Synthetic Biology Journal ›› 2020, Vol. 1 ›› Issue (1): 71-83.DOI: 10.12211/2096-8280.2020-054
• Invited Review • Previous Articles Next Articles
Research progress in bioproduction of aliphatic diamines by synthetic biotechnology
WANG Xin, WANG Jing, CHEN Kequan, OUYANG Pingkai
- College of Biotechnology and Pharmaceutical Engineering, Nanjing Tech University, State Key Laboratory of Materials-Oriented Chemical Engineering, Jiangsu National Synergetic Innovation Center for Advance Material, Nanjing 211816, Jiangsu, China
-
Received:2020-04-20Revised:2020-05-06Online:2020-07-07Published:2020-02-29 -
Contact:OUYANG Pingkai
合成生物技术制备脂肪族二元胺的研究进展
王昕, 王静, 陈可泉, 欧阳平凯
- 南京工业大学生物与制药工程学院,材料化学工程国家重点实验室,江苏省国家先进材料协同创新中心,江苏 南京 211816
-
通讯作者:欧阳平凯 -
作者简介:王昕(1988—),女,博士,副教授。研究方向为生物催化。E-mail:xinwang1988@njtech.edu.cn
欧阳平凯(1945—),男,教授,中国工程院院士。研究方向为生物催化。E-mail:ouyangpk@njtech.edu.cn -
基金资助:国家重点研发计划(2018YFA0901500)
CLC Number:
Cite this article
WANG Xin, WANG Jing, CHEN Kequan, OUYANG Pingkai. Research progress in bioproduction of aliphatic diamines by synthetic biotechnology[J]. Synthetic Biology Journal, 2020, 1(1): 71-83.
王昕, 王静, 陈可泉, 欧阳平凯. 合成生物技术制备脂肪族二元胺的研究进展[J]. 合成生物学, 2020, 1(1): 71-83.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2020-054
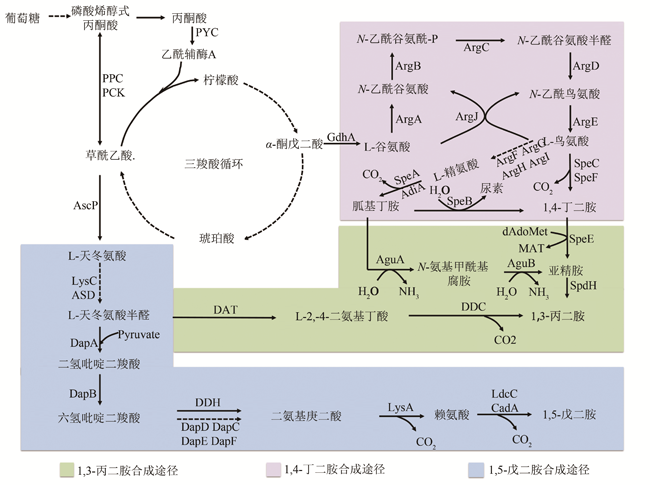
Fig. 1 The biosynthetic pathway of the aliphatic diamines with 3~5 carbon atoms (The solid arrows indicate one-step metabolic pathways, and the dotted arrows indicate multiple-step metabolic pathways. The green square represents the synthetic pathway of 1,3-propanediamine, the purple square represents the synthetic pathway of 1,4-butanediamine synthesis route, and the blue square represents the synthetic pathway of 1,5-pentanediamine)
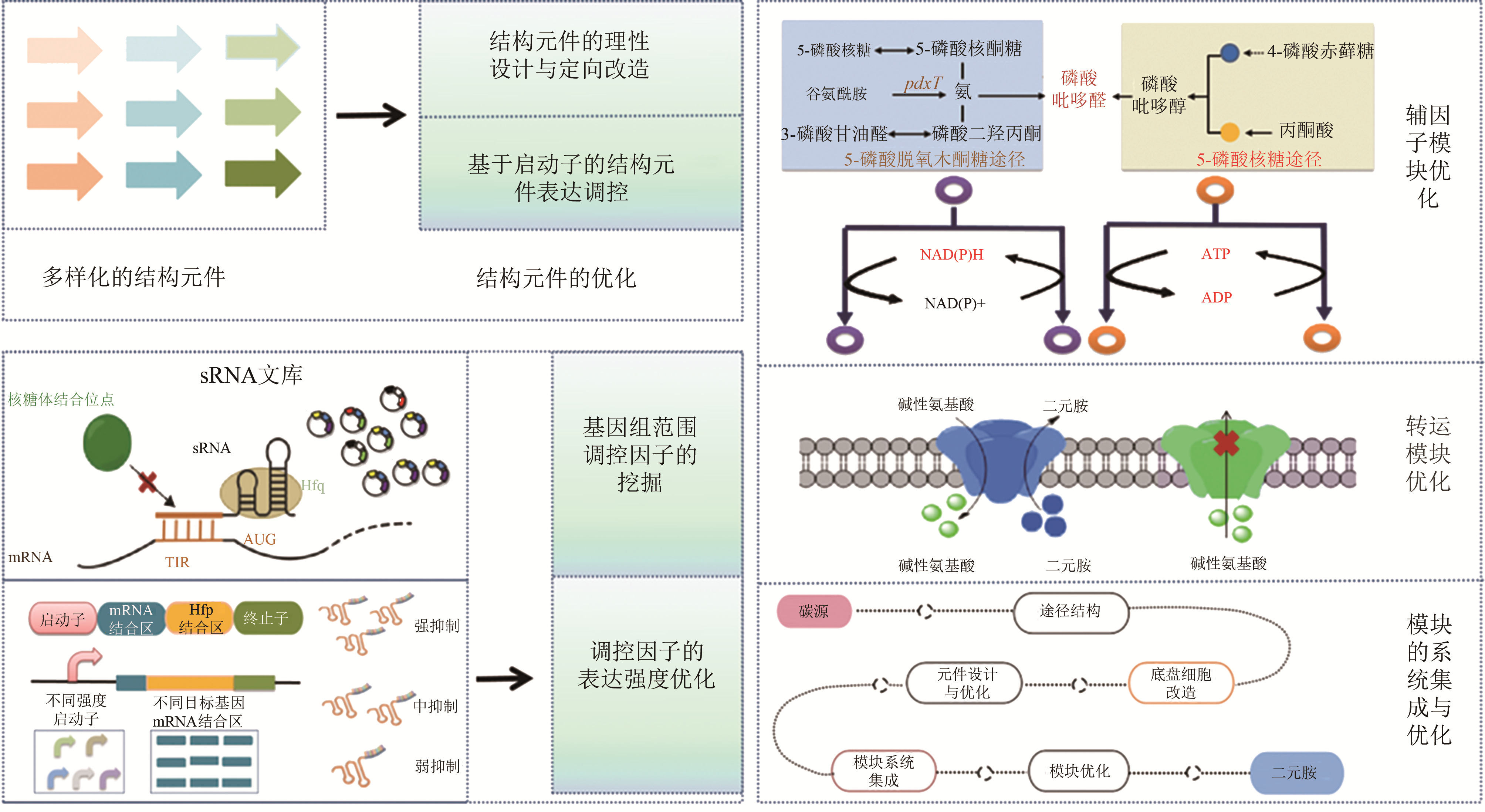
Fig. 2 The engineering strategy for the optimization of diamine synthetic cells[14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48]
| 产品 | 宿主 | 改造策略 | 产量 /g·L-1 | 产率/g·g-1 | 参考文献 |
|---|---|---|---|---|---|
1,5- 戊二胺 | 大肠杆菌 | 过表达赖氨酸脱羧酶元件cadA 过表达dapA基因增强前体赖氨酸合成 敲除1,5-戊二胺利用途径基因speE、speG、ygjG和 puuA 抑制调控元件murE的表达 | 12.6 | — | [ |
| 谷氨酸棒状杆菌 | 高强度表达E.coli 来源的赖氨酸脱羧酶元件ldcC 敲除赖氨酸分泌蛋白lysE | 103.78 | 0.303 | [ | |
1,4- 丁二胺 | 大肠杆菌 | 敲除产物降解途径基因 speE、speG、puuA 敲除前体鸟氨酸降解途径基因argI 敲除全局转录因子 rpoS 过表达合成途径基因speC、argE、argC、argB、argH、speF和argD 过表达1,4-丁二胺分泌蛋白potE 下调argF和glnA的表达水平 | 42.3 | 0.26 | [ |
| 谷氨酸棒状杆菌 | 异源表达 E. coli 来源的鸟氨酸脱羧酶speC 敲除前体鸟氨酸利用途径基因argF和argR 精确调控argF的表达水平 | 19 | 0.16 | [ | |
1,3- 丙二胺 | 大肠杆菌 | 表达不动杆菌属来源的dat和ddc组装1,3-丙二胺合成途径 过表达thrA和lysC突变体增强天冬氨酸半醛合成 过表达ppc和aspC增强前体天冬氨酸的合成 敲除pfkA增强胞内NADPH的供给 | 13.06 | 0.1 | [ |
Tab. 1 The highest production level of aliphatic diamines with 3~5 carbon atoms by different hosts
| 产品 | 宿主 | 改造策略 | 产量 /g·L-1 | 产率/g·g-1 | 参考文献 |
|---|---|---|---|---|---|
1,5- 戊二胺 | 大肠杆菌 | 过表达赖氨酸脱羧酶元件cadA 过表达dapA基因增强前体赖氨酸合成 敲除1,5-戊二胺利用途径基因speE、speG、ygjG和 puuA 抑制调控元件murE的表达 | 12.6 | — | [ |
| 谷氨酸棒状杆菌 | 高强度表达E.coli 来源的赖氨酸脱羧酶元件ldcC 敲除赖氨酸分泌蛋白lysE | 103.78 | 0.303 | [ | |
1,4- 丁二胺 | 大肠杆菌 | 敲除产物降解途径基因 speE、speG、puuA 敲除前体鸟氨酸降解途径基因argI 敲除全局转录因子 rpoS 过表达合成途径基因speC、argE、argC、argB、argH、speF和argD 过表达1,4-丁二胺分泌蛋白potE 下调argF和glnA的表达水平 | 42.3 | 0.26 | [ |
| 谷氨酸棒状杆菌 | 异源表达 E. coli 来源的鸟氨酸脱羧酶speC 敲除前体鸟氨酸利用途径基因argF和argR 精确调控argF的表达水平 | 19 | 0.16 | [ | |
1,3- 丙二胺 | 大肠杆菌 | 表达不动杆菌属来源的dat和ddc组装1,3-丙二胺合成途径 过表达thrA和lysC突变体增强天冬氨酸半醛合成 过表达ppc和aspC增强前体天冬氨酸的合成 敲除pfkA增强胞内NADPH的供给 | 13.06 | 0.1 | [ |
| 1 | JIANG Y, LOOS K. Enzymatic synthesis of biobased polyesters and polyamides [J]. Polymers, 2016, 8(7): 243. |
| 2 | WINNACKER M, RIEGER B. Biobased polyamides: recent advances in basic and applied research [J]. Macromolecular Rapid Communications, 2016, 37(17): 1391-1413. |
| 3 | GILBERT M. Aliphatic polyamides [M]. Oxford: Butterworth-Heinermann, 2017. |
| 4 | 李秀峥, 李澜鹏, 曹长海, 等. 生物基聚酰胺及其单体研究进展 [J]. 工程塑料应用, 2018(7): 138-142. |
| LI X Z, LI L P, CAO C H, et al. Research progress of bio-based polyamide and its monomer [J]. Engineering Plastics Application, 2018(7): 138-142. | |
| 5 | MA W, CAO W, ZHANG H, et al. Enhanced cadaverine production from l-lysine using recombinant Escherichia coli co-overexpressing CadA and CadB [J]. Biotechnology Letters, 2015, 37(4): 799-806. |
| 6 | KIM H T, BARITUGO K A, OH Y H, et al. High-level conversion of l-lysine into cadaverine by Escherichia coli whole cell biocatalyst expressing Hafnia alvei l-lysine decarboxylase [J]. Polymers, 2019, 11(7): 1184. |
| 7 | SEO S W, YANG J, MIN B E, et al. Synthetic biology: tools to design microbes for the production of chemicals and fuels [J]. Biotechnology Advances, 2013, 31(6): 811-817. |
| 8 | WENDISCH V F, MINDT M, PÉREZ-GARCÍA F. Biotechnological production of mono- and diamines using bacteria: recent progress, applications, and perspectives [J]. Applied Microbiology and Biotechnology, 2018, 102(8): 3583-3594. |
| 9 | LI Z, LIU J Z. Transcriptomic changes in response to putrescine production in metabolically engineered Corynebacterium glutamicum [J]. Frontiers in Microbiology, 2017, 8: 1-11. |
| 10 | SCHNEIDER J, WENDISCH V F. Putrescine production by engineered Corynebacterium glutamicum [J]. Applied Microbiology and Biotechnology, 2010, 88(4): 859-868. |
| 11 | JENSEN J V K, EBERHARDT D, WENDISCH V F. Modular pathway engineering of Corynebacterium glutamicum for production of the glutamate-derived compounds ornithine, proline, putrescine, citrulline, and arginine [J]. Journal of Biotechnology, 2015, 214: 85-94. |
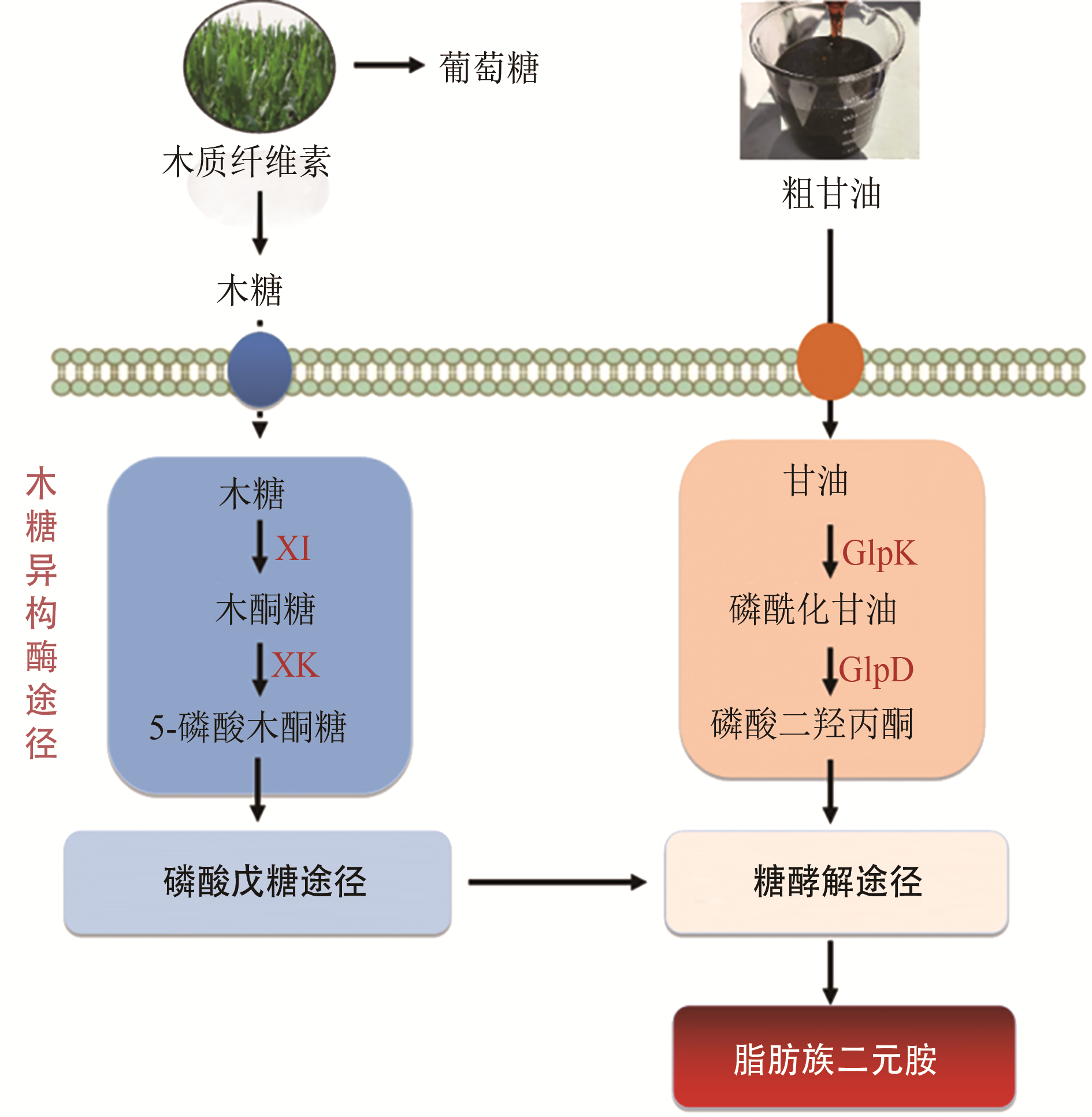
| 12 | ZHAN M, KAN B, DONG J, et al. Metabolic engineering of Corynebacterium glutamicum for improved l-arginine synthesis by enhancing NADPH supply [J]. Journal of Industrial Microbiology and Biotechnology, 2019, 46(1): 45-54. |
| 13 | CHAE T U, KIM W J, CHOI S, et al. Metabolic engineering of Escherichia coli for the production of 1,3-diaminopropane, a three carbon diamine [J]. Scientific Reports, 2015, 5: 1-13. |
| 14 | QIAN Z G, XIA X X, LEE S Y. Metabolic engineering of Escherichia coli for the production of cadaverine: a five carbon diamine [J]. Biotechnology and Bioengineering, 2011, 108(1): 93-103. |
| 15 | QIAN Z G, XIA X X, LEE S Y. Metabolic engineering of Escherichia coli for the production of putrescine: a four carbon diamine [J]. Biotechnology and Bioengineering, 2009, 104(4): 651-662. |
| 16 | MIMITSUKA T, SAWAI H, HATSU M, et al. Metabolic engineering of Corynebacterium glutamicum for cadaverine fermentation [J]. Bioscience, Biotechnology and Biochemistry, 2007, 71(9): 2130-2135. |
| 17 | KIND S, JEONG W K, SCHRÖDER H, et al. Identification and elimination of the competing N-acetyldiaminopentane pathway for improved production of diaminopentane by Corynebacterium glutamicum [J]. Applied and Environmental Microbiology, 2010, 76(15): 5175-5180. |
| 18 | NGUYEN A Q D, SCHNEIDER J, WENDISCH V F. Elimination of polyamine N-acetylation and regulatory engineering improved putrescine production by Corynebacterium glutamicum [J]. Journal of Biotechnology, 2015, 201: 75-85. |
| 19 | YAMAMOTO S, TSUZAKI Y, TOUGOU K, et al. Purification and characterization of L-2,4-diaminobutyrate decarboxylase from Acinetobacter calcoaceticus [J]. Journal of General Microbiology, 1992, 138(7): 1461-1465. |
| 20 | YAMAMOTO S, MUTOH N, IKAI H, et al. Occurrence of a novel L-2,4-diaminobutyrate decarboxylase activity in some species of Enterobacteriaceae, and purification and characterization of the enzymes of Enterobacter aerogenes and Serratia marcescens [J]. Biological and Pharmaceutical Bulletin, 1996,19(10): 1298-1303. |
| 21 | IKAI H, YAMAMOTO S. Cloning and expression in Escherichia coli of the gene encoding a novel L-2,4-diaminobutyratedecarboxylase of Acinetobacter baumannii [J] FEMS Microbiology Letters, 1994, 124(2): 225-228. |
| 22 | NAKAO H, TAKEUCHI K, SHINODA S, et al. L-2,4-diaminobutyric acid decarboxylase activity responsible for the formation of 1,3-diaminopropane in Enterobacter aerogenes [J]. FEMS Microbiology Letters, 1990, 70(1): 61-66. |
| 23 | BLETHEN S L, BOEKER E A, SNELL E E. Argenine decarboxylase from Escherichia coli. I. purification and specificity for substrates and coenzyme [J]. Journal of Biological Chemistry, 1968, 243(8):1671-1677. |
| 24 | WU W H, MORRIS D R. Biosynthetic arginine decarboxylase from Escherichia coli: purification and properties [J]. Journal of Biological Chemistry, 1973,248(5): 1687-1695. |
| 25 | GOLDEMBERG S H. Lysine decarboxylase mutants of Escherichia coli: evidence for two enzyme forms [J]. Journal of Bacteriology, 1980, 141(3): 1428-1431. |
| 26 | KIKUCHI Y, KOJIMA H, TANAKA T, et al. Characterization of a second lysine decarboxylase isolated from Escherichia coli [J]. Journal of Bacteriology, 1997, 179(14): 4486-4492. |
| 27 | LEMONNIER M, LANE D. Expression of the second lysine decarboxylate gene of Escherichia coli [J]. Microbiology, 1998, 144(3): 751-760. |
| 28 | SABO D L, BOEKER E A, BYERS B, et al. Purification and physical properties of inducible Escherichia coli lysine decarboxylase [J]. Biochemistry, 1974, 13(4): 662-670. |
| 29 | YAMAMOTO Y, MIWA Y, MIYOSHI K, et al. The Escherichia coli ldcC gene encodes another lysine decarboxylase, probably a constitutive enzyme [J]. Genes and Genetic Systems, 1997, 72(3): 167-172. |
| 30 | LI Z, SHEN Y P, JIANG X L, et al. Metabolic evolution and a comparative omics analysis of Corynebacterium glutamicum for putrescine production[J]. Journal of Industrial Microbiology and Biotechnology, 2018, 45(2): 123-139. |
| 31 | WANG C, ZHANG K, ZHONG J C, et al. Directed evolution and mutagenesis of lysine decarboxylase from Hafnia alvei AS1.1009 to improve its activity toward efficient cadaverine production[J]. Biotechnology and Bioprocess Engineering, 2015, 20(3): 439-446. |
| 32 | HONG E Y, LEE S G, PARK B J, et al. Simultaneously enhancing the stability and catalytic activity of multimeric lysine decarboxylase CadA by engineering interface regions for enzymatic production of cadaverine at high concentration of lysine [J]. Biotechnology Journal, 2017, 12(11): 1700268. |
| 33 | CHOI H, KYEONG H H, CHOI J M, et al. Rational design of ornithine decarboxylase with high catalytic activity for the production of putrescine [J]. Applied Microbiology and Biotechnology, 2014, 98(17): 7483-7490. |
| 34 | HONG E Y, KIM J Y, UPADHYAY R, et al. Rational engineering of ornithine decarboxylase with greater selectivity for ornithine over lysine through protein network analysis[J]. Journal of Biotechnology, 2018, 281: 175-182. |
| 35 | OH Y H, CHOI J W, KIM E Y, et al. Construction of synthetic promoter-based expression cassettes for the production of cadaverine in recombinant Corynebacterium glutamicum [J]. Applied Biochemistry and Biotechnology, 2015, 176(7): 2065-2075. |
| 36 | KIM H T, BARITUGO K A, OH Y H, et al. Metabolic engineering of Corynebacterium glutamicum for the high-level production of cadaverine that can be used for the synthesis of biopolyamide 510 [J]. ACS Sustainable Chemistry and Engineering, 2018, 6(4): 5296-5305. |
| 37 | SCHNEIDER J, EBERHARDT D, WENDISCH V F. Improving putrescine production by Corynebacterium glutamicum by fine-tuning ornithine transcarbamoylase activity using a plasmid addiction system [J]. Applied Microbiology and Biotechnology, 2012, 95(1): 169-178. |
| 38 | NA D, YOO S M, CHUNG H, et al. Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs[J]. Nature Biotechnology, 2013, 31(2): 170-174. |
| 39 | NOH M, YOO S M, KIM W J, et al. Gene expression knockdown by modulating synthetic small RNA expression in Escherichia coli [J]. Cell Systems, 2017, 5(4): 418-426. |
| 40 | LIANG J, HAN Q, TAN Y, et al. Current advances on structure-function relationships of pyridoxal 5′-phosphate dependent enzymes [J]. Frontiers in Molecular Biosciences, 2019(6): 4. |
| 41 | FITZPATRICK T B, AMRHEIN N, KAPPES B, et al. Two independent routes of de novo vitamin B6 biosynthesis: not that different after all [J]. Biochemical Journal, 2007, 407(1): 1-13. |
| 42 | FITZPATRICK T B, MOCCAND C, ROUX C. Vitamin B6 biosynthesis: charting the mechanistic landscape [J]. ChemBioChem, 2010, 11(9): 1185-1193. |
| 43 | ROSENBERG J, ISCHEBECK T, COMMICHAU F M. Vitamin B6 metabolism in microbes and approaches for fermentative production [J]. Biotechnology Advances, 2017, 35(1): 31-40. |
| 44 | RASCHLE T, AMRHEIN N, FITZPATRICK T B. On the two components of pyridoxal 5′-phosphate synthase from Bacillus subtilis [J]. Journal of Biological Chemistry, 2005, 280(37): 32291-32300. |
| 45 | TATSUO H, KEIKO I, MASAAKI T. Recombinant microorganism for the production of vitamin B6: US528891 [P]. 2005-10-19. |
| 46 | MA W, CAO W, ZHANG B, et al. Engineering a pyridoxal 5′-phosphate supply for cadaverine production by using Escherichia coli whole-cell biocatalysis [J]. Scientific Reports, 2015, 5: 1-10. |
| 47 | LI M, LI D, HUANG Y, et al. Improving the secretion of cadaverine in Corynebacterium glutamicum by cadaverine-lysine antiporter [J]. Journal of Industrial Microbiology and Biotechnology, 2014, 41(4): 701-709. |
| 48 | KIND S, KREYE S, WITTMANN C. Metabolic engineering of cellular transport for overproduction of the platform chemical 1,5-diaminopentane in Corynebacterium glutamicum [J]. Metabolic Engineering, 2011, 13(5): 617-627. |
| 49 | KIND S, NEUBAUER S, BECKER J, et al. From zero to hero-production of bio-based nylon from renewable resources using engineered Corynebacterium glutamicum [J]. Metabolic Engineering, 2014, 25: 113-123. |
| 50 | ARISTIDOU A, PENTTIL A M. Metabolic engineering applications to renewable resource utilization [J]. Current Opinion in Biotechnology, 2000, 11: 187-198. |
| 51 | CHOI J W, JEON E J, JEONG K J. Recent advances in engineering Corynebacterium glutamicum for utilization of hemicellulosic biomass [J]. Current Opinion in Biotechnology, 2019, 57: 17-24. |
| 52 | MEISWINKEL T M, GOPINATH V, LINDNER S N, et al. Accelerated pentose utilization by Corynebacterium glutamicum for accelerated production of lysine, glutamate, ornithine and putrescine [J]. Microbial Biotechnology, 2013, 6(2): 131-140. |
| 53 | BUSCHKE N, SCHRODER H, WITTMANN C. Metabolic engineering of Corynebacterium glutamicum for production of 1,5-diaminopentane from hemicellulose [J]. Biotechnology Journal, 2011, 6(3): 306-317. |
| 54 | BUSCHKE N, BECKER J, SCHAFER R, et al. Systems metabolic engineering of xylose-utilizing Corynebacterium glutamicum for production of 1,5-diaminopentane [J]. Biotechnology Journal, 2013, 8(5): 557-570. |
| 55 | CHEN Z, LIU D. Toward glycerol biorefinery: metabolic engineering for the production of biofuels and chemicals from glycerol [J]. Biotechnology for Biofuels, 2016, 9(1): 1-15. |
| 56 | BECKERS V, POBLETE-CASTRO I, TOMASCH J, et al. Integrated analysis of gene expression and metabolic fluxes in PHA-producing Pseudomonas putida grown on glycerol [J]. Microbial Cell Factories, 2016, 15(1): 1-18. |
| 57 | GAO C, YANG X, WANG H, et al. Robust succinic acid production from crude glycerol using engineered Yarrowia lipolytica [J]. Biotechnology for Biofuels, 2016, 9(1): 1-11. |
| 58 | MEISWINKEL T M, RITTMANN D, LINDNER S N, et al. Crude glycerol-based production of amino acids and putrescine by Corynebacterium glutamicum [J]. Bioresource Technology, 2013, 145: 254-258. |
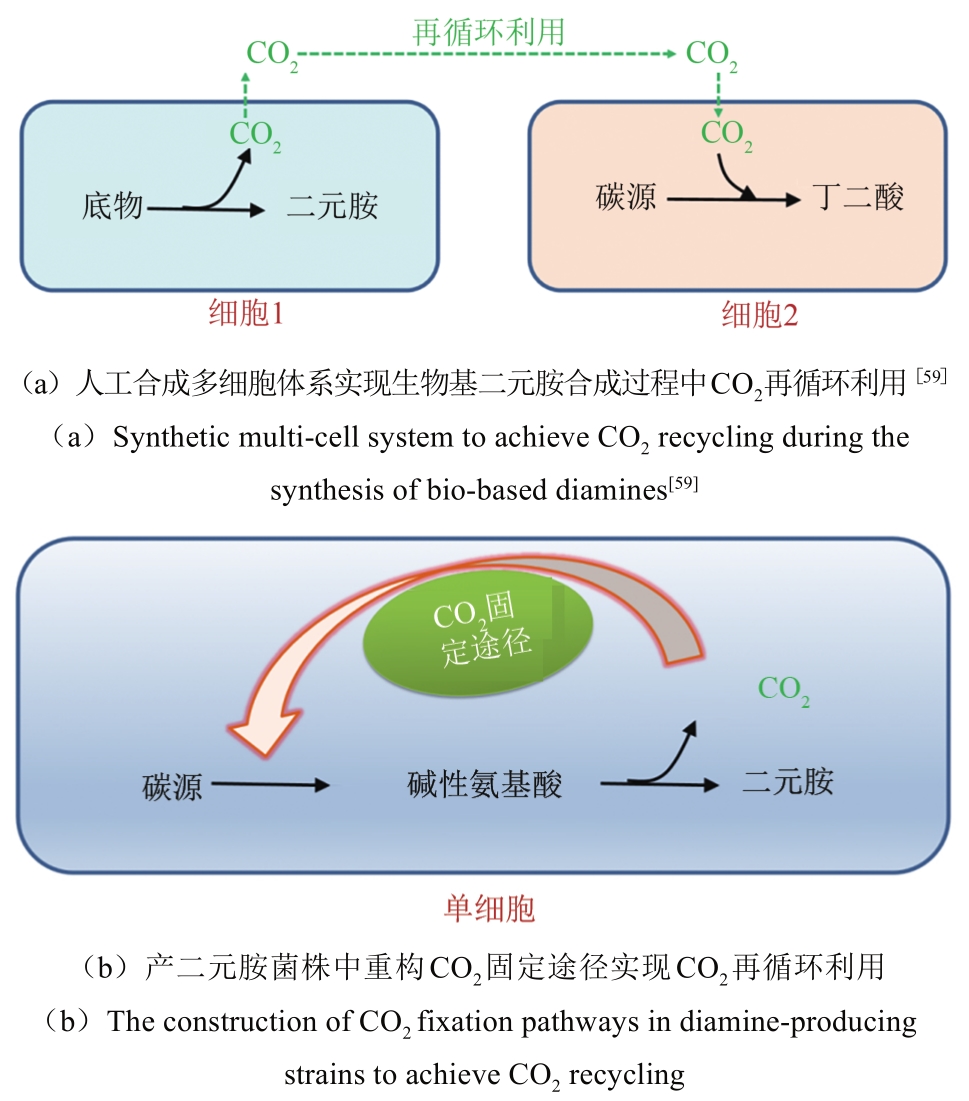
| 59 | WANG J, MAO J, TIAN W, et al. Coproduction of succinic acid and cadaverine using lysine as a neutralizer and CO2 donor with l-lysine decarboxylase overexpressed: Escherichia coli AFP111 [J]. Green Chemistry, 2018, 20(12): 2880-2887. |
| 60 | CLAASSENS N J, SOUSA D Z, SANTOS V A P M DOS, et al. Harnessing the power of microbial autotrophy [J]. Nature Reviews Microbiology, 2016, 14(11):692-706. |
| 61 | GUADALUPE-MEDINA V, WISSELINK H W, LUTTIK M A, et al. Carbon dioxide fixation by Calvin-cycle enzymes improves ethanol yield in yeast [J]. Biotechnology for Biofuels, 2013, 6(1):125. |
| 62 | HU G, ZHOU J, CHEN X, et al. Engineering synergetic CO2-fixing pathways for malate production [J]. Metabolic Engineering, 2018, 47:496-504. |
| 63 | LIMSUWUN K, JONES P G. Spermidine acetyltransferase is required to prevent spermidine toxicity at low temperatures in Escherichia coli [J]. Journal of Bacteriology, 2000, 182(19):5373-5380. |
| 64 | KATINKA M, COSSART P, SIBILLI L, et al. Nucleotide sequence of the thrA gene of Escherichia coli [J]. PNAS, 1980, 77: 5730-5733. |
| 65 | HU G, LI Y, YE C, et al. Engineering microorganisms for enhanced CO2 sequestration [J]. Trends in Biotechnology, 2019, 37(5): 532-547. |
| 66 | GONG F, ZHU H, ZHANG Y, et al. Biological carbon fixation: from natural to synthetic [J]. 2018, 28:221-227. |
| 67 | PLEGARIA J S, KERFELD C A. Engineering nanoreactors using bacterial microcompartment architectures [J]. Journal of CO2 Utilization, 2018, 51:1-7. |
| 68 | WANG S Z, ZHANG Y H, REN H, et al. Strategies and perspectives of assembling multi-enzyme systems [J]. Critical Reviews in Biotechnology, 2017, 37(8):1-14. |
| 69 | PRICE J V, CHEN L, WHITAKER W B, et al. Scaffoldless engineered enzyme assembly for enhanced methanol utilization [J]. PNAS, 2016, 113(45): 12691-12696. |
| 70 | KUMAR M, SUNDARAM S, GNANSOUNOU E, et al. Carbon dioxide capture, storage and production of biofuel and biomaterials by bacteria: a review [J]. Bioresource Technology, 2018, 247: 1059-1068. |
| 71 | TURMO A, GONZALEZ-ESQUER C R, Kerfeld C A. Carboxysomes: metabolic modules for CO2 fixation [J]. FEMS Microbiology Letters, 2017, 364(18):176. |
| [1] | GAO Ge, BIAN Qi, WANG Baojun. Synthetic genetic circuit engineering: principles, advances and prospects [J]. Synthetic Biology Journal, 2025, 6(1): 45-64. |
| [2] | LI Jiyuan, WU Guosheng. Two hypothesises for the origins of organisms from the synthetic biology perspective [J]. Synthetic Biology Journal, 2025, 6(1): 190-202. |
| [3] | JIAO Hongtao, QI Meng, SHAO Bin, JIANG Jinsong. Legal issues for the storage of DNA data [J]. Synthetic Biology Journal, 2025, 6(1): 177-189. |
| [4] | TANG Xinghua, LU Qianneng, HU Yilin. Philosophical reflections on synthetic biology in the Anthropocene [J]. Synthetic Biology Journal, 2025, 6(1): 203-212. |
| [5] | XU Huaisheng, SHI Xiaolong, LIU Xiaoguang, XU Miaomiao. Key technologies for DNA storage: encoding, error correction, random access, and security [J]. Synthetic Biology Journal, 2025, 6(1): 157-176. |
| [6] | SHI Ting, SONG Zhan, SONG Shiyi, ZHANG Yi-Heng P. Job. In vitro BioTransformation (ivBT): a new frontier of industrial biomanufacturing [J]. Synthetic Biology Journal, 2024, 5(6): 1437-1460. |
| [7] | CHAI Meng, WANG Fengqing, WEI Dongzhi. Synthesis of organic acids from lignocellulose by biotransformation [J]. Synthetic Biology Journal, 2024, 5(6): 1242-1263. |
| [8] | SHAO Mingwei, SUN Simian, YANG Shimao, CHEN Guoqiang. Bioproduction based on extremophiles [J]. Synthetic Biology Journal, 2024, 5(6): 1419-1436. |
| [9] | CHEN Yu, ZHANG Kang, QIU Yijing, CHENG Caiyun, YIN Jingjing, SONG Tianshun, XIE Jingjing. Progress of microbial electrosynthesis for conversion of CO2 [J]. Synthetic Biology Journal, 2024, 5(5): 1142-1168. |
| [10] | ZHENG Haotian, LI Chaofeng, LIU Liangxu, WANG Jiawei, LI Hengrun, NI Jun. Design, optimization and application of synthetic carbon-negative phototrophic community [J]. Synthetic Biology Journal, 2024, 5(5): 1189-1210. |
| [11] | CHEN Ziling, XIANG Yangfei. Integrated development of organoid technology and synthetic biology [J]. Synthetic Biology Journal, 2024, 5(4): 795-812. |
| [12] | CAI Bingyu, TAN Xiangtian, LI Wei. Advances in synthetic biology for engineering stem cell [J]. Synthetic Biology Journal, 2024, 5(4): 782-794. |
| [13] | XIE Huang, ZHENG Yilei, SU Yiting, RUAN Jingyi, LI Yongquan. An overview on reconstructing the biosynthetic system of actinomycetes for polyketides production [J]. Synthetic Biology Journal, 2024, 5(3): 612-630. |
| [14] | ZHA Wenlong, BU Lan, ZI Jiachen. Advances in synthetic biology for producing potent pharmaceutical ingredients of traditional Chinese medicine [J]. Synthetic Biology Journal, 2024, 5(3): 631-657. |
| [15] | HUI Zhen, TANG Xiaoyu. Applications of the CRISPR/Cas9 editing system in the study of microbial natural products [J]. Synthetic Biology Journal, 2024, 5(3): 658-671. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||