanol-dependent growth and glutamate production[J]. Metabolic Engineering, 2018, 49: 220-231. |
| 78 | ZHANG W M, ZHANG T, SONG M, et al. Metabolic engineering of Escherichia coli for high yield production of succinic acid driven by methanol[J]. ACS Synthetic Biology, 2018, 7(12): 2803-2811. |
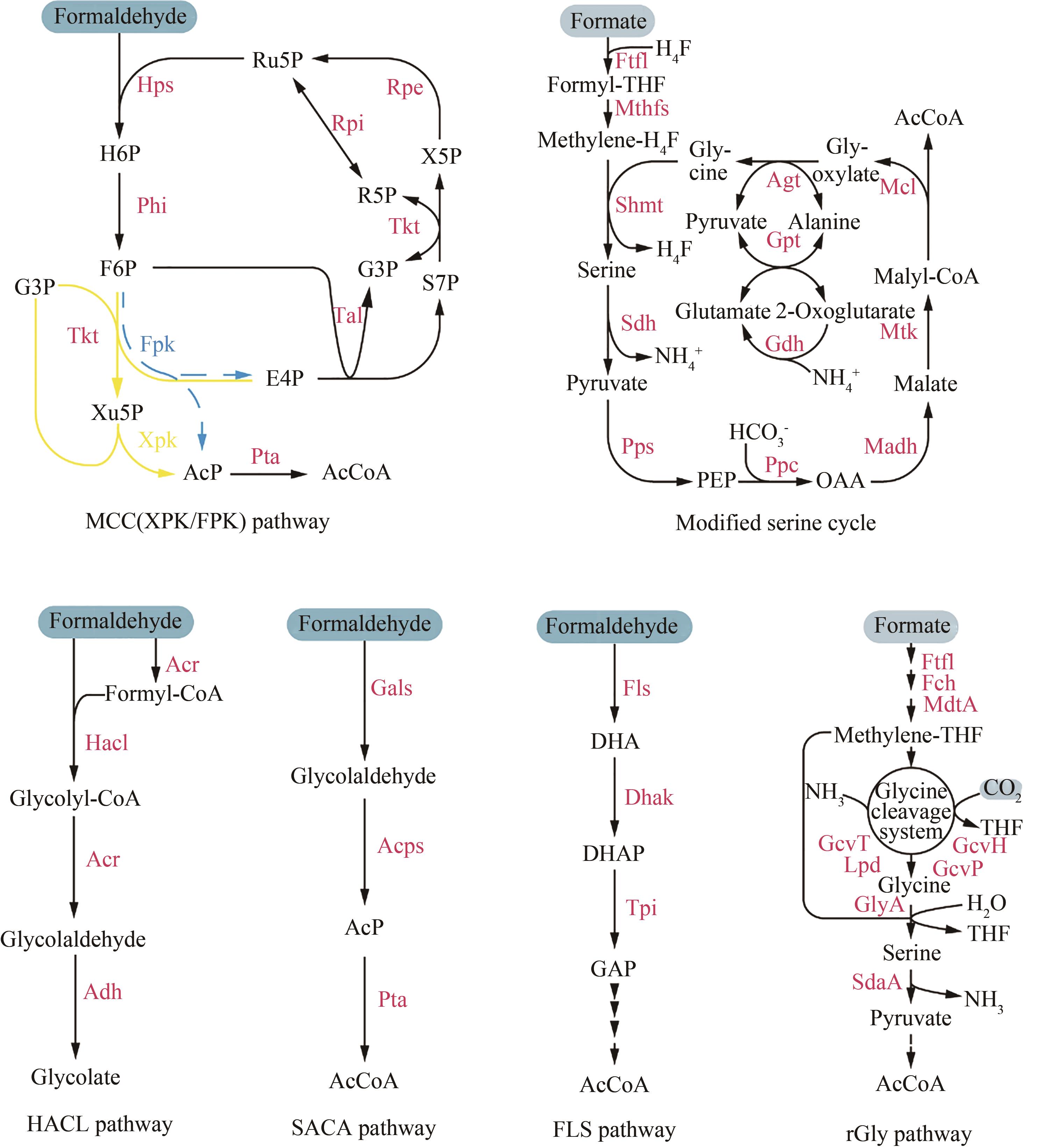
| 79 | YU H, LIAO J C. A modified serine cycle in Escherichia coli coverts methanol and CO2 to two-carbon compounds[J]. Nature Communications, 2018, 9: 3992. |
| 80 | WANG C, REN J, ZHOU L B, et al. An aldolase-catalyzed new metabolic pathway for the assimilation of formaldehyde and methanol to synthesize 2-keto-4-hydroxybutyrate and 1, 3-propanediol in Escherichia coli [J]. ACS Synthetic Biology, 2019, 8(11): 2483-2493. |
| 81 | CHOU A, CLOMBURG J M, QIAN S, et al. 2-Hydroxyacyl-CoA lyase catalyzes acyloin condensation for one-carbon bioconversion[J]. Nature Chemical Biology, 2019, 15(9): 900-906. |
| 82 | BENNETT R K, DILLON M, GERALD HAR J R, et al. Engineering Escherichia coli for methanol-dependent growth on glucose for metabolite production[J]. Metabolic Engineering, 2020, 60: 45-55. |
| 83 | CHEN C T, CHEN F Y H, BOGORAD I W, et al. Synthetic methanol auxotrophy of Escherichia coli for methanol-dependent growth and production[J]. Metabolic Engineering, 2018, 49: 257-266. |
| 84 | LEE P C, YOON Y G, SCHMIDT-DANNERT C. Investigation of cellular targeting of carotenoid pathway enzymes in Pichia pastoris [J]. Journal of Biotechnology, 2009, 140(3/4): 227-233. |
| 85 | GAO J Q, GAO N, ZHAI X X, et al. Recombination machinery engineering for precise genome editing in methylotrophic yeast Ogataea polymorpha [J]. iScience, 2021, 24(3): 102168. |
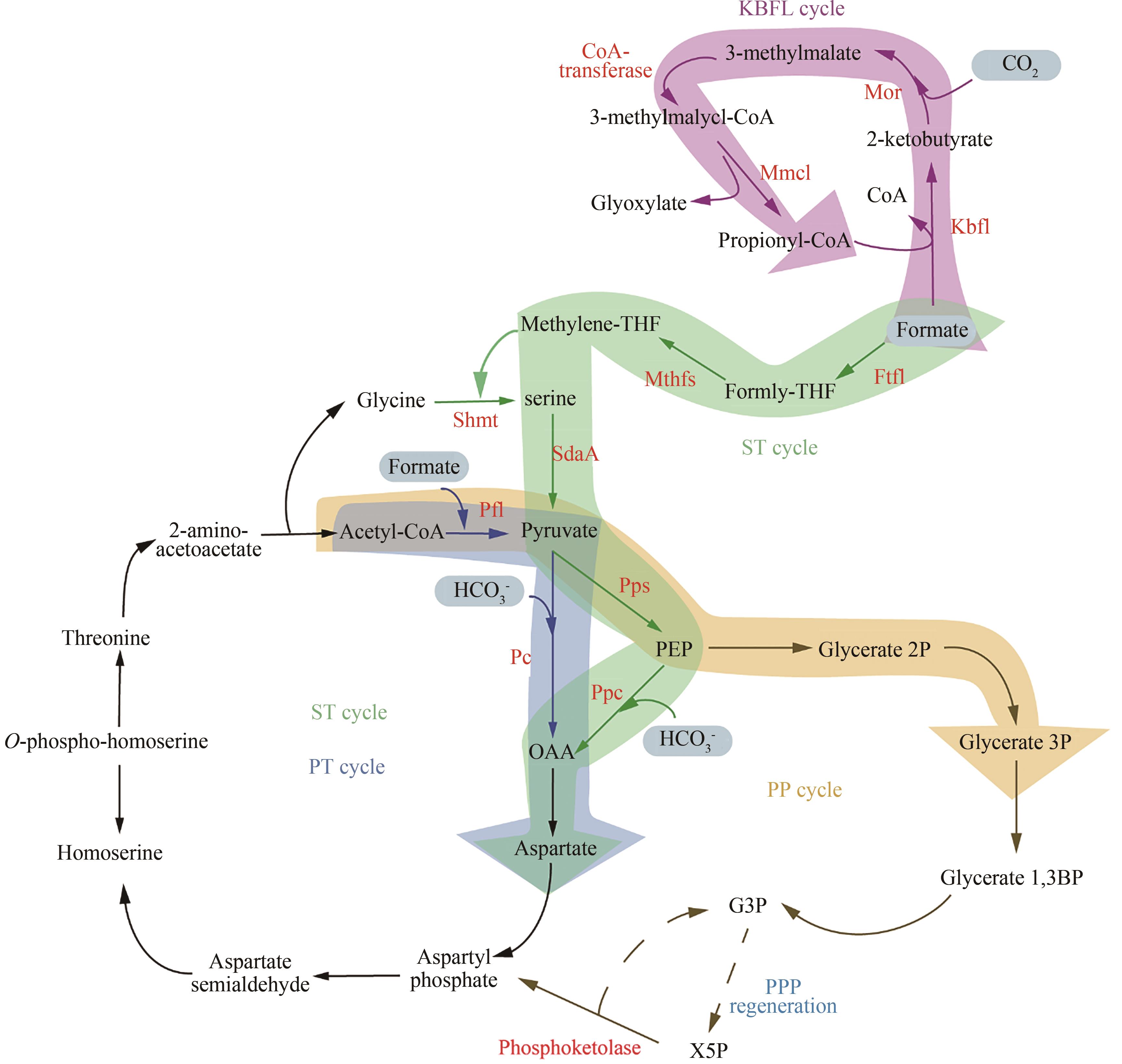
| 86 | ANTHONY C. How half a century of research was required to understand bacterial growth on C1 and C2 compounds; the story of the serine cycle and the ethylmalonyl-CoA pathway[J]. Science Progress, 2011, 94(Pt 2): 109-137. |
| 87 | ALBER B E. Biotechnological potential of the ethylmalonyl-CoA pathway[J]. Applied Microbiology and Biotechnology, 2011, 89(1): 17-25. |
| 88 | BAR-EVEN A. Formate assimilation: the metabolic architecture of natural and synthetic pathways[J]. Biochemistry, 2016, 55(28): 3851-3863. |
| 89 | KHADEM A F, POL A, WIECZOREK A, et al. Autotrophic methanotrophy in verrucomicrobia: Methylacidiphilum fumariolicum SolV uses the Calvin-Benson-Bassham cycle for carbon dioxide fixation[J]. Journal of Bacteriology, 2011, 193(17): 4438-4446. |
| 90 | GASSLER T, SAUER M, GASSER B, et al. The industrial yeast Pichia pastoris is converted from a heterotroph into an autotroph capable of growth on CO2 [J]. Nature Biotechnology, 2020, 38(2): 210-216. |
| 91 | CHEN F Y H, JUNG H W, TSUEI C Y, et al. Converting Escherichia coli to a synthetic methylotroph growing solely on methanol[J]. Cell, 2020, 182(4): 933-946.e14. |
| 92 | BANG J, AHN J H, LEE J A, et al. Synthetic formatotrophs for one-carbon biorefinery[J]. Advanced Science (Weinheim, Baden-Wurttemberg, Germany), 2021, 8(12): 2100199. |
| 93 | KIM S, LINDNER S N, ASLAN S, et al. Growth of E. coli on formate and methanol via the reductive glycine pathway[J]. Nature Chemical Biology, 2020, 16(5): 538-545. |
| 94 | ANTONOVSKY N, GLEIZER S, NOOR E, et al. Sugar synthesis from CO2 in Escherichia coli [J]. Cell, 2016, 166(1): 115-125. |
| 95 | GLEIZER S, BEN-NISSAN R, BAR-ON Y M, et al. Conversion of Escherichia coli to generate all biomass carbon from CO2 [J]. Cell, 2019, 179(6): 1255-1263. |
| 96 | WOOLSTON B M, KING J R, REITER M, et al. Improving formaldehyde consumption drives methanol assimilation in engineered E. coli [J]. Nature Communications, 2018, 9: 2387. |
| 97 | ROHLHILL J, GERALD HAR J R, ANTONIEWICZ M R, et al. Improving synthetic methylotrophy via dynamic formaldehyde regulation of pentose phosphate pathway genes and redox perturbation[J]. Metabolic Engineering, 2020, 57: 247-255. |
| 98 | SIEGEL J B, SMITH A L, POUST S, et al. Computational protein design enables a novel one-carbon assimilation pathway[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(12): 3704-3709. |
| 99 | LU X Y, LIU Y W, YANG Y Q, et al. Constructing a synthetic pathway for acetyl-coenzyme A from one-carbon through enzyme design[J]. Nature Communications, 2019, 10: 1378. |
| 100 | YISHAI O, BOUZON M, DÖRING V, et al. In vivo assimilation of one-carbon via a synthetic reductive glycine pathway in Escherichia coli [J]. ACS Synthetic Biology, 2018, 7(9): 2023-2028. |
| 101 | DÖRING V, DARII E, YISHAI O, et al. Implementation of a reductive route of one-carbon assimilation in Escherichia coli through directed evolution[J]. ACS Synthetic Biology, 2018, 7(9): 2029-2036. |
| 102 | BANG J, LEE S Y. Assimilation of formic acid and CO2 by engineered Escherichia coliequipped with reconstructed one-carbon assimilation pathways[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(40): E9271-E9279. |
| 103 | BANG J, HWANG C H, AHN J H, et al. Escherichia coli is engineered to grow on CO2 and formic acid[J]. Nature Microbiology, 2020, 5(12): 1459-1463. |
| 104 | SCHWANDER T, BORZYSKOWSKI L S VON, BURGENER S, et al. A synthetic pathway for the fixation of carbon dioxide in vitro [J]. Science, 2016, 354(6314): 900-904. |
| 105 | WITTHOFF S, MÜHLROTH A, MARIENHAGEN J, et al. C1 metabolism in Corynebacterium glutamicum: an endogenous pathway for oxidation of methanol to carbon dioxide[J]. Applied and Environmental Microbiology, 2013, 79(22): 6974-6983. |
| 106 | YASOKAWA D, MURATA S, IWAHASHI Y, et al. Toxicity of methanol and formaldehyde towards Saccharomyces cerevisiae as assessed by DNA microarray analysis[J]. Applied Biochemistry and Biotechnology, 2010, 160(6): 1685-1698. |
| 107 | DAI Z X, GU H L, ZHANG S J, et al. Metabolic construction strategies for direct methanol utilization in Saccharomyces cerevisiae [J]. Bioresource Technology, 2017, 245: 1407-1412. |
| 108 | ESPINOSA M I, WILLIAMS T C, PRETORIUS I S, et al. Benchmarking two Saccharomyces cerevisiae laboratory strains for growth and transcriptional response to methanol[J]. Synthetic and Systems Biotechnology, 2019, 4(4): 180-188. |
| 109 | ESPINOSA M I, GONZALEZ-GARCIA R A, VALGEPEA K, et al. Adaptive laboratory evolution of native methanol assimilation in Saccharomyces cerevisiae [J]. Nature Communications, 2020, 11: 5564. |
| 110 | GONZALEZ DE LA CRUZ J, MACHENS F, MESSERSCHMIDT K, et al. Core catalysis of the reductive glycine pathway demonstrated in yeast[J]. ACS Synthetic Biology, 2019, 8(5): 911-917. |
| 111 | 袁姚梦, 邢新会, 张翀. 微生物细胞工厂的设计构建:从诱变育种到全基因组定制化创制[J]. 合成生物学, 2020, 1(6): 656-673. |
| YUAN Y M, XING X H, ZHANG C. Progress and prospective of engineering microbial cell factories: from random mutagenesis to customized design in genome scale[J]. Synthetic Biology Journal, 2020, 1(6): 656-673. | |
| 112 | YANG Y K, LIU G Q, CHEN X, et al. High efficiency CRISPR/Cas9 genome editing system with an eliminable episomal sgRNA plasmid in Pichia pastoris [J]. Enzyme and Microbial Technology, 2020, 138: 109556. |
| 113 | WENINGER A, HATZL A M, SCHMID C, et al. Combinatorial optimization of CRISPR/Cas9 expression enables precision genome engineering in the methylotrophic yeast Pichia pastoris [J]. Journal of Biotechnology, 2016, 235: 139-149. |
| 114 | LIU Q, SHI X N, SONG L L, et al. CRISPR-Cas9-mediated genomic multiloci integration in Pichia pastoris [J]. Microbial Cell Factories, 2019, 18(1): 144. |
| 115 | CAI P, DUAN X P, WU X Y, et al. Recombination machinery engineering facilitates metabolic engineering of the industrial yeast Pichia pastoris [J]. Nucleic Acids Research, 2021, 49(13): 7791-7805. |
| 116 | MO X H, ZHANG H, WANG T M, et al. Establishment of CRISPR interference in Methylorubrum extorquens and application of rapidly mining a new phytoene desaturase involved in carotenoid biosynthesis[J]. Applied Microbiology and Biotechnology, 2020, 104(10): 4515-4532. |
| 117 | ZHU L P, SONG S Z, YANG S. Gene repression using synthetic small regulatory RNA in Methylorubrum extorquens [J]. Journal of Applied Microbiology, 2021, 131(6): 2861-2875. |
| 118 | NOH M, YOO S M, YANG D, et al. Broad-spectrum gene repression using scaffold engineering of synthetic sRNAs[J]. ACS Synthetic Biology, 2019, 8(6): 1452-1461. |
| 119 | HU G P, LI Z H, MA D L, et al. Light-driven CO2 sequestration in Escherichia coli to achieve theoretical yield of chemicals[J]. Nature Catalysis, 2021, 4(5): 395-406. |
| 120 | STINGELE J, SCHWARZ M S, BLOEMEKE N, et al. A DNA-dependent protease involved in DNA-protein crosslink repair[J]. Cell, 2014, 158(2): 327-338. |
| 121 | 张卉, 袁姚梦, 张翀, 等. 合成甲基营养细胞工厂同化甲醇的研究进展及未来展望[J]. 合成生物学, 2021, 2(2): 222-233. |
| ZHANG H, YUAN Y M, ZHANG C, et al. Research progresses and future prospects of synthetic methylotrophic cell factory for methanol assimilation[J]. Synthetic Biology Journal, 2021, 2(2): 222-233. | |
| 122 | HAMMER S K, AVALOS J L. Harnessing yeast organelles for metabolic engineering[J]. Nature Chemical Biology, 2017, 13(8): 823-832. |
| [1] | GUO Shuyuan, ZHANG Qiannan, Gulikezi· MAIMAITIREXIATI, YANG Yiqun, YU Tao. Advances in microbial production of liquid biofuels [J]. Synthetic Biology Journal, 2025, 6(1): 18-44. |
| [2] | SHI Ting, SONG Zhan, SONG Shiyi, ZHANG Yi-Heng P. Job. In vitro BioTransformation (ivBT): a new frontier of industrial biomanufacturing [J]. Synthetic Biology Journal, 2024, 5(6): 1437-1460. |
| [3] | SHAO Mingwei, SUN Simian, YANG Shimao, CHEN Guoqiang. Bioproduction based on extremophiles [J]. Synthetic Biology Journal, 2024, 5(6): 1419-1436. |
| [4] | ZHAO Liang, LI Zhenshuai, FU Liping, LYU Ming, WANG Shi’an, ZHANG Quan, LIU Licheng, LI Fuli, LIU Ziyong. Progress in biomanufacturing of lipids and single cell protein from one-carbon compounds [J]. Synthetic Biology Journal, 2024, 5(6): 1300-1318. |
| [5] | ZHU Fanghuan, CEN Xuecong, CHEN Zhen. Research progress of diols production by microbes [J]. Synthetic Biology Journal, 2024, 5(6): 1367-1385. |
| [6] | YU Wei, GAO Jiaoqi, ZHOU Yongjin. Bioconversion of one carbon feedstocks for producing organic acids [J]. Synthetic Biology Journal, 2024, 5(5): 1169-1188. |
| [7] | CHEN Xiwei, ZHANG Huaran, ZOU Yi. Biosynthesis and metabolic engineering of fungal non-ribosomal peptides [J]. Synthetic Biology Journal, 2024, 5(3): 571-592. |
| [8] | XIE Huang, ZHENG Yilei, SU Yiting, RUAN Jingyi, LI Yongquan. An overview on reconstructing the biosynthetic system of actinomycetes for polyketides production [J]. Synthetic Biology Journal, 2024, 5(3): 612-630. |
| [9] | ZHAO Jingyu, ZHANG Jian, QI Qingsheng, WANG Qian. Research progress in biosensors based on bacterial two-component systems [J]. Synthetic Biology Journal, 2024, 5(1): 38-52. |
| [10] | LIU Weisong, ZHANG Kuncheng, CUI Huijuan, ZHU Zhiguang, ZHANG Yiheng, ZHANG Lingling. Electro-assisted carbon dioxide biotransformation [J]. Synthetic Biology Journal, 2023, 4(6): 1191-1222. |
| [11] | SUN Huili, CUI Jinyu, LUAN Guodong, LYU Xuefeng. Progress of cyanobacterial synthetic biotechnology for efficient light-driven carbon fixation and ethanol production [J]. Synthetic Biology Journal, 2023, 4(6): 1161-1177. |
| [12] | YAN Xiongying, WANG Zhen, LOU Jiyun, ZHANG Haoyu, HUANG Xingyu, WANG Xia, YANG Shihui. Progress in the construction of microbial cell factories for efficient biofuel production [J]. Synthetic Biology Journal, 2023, 4(6): 1082-1121. |
| [13] | MING Yang, CHEN Bin, HUANG Xiaoqiang. Recent advances in photoenzymatic synthesis [J]. Synthetic Biology Journal, 2023, 4(4): 651-675. |
| [14] | CHENG Zhenzhen, ZHANG Jian, GAO Cong, LIU Liming, CHEN Xiulai. Progress in metabolic engineering of microorganisms for the utilization of formate [J]. Synthetic Biology Journal, 2023, 4(4): 756-778. |
| [15] | LIU Jiayu, YANG Zhihan, YANG Lei, ZHU Liying, ZHU Zhengming, JIANG Ling. Advances in the development of Clostridium tyrobutyricum cell factories driven by synthetic biotechnology [J]. Synthetic Biology Journal, 2022, 3(6): 1174-1200. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||