Synthetic Biology Journal ›› 2022, Vol. 3 ›› Issue (5): 932-952.DOI: 10.12211/2096-8280.2021-104
• Invited Review • Previous Articles Next Articles
Challenges and opportunities in the research of Synechococcus chassis under the context of carbon peak and neutrality
TAO Fei1( ), SUN Tao2, WANG Yu3, WEI Ting4, NI Jun1, XU Ping1
), SUN Tao2, WANG Yu3, WEI Ting4, NI Jun1, XU Ping1
- 1.State Key Laboratory of Microbial Metabolism,School of Life Sciences & Biotechnology,Shanghai Jiao Tong University,Shanghai 200240,China
2.Center for Biosafety Research and Strategy,Tianjin University,Tianjin 300072,China
3.Key Laboratory of Systems Microbial Biotechnology,Tianjin Institute of Industrial Biotechnology,Chinese Academy of Sciences,National Center of Technology Innovation for Synthetic Biology,Tianjin 300308,China
4.CAS Key Laboratory for Quantitative Engineering Biology,Shenzhen Institute of Synthetic Biology,Shenzhen Institute of Advanced Technology,Chinese Academy of Sciences,Shenzhen 518055,Guangdong,China
-
Received:2021-11-18Revised:2021-12-22Online:2022-11-16Published:2022-10-31 -
Contact:TAO Fei
“双碳”背景下聚球藻底盘研究的挑战与机遇
陶飞1( ), 孙韬2, 王钰3, 魏婷4, 倪俊1, 许平1
), 孙韬2, 王钰3, 魏婷4, 倪俊1, 许平1
- 1.上海交通大学生命科学技术学院,微生物代谢国家重点实验室,上海 200240
2.天津大学生物安全战略研究中心,天津 300072
3.中国科学院系统微生物工程重点实验室,中国科学院天津工业生物技术研究所,国家合成生物技术创新中心,天津 300308
4.中国科学院深圳先进技术研究院,深圳合成生物学创新研究院,中国科学院定量工程生物学重点实验室,广东 深圳 518055
-
通讯作者:陶飞 -
作者简介:陶飞 (1983—),男,研究员,博士生导师。主要从事微生物合成生物学研究,方向包括光驱动细胞工厂、智能代谢重编程、生物传感等。 E-mail:taofei@sjtu.edu.cn -
基金资助:国家重点研发计划(2018YFA0903600)
CLC Number:
Cite this article
TAO Fei, SUN Tao, WANG Yu, WEI Ting, NI Jun, XU Ping. Challenges and opportunities in the research of Synechococcus chassis under the context of carbon peak and neutrality[J]. Synthetic Biology Journal, 2022, 3(5): 932-952.
陶飞, 孙韬, 王钰, 魏婷, 倪俊, 许平. “双碳”背景下聚球藻底盘研究的挑战与机遇[J]. 合成生物学, 2022, 3(5): 932-952.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2021-104
| Strain | Type | Engineered CRISPR system | Application | Ref. |
|---|---|---|---|---|
| Synechococcus elongatus PCC 7942 | Cas9 | One plasmid pCas9-NSI expresses Streptococcus pyogenes Cas9, tracrRNA and crRNA under the native promoter | Simultaneous glgc knock-out and gltA/ppc knock-in with a 100% efficiency after 3 passages of antibiotic selection | [ |
| One plasmid harbors the editing template | ||||
| Cas12a | One plasmid pSL2680 expresses Francisella novicida Cas12a under a lac promoter and an endogenous CRISPR array under a J23119 promoter | rpaA, atpA, and ppnK point mutations, no efficiency data were provided | [ | |
| Synechococcus elongatus UTEX 2973 | Cas9 | One plasmid pSL2546 expresses S. pyogenes Cas9 under a rpsL(XC) promoter and tracrRNA and sgRNA under a gapdhp(EL) promoter and harbors the editing template | nblA deletion with a 100% efficiency in the first patch | [ |
| Cas12a | One plasmid pSL2680 expresses F. novicida Cas12a under a lac promoter and an endogenous CRISPR array under a J23119 promoter | psbA S264A point mutation with 75% efficiency, eyfp knock-in with a 60% efficiency, and nblA deletion with a 90% efficiency after 3~4 passages of antibiotic selection | [ |
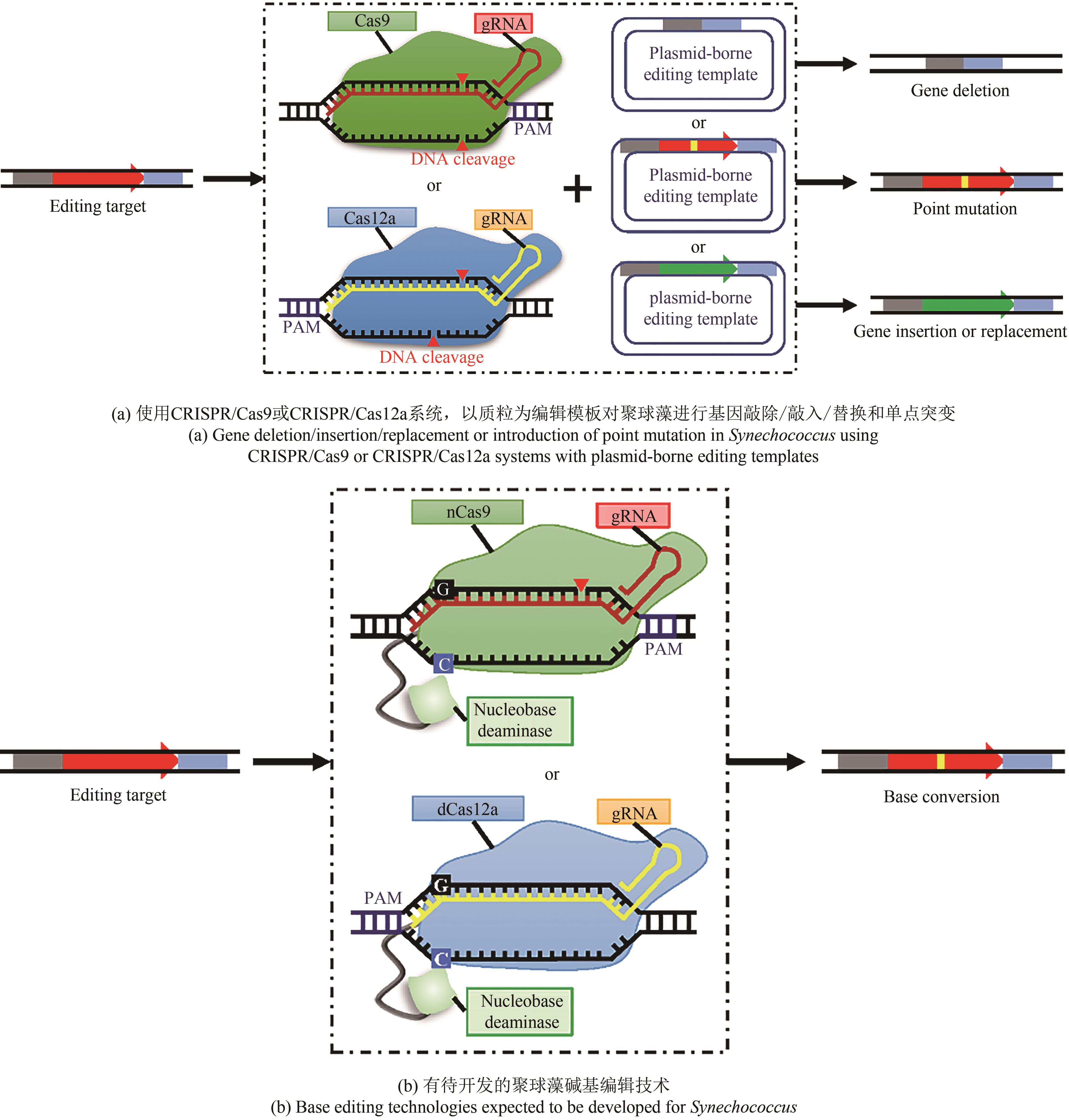
Tab. 1 CRISPR-based genome editing technologies for Synechococcus
| Strain | Type | Engineered CRISPR system | Application | Ref. |
|---|---|---|---|---|
| Synechococcus elongatus PCC 7942 | Cas9 | One plasmid pCas9-NSI expresses Streptococcus pyogenes Cas9, tracrRNA and crRNA under the native promoter | Simultaneous glgc knock-out and gltA/ppc knock-in with a 100% efficiency after 3 passages of antibiotic selection | [ |
| One plasmid harbors the editing template | ||||
| Cas12a | One plasmid pSL2680 expresses Francisella novicida Cas12a under a lac promoter and an endogenous CRISPR array under a J23119 promoter | rpaA, atpA, and ppnK point mutations, no efficiency data were provided | [ | |
| Synechococcus elongatus UTEX 2973 | Cas9 | One plasmid pSL2546 expresses S. pyogenes Cas9 under a rpsL(XC) promoter and tracrRNA and sgRNA under a gapdhp(EL) promoter and harbors the editing template | nblA deletion with a 100% efficiency in the first patch | [ |
| Cas12a | One plasmid pSL2680 expresses F. novicida Cas12a under a lac promoter and an endogenous CRISPR array under a J23119 promoter | psbA S264A point mutation with 75% efficiency, eyfp knock-in with a 60% efficiency, and nblA deletion with a 90% efficiency after 3~4 passages of antibiotic selection | [ |

Fig. 3 Schematic design of phage-assisted continuous evolution (PACE) MP—mutagenesis plasmid; AP—accessory plasmid; SP—selection phage or its genome inside the infected host cell; POI—protein of interest to be evolved
| 产物 | 产量 | 宿主 | 参考文献 |
|---|---|---|---|
| 2, 3-丁二醇 | 2.38 g/L | S. elongatus PCC 7942 | [ |
| 异丁醇 | 550 mg/L | S. elongatus PCC 7942 | [ |
| 3-羟基丙酸 | 659 mg/L | S. elongatus PCC 7942 | [ |
| D-乳酸 | 798 mg/L | S. elongatus PCC 7942 | [ |
| 甘油 | 1.24 g/L | S. elongatus PCC 7942 | [ |
| β-聚羟基丁酸 | 420 mg/L | S. elongatus UTEX2973 | [ |
| 咖啡酸 | 4.7 mg/L | S. elongatus PCC 7942 | [ |
| 对香豆酸 | 128.2 mg/L | S. elongatus PCC 7942 | [ |
| 阿魏酸 | 6.3 mg/L | S. elongatus PCC 7942 | [ |
| 柚皮素 | 4.6 mg/L | S. elongatus PCC 7942 | [ |
| 白藜芦醇 | 7.1 mg/L | S. elongatus PCC 7942 | [ |
| 双去甲氧基姜黄素 | 4.1 mg/L | S. elongatus PCC 7942 | [ |
| 异戊二烯 | 60 mg/(L·d) | S. elongatus PCC 7942 | [ |
| 柠檬烯 | 50 μg/(L·h) | Synechococcus sp. PCC 7002 | [ |
| 红没药烯 | 7.5 μg/(L·h) | Synechococcus sp. PCC 7002 | [ |
| 乙烯 | 512 μg/(L·OD·h) | S. elongatus PCC 7942 | [ |
| 甘露醇 | 0.63 g/(L·d) | Synechococcus sp. PCC 7002 | [ |
Tab. 2 Chemical production using Synechococcus strain as chassis
| 产物 | 产量 | 宿主 | 参考文献 |
|---|---|---|---|
| 2, 3-丁二醇 | 2.38 g/L | S. elongatus PCC 7942 | [ |
| 异丁醇 | 550 mg/L | S. elongatus PCC 7942 | [ |
| 3-羟基丙酸 | 659 mg/L | S. elongatus PCC 7942 | [ |
| D-乳酸 | 798 mg/L | S. elongatus PCC 7942 | [ |
| 甘油 | 1.24 g/L | S. elongatus PCC 7942 | [ |
| β-聚羟基丁酸 | 420 mg/L | S. elongatus UTEX2973 | [ |
| 咖啡酸 | 4.7 mg/L | S. elongatus PCC 7942 | [ |
| 对香豆酸 | 128.2 mg/L | S. elongatus PCC 7942 | [ |
| 阿魏酸 | 6.3 mg/L | S. elongatus PCC 7942 | [ |
| 柚皮素 | 4.6 mg/L | S. elongatus PCC 7942 | [ |
| 白藜芦醇 | 7.1 mg/L | S. elongatus PCC 7942 | [ |
| 双去甲氧基姜黄素 | 4.1 mg/L | S. elongatus PCC 7942 | [ |
| 异戊二烯 | 60 mg/(L·d) | S. elongatus PCC 7942 | [ |
| 柠檬烯 | 50 μg/(L·h) | Synechococcus sp. PCC 7002 | [ |
| 红没药烯 | 7.5 μg/(L·h) | Synechococcus sp. PCC 7002 | [ |
| 乙烯 | 512 μg/(L·OD·h) | S. elongatus PCC 7942 | [ |
| 甘露醇 | 0.63 g/(L·d) | Synechococcus sp. PCC 7002 | [ |
| 1 | PALINKAS L A, WONG M. Global climate change and mental health[J]. Current Opinion in Psychology, 2020, 32: 12-16. |
| 2 | 丁仲礼. 中国碳中和框架路线图研究[J]. 中国工业和信息化, 2021(8): 54-61. |
| DING Z L. Research on the roadmap and framework of China's carbon neutral[J]. China Industry & Information Technology, 2021(8): 54-61. | |
| 3 | 王永中. 碳达峰、碳中和目标与中国的新能源革命[J]. 人民论坛⋅学术前沿, 2021(14): 88-96. |
| WANG Y Z. The targets of carbon peak and carbon neutralization and China's new energy revolution[J]. Frontiers, 2021(14): 88-96. | |
| 4 | 胡鞍钢. 中国实现2030年前碳达峰目标及主要途径[J]. 北京工业大学学报(社会科学版), 2021, 21(3): 1-15. |
| HU A G. China's goal of achieving carbon peak by 2030 and its main approaches[J]. Journal of Beijing University of Technology (Social Sciences Edition), 2021, 21(3): 1-15. | |
| 5 | 王灿, 张雅欣. 碳中和愿景的实现路径与政策体系[J]. 中国环境管理, 2020, 12(6): 58-64. |
| WANG C, ZHANG Y X. Implementation pathway and policy system of carbon neutrality vision[J]. Chinese Journal of Environmental Management, 2020, 12(6): 58-64. | |
| 6 | FUCHSMAN C A, PALEVSKY H I, WIDNER B, et al. Cyanobacteria and cyanophage contributions to carbon and nitrogen cycling in an oligotrophic oxygen-deficient zone[J]. The ISME Journal, 2019, 13(11): 2714-2726. |
| 7 | SÁNCHEZ-BARACALDO P, BIANCHINI G, WILSON J D, et al. Cyanobacteria and biogeochemical cycles through Earth history[J]. Trends in Microbiology, 2022, 30(2): 143-157. |
| 8 | AL-HAJ L, LUI Y T, ABED R M M, et al. Cyanobacteria as chassis for industrial biotechnology: progress and prospects[J]. Life, 2016, 6(4): 42. |
| 9 | XU X M, RISOUL V, BYRNE D, et al. HetL, HetR and PatS form a reaction-diffusion system to control pattern formation in the cyanobacterium Nostoc PCC 7120[J]. eLife, 2020, 9: e59190. |
| 10 | KIENINGER A K, MALDENER I. Cell-cell communication through septal junctions in filamentous cyanobacteria[J]. Current Opinion in Microbiology, 2021, 61: 35-41. |
| 11 | ZHAO L S, HUOKKO T, WILSON S, et al. Structural variability, coordination and adaptation of a native photosynthetic machinery[J]. Nature Plants, 2020, 6(7): 869-882. |
| 12 | PEMBROKE J T, RYAN M P. Cyanobacterial biofuel production: current development, challenges and future needs[M]// Biofuel and biorefinery technologies. Cham: Springer International Publishing, 2020: 35-62. |
| 13 | KOCH M, BRUCKMOSER J, SCHOLL J, et al. Maximizing PHB content in Synechocystis sp. PCC 6803: a new metabolic engineering strategy based on the regulator PirC[J]. Microbial Cell Factories, 2020, 19(1): 231. |
| 14 | CASE A E, ATSUMI S. Cyanobacterial chemical production[J]. Journal of Biotechnology, 2016, 231: 106-114. |
| 15 | WANG F, GAO Y Y, YANG G. Recent advances in synthetic biology of cyanobacteria for improved chemicals production[J]. Bioengineered, 2020, 11(1): 1208-1220. |
| 16 | ZAHRA Z, CHOO D H, LEE H, et al. Cyanobacteria: review of current potentials and applications[J]. Environments, 2020, 7(2): 13. |
| 17 | LI C, TAO F, XU P. Carbon flux trapping: highly efficient production of polymer-grade D-lactic acid with a thermophilic D-lactate dehydrogenase[J]. ChemBioChem: a European Journal of Chemical Biology, 2016, 17(16): 1491-1494. |
| 18 | LI C, TAO F, NI J, et al. Enhancing the light-driven production of D-lactate by engineering cyanobacterium using a combinational strategy[J]. Scientific Reports, 2015, 5: 9777. |
| 19 | ZHU Z, JIANG J H, FA Y. Overcoming the biological contamination in microalgae and cyanobacteria mass cultivations for photosynthetic biofuel production[J]. Molecules, 2020, 25(22): 5220. |
| 20 | MAZARD S, PENESYAN A, OSTROWSKI M, et al. Tiny microbes with a big impact: the role of cyanobacteria and their metabolites in shaping our future[J]. Marine Drugs, 2016, 14(5): 97. |
| 21 | ONGENA J, OOST G V. Energy for future centuries: prospects for fusion power as a future energy source[J]. Fusion Science and Technology, 2012, 61(2T): 3-16. |
| 22 | KWOK S. Abiotic synthesis of complex organics in the Universe[J]. Nature Astronomy, 2017, 1(10): 642-643. |
| 23 | WOODLEY J M. Towards the sustainable production of bulk-chemicals using biotechnology[J]. New Biotechnology, 2020, 59: 59-64. |
| 24 | VAVITSAS K, KUGLER A, SATTA A, et al. Doing synthetic biology with photosynthetic microorganisms[J]. Physiologia Plantarum, 2021, 173(2): 624-638. |
| 25 | CHAUVAT F, CASSIER-CHAUVAT C. Chapter Six - Genomics of cyanobacteria: new insights and lessons for shaping our future-A follow-up of volume 65: genomics of cyanobacteria[M] Advances in botanical research (Volume 100), 2021: 213-235. |
| 26 | LEWIS N S, NOCERA D G. Powering the planet: chemical challenges in solar energy utilization[J]. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(43): 15729-15735. |
| 27 | AHMED S D, AL-ISMAIL F S M, SHAFIULLAH M, et al. Grid integration challenges of wind energy: a review[J]. IEEE Access, 8: 10857-10878. |
| 28 | OBI M, BASS R. Trends and challenges of grid-connected photovoltaic systems - a review[J]. Renewable and Sustainable Energy Reviews, 2016, 58: 1082-1094. |
| 29 | CHEN R S, CHENG Y F, HAN S Y, et al. Whole genome sequencing and comparative transcriptome analysis of a novel seawater adapted, salt-resistant rice cultivar-sea rice 86[J]. BMC Genomics, 2017, 18(1): 655. |
| 30 | MELANDRI G, ABDELGAWAD H, FLOKOVÁ K, et al. Drought tolerance in selected aerobic and upland rice varieties is driven by different metabolic and antioxidative responses[J]. Planta, 2021, 254(1): 13. |
| 31 | HUANG Q S, JIANG F H, WANG L Z, et al. Design of photobioreactors for mass cultivation of photosynthetic organisms[J]. Engineering, 2017, 3(3): 318-329. |
| 32 | CASSIER-CHAUVAT C, DIVE V, CHAUVAT F. Cyanobacteria: photosynthetic factories combining biodiversity, radiation resistance, and genetics to facilitate drug discovery[J]. Applied Microbiology and Biotechnology, 2017, 101(4): 1359-1364. |
| 33 | VASILE N S, CORDARA A, USAI G, et al. Computational analysis of dynamic light exposure of unicellular algal cells in a flat-panel photobioreactor to support light-induced CO2 bioprocess development[J]. Frontiers in Microbiology, 2021, 12: 639482. |
| 34 | SAWA M, FANTUZZI A, BOMBELLI P, et al. Electricity generation from digitally printed cyanobacteria[J]. Nature Communications, 2017, 8: 1327. |
| 35 | SCHUERGERS N, WERLANG C, AJO-FRANKLIN C M, et al. A synthetic biology approach to engineering living photovoltaics[J]. Energy & Environmental Science, 2017, 10(5): 1102-1115. |
| 36 | QIAO Y, JIANG K Z, DENG H, et al. A high-energy-density and long-life lithium-ion battery via reversible oxide-peroxide conversion[J]. Nature Catalysis, 2019, 2(11): 1035-1044. |
| 37 | LIU X F, MIAO R, LINDBERG P, et al. Modular engineering for efficient photosynthetic biosynthesis of 1-butanol from CO2 in cyanobacteria[J]. Energy & Environmental Science, 2019, 12(9): 2765-2777. |
| 38 | ALDRIDGE S. Industry backs biocatalysis for greener manufacturing[J]. Nature Biotechnology, 2013, 31(2): 95-96. |
| 39 | GAO X, GAO F, LIU D, et al. Engineering the methylerythritol phosphate pathway in cyanobacteria for photosynthetic isoprene production from CO2 [J]. Energy & Environmental Science, 2016, 9(4): 1400-1411. |
| 40 | ZHU T, XIE X M, LI Z M, et al. Enhancing photosynthetic production of ethylene in genetically engineered Synechocystis sp. PCC 6803[J]. Green Chemistry, 2015, 17(1): 421-434. |
| 41 | WANG Y, TAO F, NI J, et al. Production of C3 platform chemicals from CO2 by genetically engineered cyanobacteria[J]. Green Chemistry, 2015, 17(5): 3100-3110. |
| 42 | NI J, TAO F, WANG Y, et al. A photoautotrophic platform for the sustainable production of valuable plant natural products from CO2 [J]. Green Chemistry, 2016, 18(12): 3537-3548. |
| 43 | NI J, LIU H Y, TAO F, et al. Remodeling of the photosynthetic chain promotes direct CO2 conversion into valuable aromatic compounds[J]. Angewandte Chemie International Edition, 2018, 57(49): 15990-15994. |
| 44 | LIU D, LIBERTON M, HENDRY J I, et al. Engineering biology approaches for food and nutrient production by cyanobacteria[J]. Current Opinion in Biotechnology, 2021, 67: 1-6. |
| 45 | ATAEIAN M, LIU Y H, CANON-RUBIO K A, et al. Direct capture and conversion of CO2 from air by growing a cyanobacterial consortium at pH up to 11.2[J]. Biotechnology and Bioengineering, 2019, 116(7): 1604-1611. |
| 46 | UPENDAR G, SINGH S, CHAKRABARTY J, et al. Sequestration of carbon dioxide and production of biomolecules using cyanobacteria[J]. Journal of Environmental Management, 2018, 218: 234-244. |
| 47 | KUMAR K, DASGUPTA C N, NAYAK B, et al. Development of suitable photobioreactors for CO2 sequestration addressing global warming using green algae and cyanobacteria[J]. Bioresource Technology, 2011, 102(8): 4945-4953. |
| 48 | MUÑOZ-ROJAS M, ROMÁN J R, RONCERO-RAMOS B, et al. Cyanobacteria inoculation enhances carbon sequestration in soil substrates used in dryland restoration[J]. Science of the Total Environment, 2018, 636: 1149-1154. |
| 49 | PANDIT S, NAYAK B K, DAS D. Microbial carbon capture cell using cyanobacteria for simultaneous power generation, carbon dioxide sequestration and wastewater treatment[J]. Bioresource Technology, 2012, 107: 97-102. |
| 50 | BURNAP R L. Systems and photosystems: cellular limits of autotrophic productivity in cyanobacteria[J]. Frontiers in Bioengineering and Biotechnology, 2015, 3: 1. |
| 51 | AHMAD M AL, NATOUR Z AL, ATTOUB S, et al. Monitoring of the budding yeast cell cycle using electrical parameters[J]. IEEE Access, 6: 19231-19237. |
| 52 | GIBSON B, WILSON D J, FEIL E, et al. The distribution of bacterial doubling times in the wild[J]. Proceedings Biological Sciences, 2018, 285(1880): 20180789. |
| 53 | O'DONNELL M, LANGSTON L, STILLMAN B. Principles and concepts of DNA replication in bacteria, archaea, and eukarya[J]. Cold Spring Harbor Perspectives in Biology, 2013, 5(7): a010108. |
| 54 | FOSSUM S, CROOKE E, SKARSTAD K. Organization of sister origins and replisomes during multifork DNA replication in Escherichia coli [J]. The EMBO Journal, 2007, 26(21): 4514-4522. |
| 55 | WŁODARCZYK A, SELÃO T T, NORLING B, et al. Newly discovered Synechococcus sp. PCC 11901 is a robust cyanobacterial strain for high biomass production[J]. Communications Biology, 2020, 3: 215. |
| 56 | YU J J, LIBERTON M, CLIFTEN P F, et al. Synechococcus elongatus UTEX 2973, a fast growing cyanobacterial chassis for biosynthesis using light and CO2 [J]. Scientific Reports, 2015, 5: 8132. |
| 57 | UNGERER J, LIN P C, CHEN H Y, et al. Adjustments to photosystem stoichiometry and electron transfer proteins are key to the remarkably fast growth of the cyanobacterium Synechococcus elongatus UTEX 2973[J]. mBio, 2018, 9(1): e02327-e02317. |
| 58 | JAHN M, VIALAS V, KARLSEN J, et al. Growth of cyanobacteria is constrained by the abundance of light and carbon assimilation proteins[J]. Cell Reports, 2018, 25(2): 478-486.e8. |
| 59 | GEISZ J F, FRANCE R M, SCHULTE K L, et al. Six-junction III-V solar cells with 47.1% conversion efficiency under 143 Suns concentration[J]. Nature Energy, 2020, 5(4): 326-335. |
| 60 | GAN F, ZHANG S Y, ROCKWELL N C, et al. Extensive remodeling of a cyanobacterial photosynthetic apparatus in far-red light[J]. Science, 2014, 345(6202): 1312-1317. |
| 61 | TEODOR A H, BRUCE B D. Putting photosystem I to work: truly green energy[J]. Trends in Biotechnology, 2020, 38(12): 1329-1342. |
| 62 | FANG X, KALATHIL S, REISNER E. Semi-biological approaches to solar-to-chemical conversion[J]. Chemical Society Reviews, 2020, 49(14): 4926-4952. |
| 63 | LIPS D, SCHUURMANS J M, BRANCO DOS SANTOS F, et al. Many ways towards 'solar fuel': quantitative analysis of the most promising strategies and the main challenges during scale-up[J]. Energy & Environmental Science, 2018, 11(1): 10-22. |
| 64 | KIRST H, FORMIGHIERI C, MELIS A. Maximizing photosynthetic efficiency and culture productivity in cyanobacteria upon minimizing the phycobilisome light-harvesting antenna size[J]. Biochimica et Biophysica Acta (BBA)-Bioenergetics, 2014, 1837(10): 1653-1664. |
| 65 | HIROSE Y, SONG C H, WATANABE M, et al. Diverse chromatic acclimation processes regulating phycoerythrocyanin and rod-shaped phycobilisome in cyanobacteria[J]. Molecular Plant, 2019, 12(8): 1167-1169. |
| 66 | LUIMSTRA V M, SCHUURMANS J M, VERSCHOOR A M, et al. Blue light reduces photosynthetic efficiency of cyanobacteria through an imbalance between photosystems I and II[J]. Photosynthesis Research, 2018, 138(2): 177-189. |
| 67 | GONG F Y, ZHU H W, ZHANG Y P, et al. Biological carbon fixation: from natural to synthetic[J]. Journal of CO2 Utilization, 2018, 28: 221-227. |
| 68 | LIANG F Y, LINDBERG P, LINDBLAD P. Engineering photoautotrophic carbon fixation for enhanced growth and productivity[J]. Sustainable Energy & Fuels, 2018, 2(12): 2583-2600. |
| 69 | BARATI B, ZENG K, BAEYENS J, et al. Recent progress in genetically modified microalgae for enhanced carbon dioxide sequestration[J]. Biomass and Bioenergy, 2021, 145: 105927. |
| 70 | YU H, LI X Q, DUCHOUD F, et al. Augmenting the Calvin-Benson-Bassham cycle by a synthetic malyl-CoA-glycerate carbon fixation pathway[J]. Nature Communications, 2018, 9: 2008. |
| 71 | WEI L, SHEN C, HAJJAMI M EL, et al. Knockdown of carbonate anhydrase elevates Nannochloropsis productivity at high CO2 level[J]. Metabolic Engineering, 2019, 54: 96-108. |
| 72 | CLARK R L, GORDON G C, BENNETT N R, et al. High-CO2 requirement as a mechanism for the containment of genetically modified cyanobacteria[J]. ACS Synthetic Biology, 2018, 7(2): 384-391. |
| 73 | TUSCHHOFF E J, HUTTER C R, GLOR R E. Improving sustainable use of genetic resources in biodiversity archives[J]. PeerJ, 2020, 8: e8369. |
| 74 | GRÉBERT T, DORÉ H, PARTENSKY F, et al. Light color acclimation is a key process in the global ocean distribution of Synechococcus cyanobacteria [J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(9): E2010-E2019. |
| 75 | XIA X M, LIU H B, CHOI D, et al. Variation of Synechococcus pigment genetic diversity along two turbidity gradients in the China seas[J]. Microbial Ecology, 2018, 75(1): 10-21. |
| 76 | CALLIERI C. Synechococcus plasticity under environmental changes[J]. FEMS Microbiology Letters, 2017, 364(23): fnx229. |
| 77 | PATTHARAPRACHAYAKUL N, LEE M, INCHAROENSAKDI A, et al. Current understanding of the cyanobacterial CRISPR-Cas systems and development of the synthetic CRISPR-Cas systems for cyanobacteria[J]. Enzyme and Microbial Technology, 2020, 140: 109619. |
| 78 | BEHLER J, VIJAY D, HESS W R, et al. CRISPR-based technologies for metabolic engineering in cyanobacteria[J]. Trends in Biotechnology, 2018, 36(10): 996-1010. |
| 79 | 李洋, 申晓林, 孙新晓, 等. CRISPR基因编辑技术在微生物合成生物学领域的研究进展[J]. 合成生物学, 2021, 2(1): 106-120. |
| LI Y, SHEN X L, SUN X X, et al. Advances of CRISPR gene editing in microbial synthetic biology[J]. Synthetic Biology Journal, 2021, 2(1): 106-120. | |
| 80 | LI H, SHEN C R, HUANG C H, et al. CRISPR-Cas9 for the genome engineering of cyanobacteria and succinate production[J]. Metabolic Engineering, 2016, 38: 293-302. |
| 81 | UNGERER J, WENDT K E, HENDRY J I, et al. Comparative genomics reveals the molecular determinants of rapid growth of the cyanobacterium Synechococcus elongatus UTEX 2973[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(50): E11761-E11770. |
| 82 | WENDT K E, UNGERER J, COBB R E, et al. CRISPR/Cas9 mediated targeted mutagenesis of the fast growing cyanobacterium Synechococcus elongatus UTEX 2973[J]. Microbial Cell Factories, 2016, 15(1): 115. |
| 83 | UNGERER J, PAKRASI H B. Cpf1 is a versatile tool for CRISPR genome editing across diverse species of cyanobacteria[J]. Scientific Reports, 2016, 6: 39681. |
| 84 | SUN T, LI S B, SONG X Y, et al. Toolboxes for cyanobacteria: recent advances and future direction[J]. Biotechnology Advances, 2018, 36(4): 1293-1307. |
| 85 | LIU J, WANG Y, LU Y J, et al. Development of a CRISPR/Cas9 genome editing toolbox for Corynebacterium glutamicum [J]. Microbial Cell Factories, 2017, 16(1): 205. |
| 86 | KOMOR A C, KIM Y B, PACKER M S, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage[J]. Nature, 2016, 533(7603): 420-424. |
| 87 | NISHIDA K, ARAZOE T, YACHIE N, et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems[J]. Science, 2016, 353(6305): aaf8729. |
| 88 | ZHAO D D, LI J, LI S W, et al. Glycosylase base editors enable C-to-A and C-to-G base changes[J]. Nature Biotechnology, 2021, 39(1): 35-40. |
| 89 | GAUDELLI N M, KOMOR A C, REES H A, et al. Programmable base editing of A·T to G·C in genomic DNA without DNA cleavage[J]. Nature, 2017, 551(7681): 464-471. |
| 90 | KURT I C, ZHOU R H, IYER S, et al. CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells[J]. Nature Biotechnology, 2021, 39(1): 41-46. |
| 91 | WANG Y, LIU Y, ZHENG P, et al. Microbial base editing: a powerful emerging technology for microbial genome engineering[J]. Trends in Biotechnology, 2021, 39(2): 165-180. |
| 92 | WANG Y, LIU Y, LI J W, et al. Expanding targeting scope, editing window, and base transition capability of base editing in Corynebacterium glutamicum [J]. Biotechnology and Bioengineering, 2019, 116(11): 3016-3029. |
| 93 | WANG Y, LIU Y, LIU J, et al. MACBETH: multiplex automated Corynebacterium glutamicum base editing method[J]. Metabolic Engineering, 2018, 47: 200-210. |
| 94 | WANG Y, CHENG H J, LIU Y, et al. In-situ generation of large numbers of genetic combinations for metabolic reprogramming via CRISPR-guided base editing[J]. Nature Communications, 2021, 12: 678. |
| 95 | HUANG C H, SHEN C R, LI H, et al. CRISPR interference (CRISPRi) for gene regulation and succinate production in cyanobacterium S. elongatus PCC 7942[J]. Microbial Cell Factories, 2016, 15(1): 196. |
| 96 | KNOOT C J, BISWAS S, PAKRASI H B. Tunable repression of key photosynthetic processes using Cas12a CRISPR interference in the fast-growing cyanobacterium Synechococcus sp. UTEX 2973[J]. ACS Synthetic Biology, 2020, 9(1): 132-143. |
| 97 | CHOI S Y, WOO H M. CRISPRi-dCas12a: a dCas12a-mediated CRISPR interference for repression of multiple genes and metabolic engineering in cyanobacteria[J]. ACS Synthetic Biology, 2020, 9(9): 2351-2361. |
| 98 | LIU Y, WAN X Y, WANG B J. Engineered CRISPRa enables programmable eukaryote-like gene activation in bacteria[J]. Nature Communications, 2019, 10: 3693. |
| 99 | YAO L, SHABESTARY K, BJÖRK S M, et al. Pooled CRISPRi screening of the cyanobacterium Synechocystis sp PCC 6803 for enhanced industrial phenotypes[J]. Nature Communications, 2020, 11: 1666. |
| 100 | SWANSON W J. Adaptive evolution of genes and gene families[J]. Current Opinion in Genetics & Development, 2003, 13(6): 617-622. |
| 101 | BILLIS K, BILLINI M, TRIPP H J, et al. Comparative transcriptomics between Synechococcus PCC 7942 and Synechocystis PCC 6803 provide insights into mechanisms of stress acclimation[J]. PLoS One, 2014, 9(10): e109738. |
| 102 | DANN M, ORTIZ E M, THOMAS M, et al. Enhancing photosynthesis at high light levels by adaptive laboratory evolution[J]. Nature Plants, 2021, 7(5): 681-695. |
| 103 | YOSHIKAWA K, OGAWA K, TOYA Y, et al. Mutations in hik26 and slr1916 lead to high-light stress tolerance in Synechocystis sp. PCC6803[J]. Communications Biology, 2021, 4: 343. |
| 104 | SRIVASTAVA V, AMANNA R, ROWDEN S J L, et al. Adaptive laboratory evolution of the fast-growing cyanobacterium Synechococcus elongatus PCC 11801 for improved solvent tolerance[J]. Journal of Bioscience and Bioengineering, 2021, 131(5): 491-500. |
| 105 | WANG Y X, SHI M L, NIU X F, et al. Metabolomic basis of laboratory evolution of butanol tolerance in photosynthetic Synechocystis sp. PCC 6803[J]. Microbial Cell Factories, 2014, 13: 151. |
| 106 | XU C X, SUN T, LI S B, et al. Adaptive laboratory evolution of cadmium tolerance in Synechocystis sp. PCC 6803[J]. Biotechnology for Biofuels, 2018, 11: 205. |
| 107 | TILLICH U M, WOLTER N, FRANKE P, et al. Screening and genetic characterization of thermo-tolerant Synechocystis sp. PCC6803 strains created by adaptive evolution[J]. BMC Biotechnology, 2014, 14: 66. |
| 108 | HU L, HE J Y, DONG M J, et al. Divergent metabolic and transcriptomic responses of Synechocystis sp. PCC 6803 to salt stress after adaptive laboratory evolution[J]. Algal Research, 2020, 47: 101856. |
| 109 | UCHIYAMA J, KANESAKI Y, IWATA N, et al. Genomic analysis of parallel-evolved cyanobacterium Synechocystis sp. PCC 6803 under acid stress[J]. Photosynthesis Research, 2015, 125(1/2): 243-254. |
| 110 | DURÃO P, AIGNER H, NAGY P, et al. Opposing effects of folding and assembly chaperones on evolvability of Rubisco[J]. Nature Chemical Biology, 2015, 11(2): 148-155. |
| 111 | PARIKH M R, GREENE D N, WOODS K K, et al. Directed evolution of RuBisCO hypermorphs through genetic selection in engineered E. coli [J]. Protein Engineering, Design and Selection, 2006, 19(3): 113-119. |
| 112 | BAI S Y, WALLIS J G, DENOLF P, et al. Directed evolution increases desaturation of a cyanobacterial fatty acid desaturase in eukaryotic expression systems[J]. Biotechnology and Bioengineering, 2016, 113(7): 1522-1530. |
| 113 | CARLSON J C, BADRAN A H, GUGGIANA-NILO D A, et al. Negative selection and stringency modulation in phage-assisted continuous evolution[J]. Nature Chemical Biology, 2014, 10(3): 216-222. |
| 114 | ESVELT K M, CARLSON J C, LIU D R. A system for the continuous directed evolution of biomolecules[J]. Nature, 2011, 472(7344): 499-503. |
| 115 | MILLER S M, WANG T N, LIU D R. Phage-assisted continuous and non-continuous evolution[J]. Nature Protocols, 2020, 15(12): 4101-4127. |
| 116 | WU H, WEI T, YU B B, et al. A single mutation attenuates both the transcription termination and RNA-dependent RNA polymerase activity of T7 RNA polymerase[J]. RNA Biology, 2021, 18(sup1): 451-466. |
| 117 | JOHNSTON C W, BADRAN A H, COLLINS J J. Continuous bioactivity-dependent evolution of an antibiotic biosynthetic pathway[J]. Nature Communications, 2020, 11: 4202. |
| 118 | MORRISON M S, PODRACKY C J, LIU D R. The developing toolkit of continuous directed evolution[J]. Nature Chemical Biology, 2020, 16(6): 610-619. |
| 119 | LAN E I, LIAO J C. ATP drives direct photosynthetic production of 1-butanol in cyanobacteria[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(16): 6018-6023. |
| 120 | ATSUMI S, HIGASHIDE W, LIAO J C. Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde[J]. Nature Biotechnology, 2009, 27(12): 1177-1180. |
| 121 | HIROKAWA Y, GOTO R, UMETANI Y, et al. Construction of a novel D-lactate producing pathway from dihydroxyacetone phosphate of the Calvin cycle in cyanobacterium, Synechococcus elongatus PCC 7942[J]. Journal of Bioscience and Bioengineering, 2017, 124(1): 54-61. |
| 122 | KOBAYASHI I, WATANABE S, KANESAKI Y, et al. Conserved two-component Hik34-Rre1 module directly activates heat-stress inducible transcription of major chaperone and other genes in Synechococcus elongatus PCC 7942[J]. Molecular Microbiology, 2017, 104(2): 260-277. |
| 123 | YASUDA A, INAMI D, RpaB HANAOKA M., an essential response regulator for high-light stress, is extensively involved in transcriptional regulation under light-intensity upshift conditions in Synechococcus elongatus PCC 7942[J]. The Journal of General and Applied Microbiology, 2020, 66(2): 73-79. |
| 124 | GUYET U, NGUYEN N A, DORÉ H, et al. Synergic effects of temperature and irradiance on the physiology of the marine Synechococcus strain WH7803[J]. Frontiers in Microbiology, 2020, 11: 1707. |
| 125 | MUELLER T J, UNGERER J L, PAKRASI H B, et al. Identifying the metabolic differences of a fast-growth phenotype in Synechococcus UTEX 2973[J]. Scientific Reports, 2017, 7: 41569. |
| 126 | TAN X M, HOU S W, SONG K, et al. The primary transcriptome of the fast-growing cyanobacterium Synechococcus elongatus UTEX 2973[J]. Biotechnology for Biofuels, 2018, 11: 218. |
| 127 | JAISWAL D, SENGUPTA A, SENGUPTA S, et al. A novel cyanobacterium Synechococcus elongatus PCC 11802 has distinct genomic and metabolomic characteristics compared to its neighbor PCC 11801[J]. Scientific Reports, 2020, 10: 191. |
| 128 | CUI J Y, SUN T, LI S B, et al. Improved salt tolerance and metabolomics analysis of Synechococcus elongatus UTEX 2973 by overexpressing Mrp antiporters[J]. Frontiers in Bioengineering and Biotechnology, 2020, 8: 500. |
| 129 | CUI J Y, SUN T, CHEN L, et al. Salt-tolerant Synechococcus elongatus UTEX 2973 obtained via engineering of heterologous synthesis of compatible solute glucosylglycerol[J]. Frontiers in Microbiology, 2021, 12: 650217. |
| 130 | LOU W J, TAN X M, SONG K, et al. A specific single nucleotide polymorphism in the ATP synthase gene significantly improves environmental stress tolerance of Synechococcus elongatus PCC 7942[J]. Applied and Environmental Microbiology, 2018, 84(18): e01222-e01218. |
| 131 | OLIVER J W K, MACHADO I M P, YONEDA H, et al. Cyanobacterial conversion of carbon dioxide to 2,3-butanediol[J]. Proceeding of the National Academy of Sciences of the United States of America, 2013, 110(4): 1249-1254. |
| 132 | LI X Q, SHEN C R, LIAO J C. Isobutanol production as an alternative metabolic sink to rescue the growth deficiency of the glycogen mutant of Synechococcus elongatus PCC 7942[J]. Photosynthesis Research, 2014, 120(3): 301-310. |
| 133 | LAN E I, CHUANG D S, SHEN C R, et al. Metabolic engineering of cyanobacteria for photosynthetic 3-hydroxypropionic acid production from CO2 using Synechococcus elongatus PCC 7942[J]. Metabolic Engineering, 2015, 31: 163-170. |
| 134 | ROH H, LEE J S, CHOI H I, et al. Improved CO2-derived polyhydroxybutyrate (PHB) production by engineering fast-growing cyanobacterium Synechococcus elongatus UTEX 2973 for potential utilization of flue gas[J]. Bioresource Technology, 2021, 327: 124789. |
| 135 | SHARKEY T D, WIBERLEY A E, DONOHUE A R. Isoprene emission from plants: why and how[J]. Annals of Botany, 2007, 101(1): 5-18. |
| 136 | DAVIES F K, WORK V H, BELIAEV A S, et al. Engineering limonene and bisabolene production in wild type and a glycogen-deficient mutant of Synechococcus sp. PCC 7002[J]. Frontiers in Bioengineering and Biotechnology, 2014, 2: 21. |
| 137 | TAKAHAMA K, MATSUOKA M, NAGAHAMA K, et al. Construction and analysis of a recombinant cyanobacterium expressing a chromosomally inserted gene for an ethylene-forming enzyme at the psbAI locus[J]. Journal of Bioscience and Bioengineering, 2003, 95(3): 302-305. |
| 138 | JACOBSEN J H, FRIGAARD N U. Engineering of photosynthetic mannitol biosynthesis from CO2 in a cyanobacterium[J]. Metabolic Engineering, 2014, 21: 60-70. |
| 139 | SMITH M J, FRANCIS M B. A designed A. vinelandii-S. elongatus coculture for chemical photoproduction from air, water, phosphate, and trace metals[J]. ACS Synthetic Biology, 2016, 5(9): 955-961. |
| 140 | WEISS T L, YOUNG E J, DUCAT D C. A synthetic, light-driven consortium of cyanobacteria and heterotrophic bacteria enables stable polyhydroxybutyrate production[J]. Metabolic Engineering, 2017, 44: 236-245. |
| 141 | VELMURUGAN R, INCHAROENSAKDI A. Metal oxide mediated extracellular NADPH regeneration improves ethanol production by engineered Synechocystis sp. PCC 6803[J]. Frontiers in Bioengineering and Biotechnology, 2019, 7: 148. |
| [1] | GAO Ge, BIAN Qi, WANG Baojun. Synthetic genetic circuit engineering: principles, advances and prospects [J]. Synthetic Biology Journal, 2025, 6(1): 45-64. |
| [2] | LI Jiyuan, WU Guosheng. Two hypothesises for the origins of organisms from the synthetic biology perspective [J]. Synthetic Biology Journal, 2025, 6(1): 190-202. |
| [3] | JIAO Hongtao, QI Meng, SHAO Bin, JIANG Jinsong. Legal issues for the storage of DNA data [J]. Synthetic Biology Journal, 2025, 6(1): 177-189. |
| [4] | TANG Xinghua, LU Qianneng, HU Yilin. Philosophical reflections on synthetic biology in the Anthropocene [J]. Synthetic Biology Journal, 2025, 6(1): 203-212. |
| [5] | XU Huaisheng, SHI Xiaolong, LIU Xiaoguang, XU Miaomiao. Key technologies for DNA storage: encoding, error correction, random access, and security [J]. Synthetic Biology Journal, 2025, 6(1): 157-176. |
| [6] | SHI Ting, SONG Zhan, SONG Shiyi, ZHANG Yi-Heng P. Job. In vitro BioTransformation (ivBT): a new frontier of industrial biomanufacturing [J]. Synthetic Biology Journal, 2024, 5(6): 1437-1460. |
| [7] | CHAI Meng, WANG Fengqing, WEI Dongzhi. Synthesis of organic acids from lignocellulose by biotransformation [J]. Synthetic Biology Journal, 2024, 5(6): 1242-1263. |
| [8] | SHAO Mingwei, SUN Simian, YANG Shimao, CHEN Guoqiang. Bioproduction based on extremophiles [J]. Synthetic Biology Journal, 2024, 5(6): 1419-1436. |
| [9] | CHEN Yu, ZHANG Kang, QIU Yijing, CHENG Caiyun, YIN Jingjing, SONG Tianshun, XIE Jingjing. Progress of microbial electrosynthesis for conversion of CO2 [J]. Synthetic Biology Journal, 2024, 5(5): 1142-1168. |
| [10] | ZHENG Haotian, LI Chaofeng, LIU Liangxu, WANG Jiawei, LI Hengrun, NI Jun. Design, optimization and application of synthetic carbon-negative phototrophic community [J]. Synthetic Biology Journal, 2024, 5(5): 1189-1210. |
| [11] | CHEN Ziling, XIANG Yangfei. Integrated development of organoid technology and synthetic biology [J]. Synthetic Biology Journal, 2024, 5(4): 795-812. |
| [12] | CAI Bingyu, TAN Xiangtian, LI Wei. Advances in synthetic biology for engineering stem cell [J]. Synthetic Biology Journal, 2024, 5(4): 782-794. |
| [13] | XIE Huang, ZHENG Yilei, SU Yiting, RUAN Jingyi, LI Yongquan. An overview on reconstructing the biosynthetic system of actinomycetes for polyketides production [J]. Synthetic Biology Journal, 2024, 5(3): 612-630. |
| [14] | ZHA Wenlong, BU Lan, ZI Jiachen. Advances in synthetic biology for producing potent pharmaceutical ingredients of traditional Chinese medicine [J]. Synthetic Biology Journal, 2024, 5(3): 631-657. |
| [15] | HUI Zhen, TANG Xiaoyu. Applications of the CRISPR/Cas9 editing system in the study of microbial natural products [J]. Synthetic Biology Journal, 2024, 5(3): 658-671. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||