Synthetic Biology Journal ›› 2020, Vol. 1 ›› Issue (4): 440-453.DOI: 10.12211/2096-8280.2020-029
• Invited Review • Previous Articles Next Articles
Application of dynamic regulation strategies in metabolic engineering
Yu Zheng, SHEN Xiaolin, Sun Xinxiao, Wang Jia, Yuan Qipeng
- State Key Laboratory of Chemical Resource Engineering,Beijing University of Chemical Technology,Beijing 100029,China
-
Received:2020-03-19Revised:2020-04-29Online:2020-10-09Published:2020-08-31 -
Contact:Wang Jia, Yuan Qipeng
动态调控策略在代谢工程中的应用研究进展
于政, 申晓林, 孙新晓, 王佳, 袁其朋
- 北京化工大学化工资源有效利用国家重点实验室,北京 100029
-
通讯作者:王佳,袁其朋 -
作者简介:于政(1998—),男,硕士研究生。研究方向为代谢工程及合成生物学。E-mail:2016018277@mail.buct.edu.cn
王佳(1989—),女,博士,副教授。研究方向为代谢工程及合成生物学。E-mail:wangjia@mail.buct.edu.cn
袁其朋(1969—),男,博士生导师,教授。研究方向为生物化工。E-mail:yuanqp@mail.buct.edu.cn -
基金资助:国家自然科学基金(21908003);国家重点研发计划(2018YFA0903000)
CLC Number:
Cite this article
Yu Zheng, SHEN Xiaolin, Sun Xinxiao, Wang Jia, Yuan Qipeng. Application of dynamic regulation strategies in metabolic engineering[J]. Synthetic Biology Journal, 2020, 1(4): 440-453.
于政, 申晓林, 孙新晓, 王佳, 袁其朋. 动态调控策略在代谢工程中的应用研究进展[J]. 合成生物学, 2020, 1(4): 440-453.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2020-029
| 调控类型 | 输入信号 | 调控元件 | 菌株 | 产物 | 调控效果 |
|---|---|---|---|---|---|
| 转录因子 | 乙酰磷酸 | NRI | 大肠杆菌 | 番茄红素 | 50%[ |
| 丙二酰辅酶A | FapR | 大肠杆菌 | 脂肪酸 | 2.1倍[ | |
| 丙二酰辅酶A | FapR | 大肠杆菌 | 脂肪酸 | 34%[ | |
| 衣康酸 | ItcR | 大肠杆菌 | 衣康酸 | 0.78mmol/L[ | |
| 阿魏酸、香草醛 | HucR | 大肠杆菌 | 香草醛 | 11mmol/L[ | |
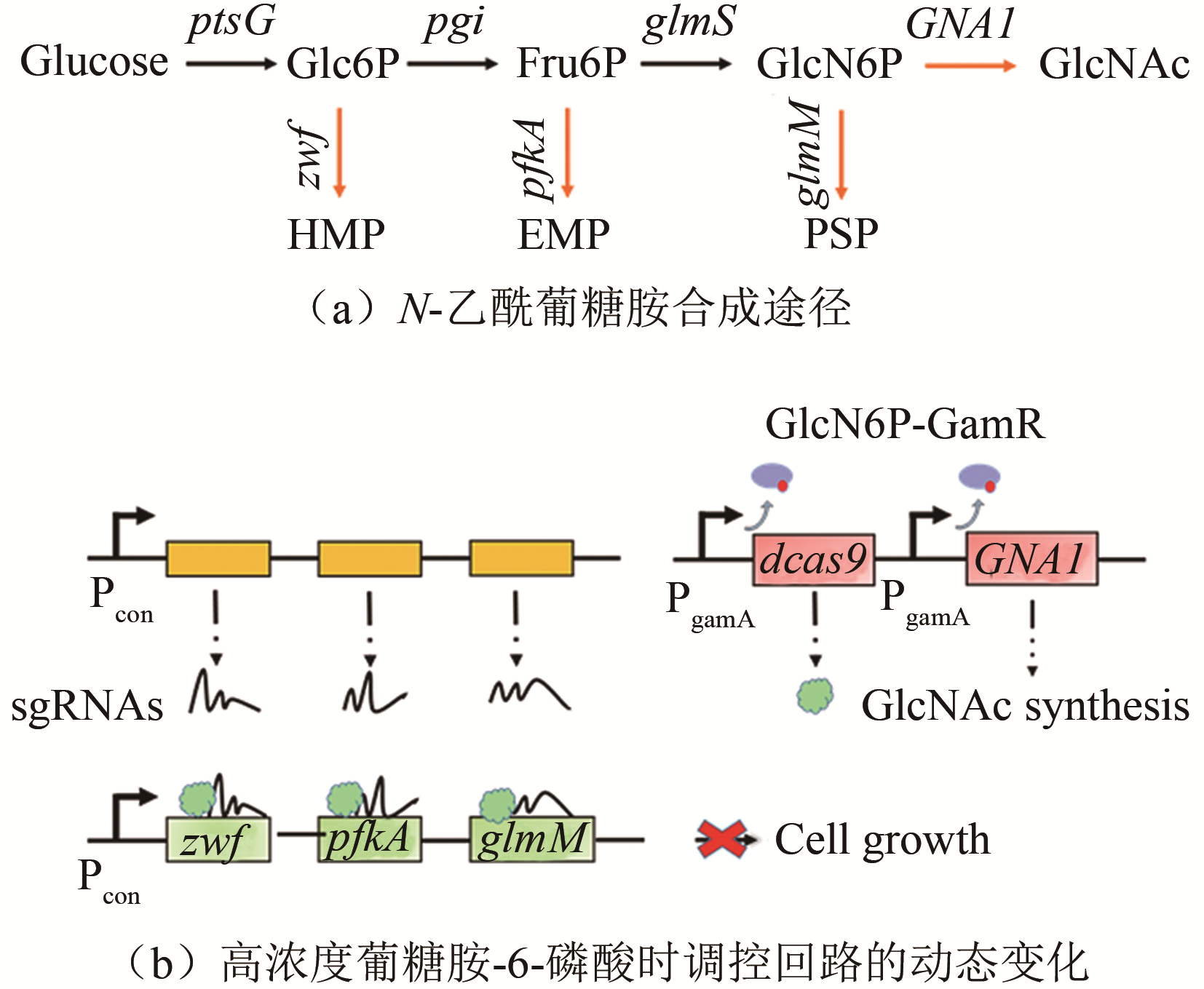
| 葡糖胺-6-磷酸 | GamR | 枯草芽孢杆菌 | N-乙酰葡糖胺 | 131.6 g/L[ | |
| 黏糠酸 | CatR | 大肠杆菌 | 黏糠酸 | 16.3倍[ | |
| 核糖开关 | 茶碱 | 茶碱核糖开关 | 大肠杆菌 | — | 3倍[ |
| 硫胺素焦磷酸 | 硫胺素焦磷酸核糖开关 | 米曲霉 | — | 4.7倍[ | |
| 赖氨酸 | 赖氨酸核糖开关 | 谷氨酸棒状杆菌 | 赖氨酸 | 63%[ | |
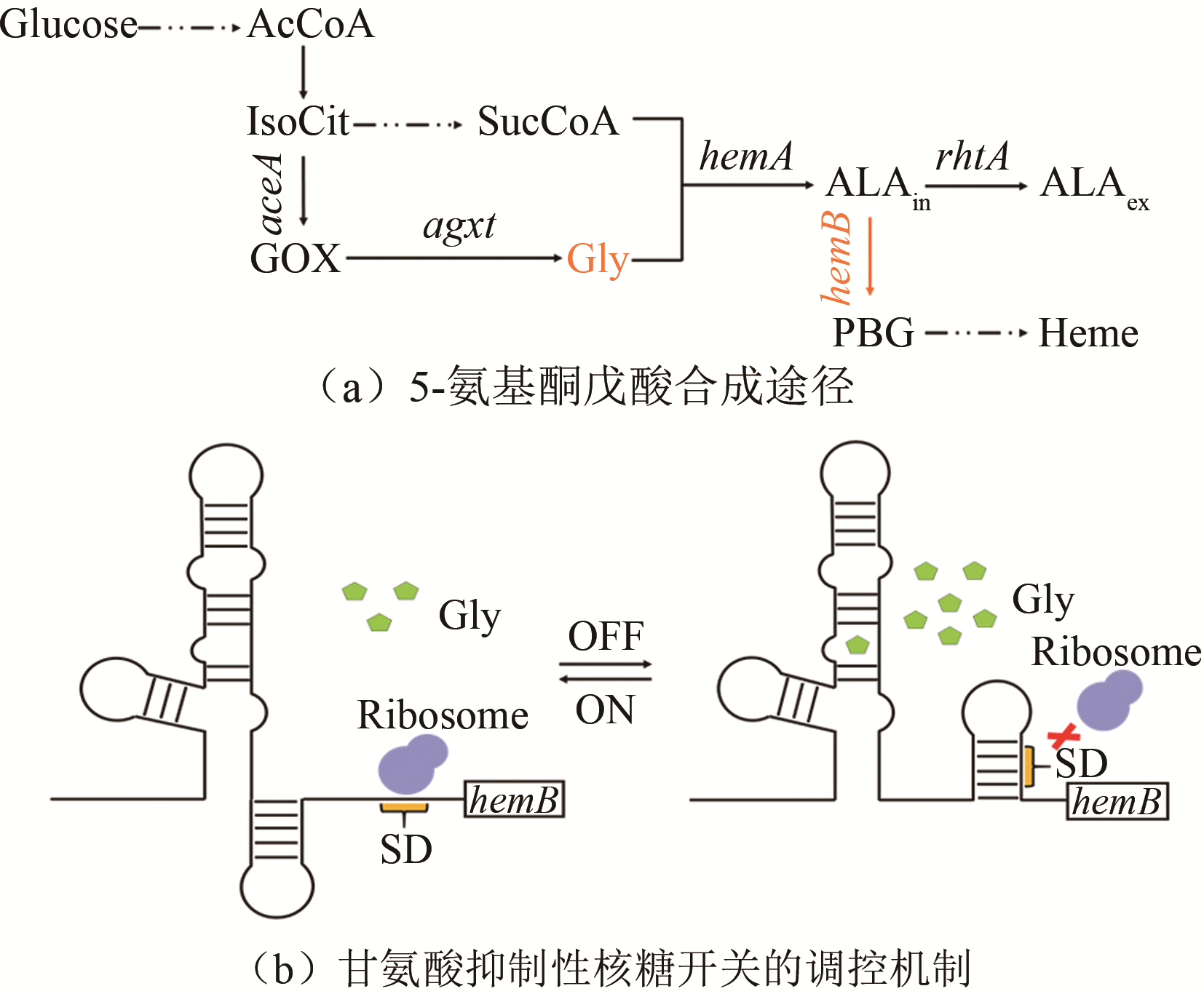
| 甘氨酸 | 甘氨酸核糖开关 | 大肠杆菌 | 5-氨基酮戊酸 | 11%[ | |
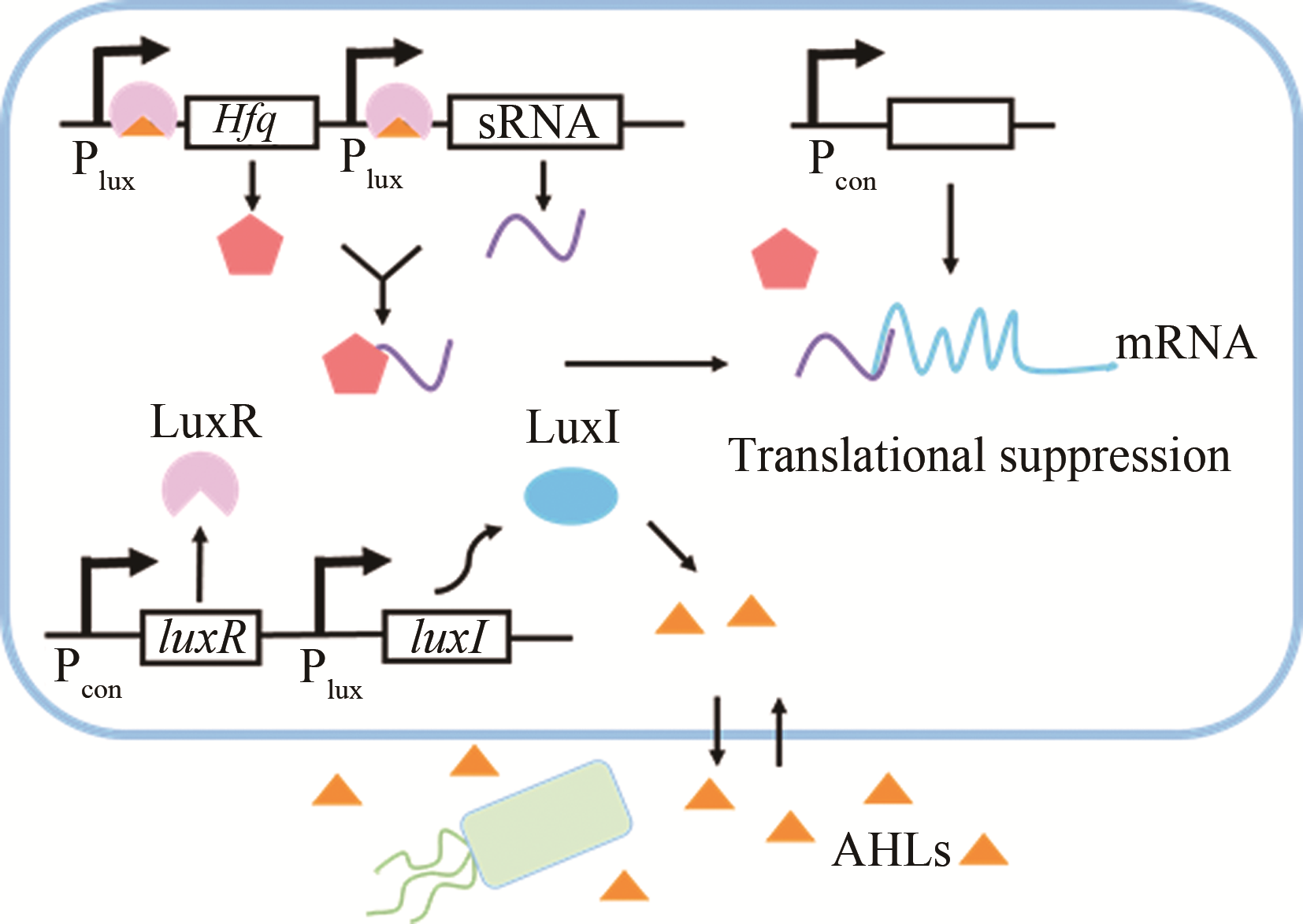
| 群体感应 | AHLs | luxI/luxR | 大肠杆菌 | 红没药烯 | 44%[ |
| AHLs | luxI/luxR | 大肠杆菌 | — | 6%~76%[ | |
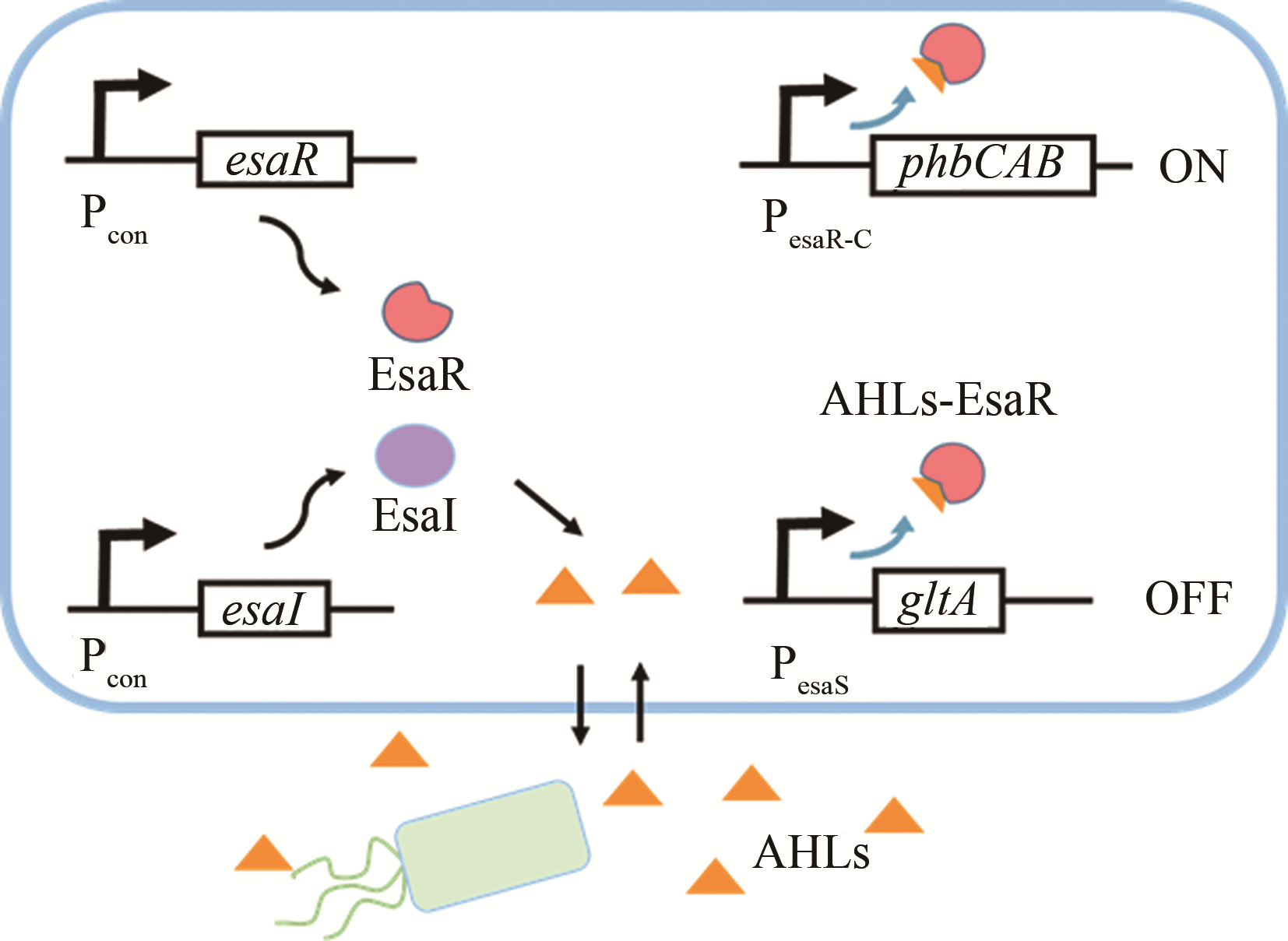
| AHLs | esaI/esaR | 大肠杆菌 | 4-羟基苯乙酸 | 46.4%[ | |
| AHLs | esaI/esaR | 大肠杆菌 | 聚-β-羟丁酸 | 6倍[ | |
| AHLs | esaI/esaR | 大肠杆菌 | 葡萄糖二酸 | 2g/L[ | |
| AHLs | lux、esa | 大肠杆菌 | 水杨酸、柚皮素 | 1.8、6倍[ | |
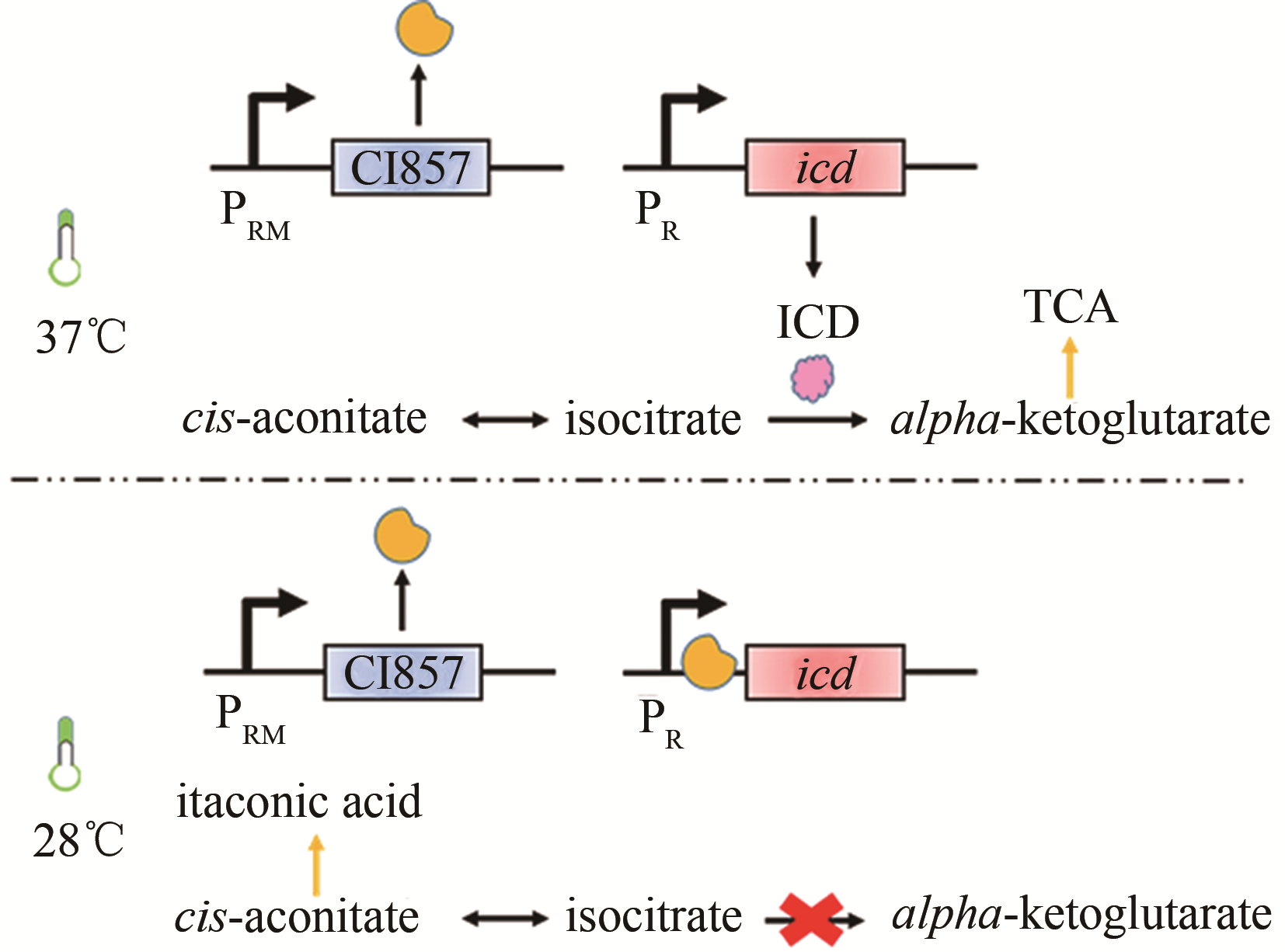
| 发酵条件 | 温度 | CI857 | 大肠杆菌 | 衣康酸 | 47g/L[ |
| 温度 | Gal4M9 | 酵母 | 番茄红素 | 1.12g/L[ | |
| 溶氧 | Pnar | 大肠杆菌 | D-乳酸 | 113.12g/L[ | |
| pH | Pgas | 黑曲霉 | 衣康酸 | 4.92g/L[ | |
| pH | CadCΔ | 大肠杆菌 | 乙二醇 | 170%[ | |
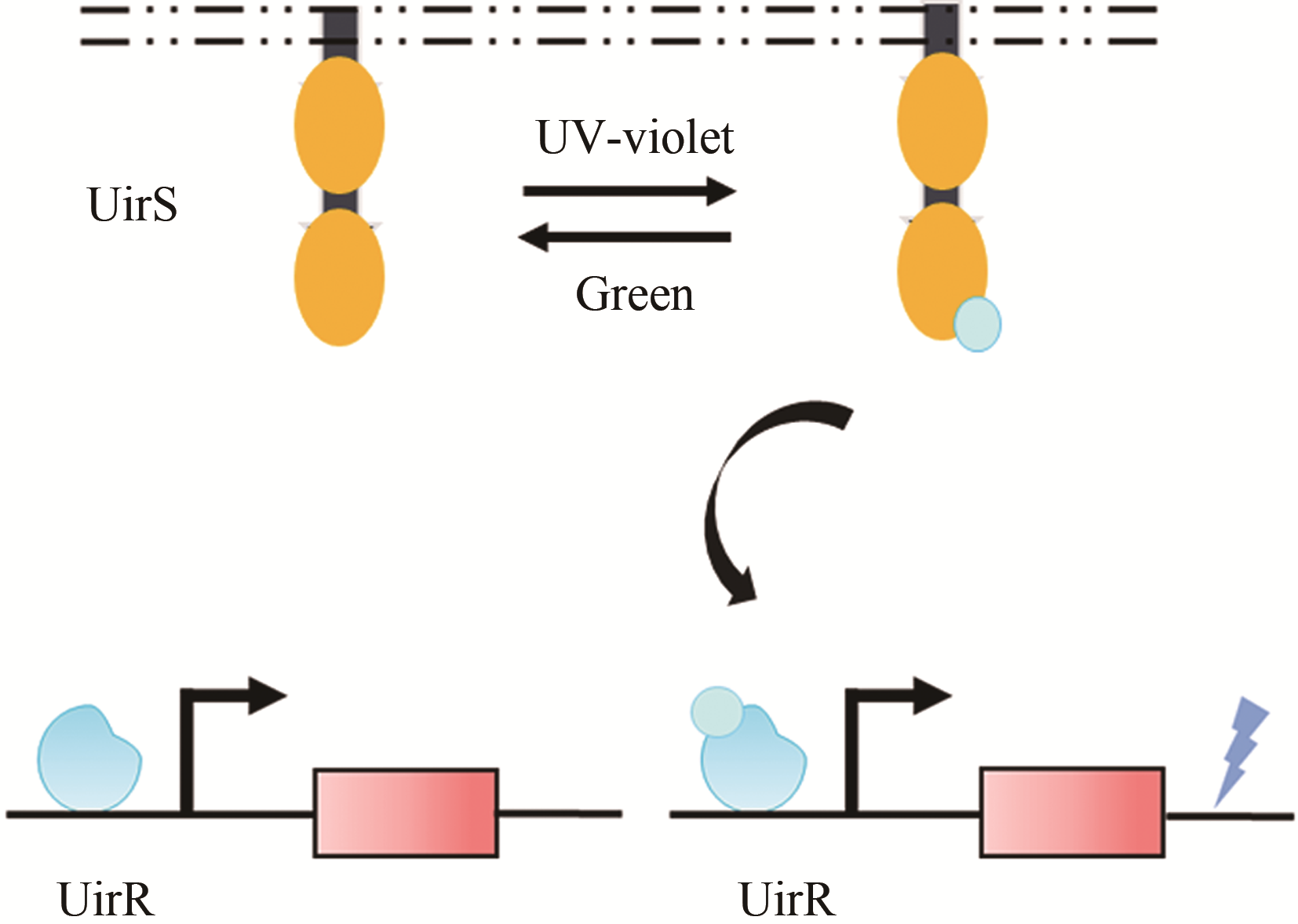
| 光 | UirS/UirR | 大肠杆菌 | — | 6.24倍[ | |
| 光 | Magnets | 大肠杆菌 | — | 300倍[ | |
| 光 | OptoEXP | 酵母 | 异丁醇 | 4倍[ | |
| 葡萄糖 | PHXT1 | 酵母 | 3-羟基丙酸 | 10倍[ | |
| 蛋白水平 | 赖氨酸 | 高丝氨酸脱氢酶 | 大肠杆菌 | 赖氨酸 | —[ |
| — | 振荡器 | 大肠杆菌 | D-木糖酸 | 199.44g/L[ | |
| — | SsrA | 大肠杆菌 | 肌醇 | 2倍[ |
Tab. 1 Applications of dynamic regulation elements in metabolic engineering
| 调控类型 | 输入信号 | 调控元件 | 菌株 | 产物 | 调控效果 |
|---|---|---|---|---|---|
| 转录因子 | 乙酰磷酸 | NRI | 大肠杆菌 | 番茄红素 | 50%[ |
| 丙二酰辅酶A | FapR | 大肠杆菌 | 脂肪酸 | 2.1倍[ | |
| 丙二酰辅酶A | FapR | 大肠杆菌 | 脂肪酸 | 34%[ | |
| 衣康酸 | ItcR | 大肠杆菌 | 衣康酸 | 0.78mmol/L[ | |
| 阿魏酸、香草醛 | HucR | 大肠杆菌 | 香草醛 | 11mmol/L[ | |
| 葡糖胺-6-磷酸 | GamR | 枯草芽孢杆菌 | N-乙酰葡糖胺 | 131.6 g/L[ | |
| 黏糠酸 | CatR | 大肠杆菌 | 黏糠酸 | 16.3倍[ | |
| 核糖开关 | 茶碱 | 茶碱核糖开关 | 大肠杆菌 | — | 3倍[ |
| 硫胺素焦磷酸 | 硫胺素焦磷酸核糖开关 | 米曲霉 | — | 4.7倍[ | |
| 赖氨酸 | 赖氨酸核糖开关 | 谷氨酸棒状杆菌 | 赖氨酸 | 63%[ | |
| 甘氨酸 | 甘氨酸核糖开关 | 大肠杆菌 | 5-氨基酮戊酸 | 11%[ | |
| 群体感应 | AHLs | luxI/luxR | 大肠杆菌 | 红没药烯 | 44%[ |
| AHLs | luxI/luxR | 大肠杆菌 | — | 6%~76%[ | |
| AHLs | esaI/esaR | 大肠杆菌 | 4-羟基苯乙酸 | 46.4%[ | |
| AHLs | esaI/esaR | 大肠杆菌 | 聚-β-羟丁酸 | 6倍[ | |
| AHLs | esaI/esaR | 大肠杆菌 | 葡萄糖二酸 | 2g/L[ | |
| AHLs | lux、esa | 大肠杆菌 | 水杨酸、柚皮素 | 1.8、6倍[ | |
| 发酵条件 | 温度 | CI857 | 大肠杆菌 | 衣康酸 | 47g/L[ |
| 温度 | Gal4M9 | 酵母 | 番茄红素 | 1.12g/L[ | |
| 溶氧 | Pnar | 大肠杆菌 | D-乳酸 | 113.12g/L[ | |
| pH | Pgas | 黑曲霉 | 衣康酸 | 4.92g/L[ | |
| pH | CadCΔ | 大肠杆菌 | 乙二醇 | 170%[ | |
| 光 | UirS/UirR | 大肠杆菌 | — | 6.24倍[ | |
| 光 | Magnets | 大肠杆菌 | — | 300倍[ | |
| 光 | OptoEXP | 酵母 | 异丁醇 | 4倍[ | |
| 葡萄糖 | PHXT1 | 酵母 | 3-羟基丙酸 | 10倍[ | |
| 蛋白水平 | 赖氨酸 | 高丝氨酸脱氢酶 | 大肠杆菌 | 赖氨酸 | —[ |
| — | 振荡器 | 大肠杆菌 | D-木糖酸 | 199.44g/L[ | |
| — | SsrA | 大肠杆菌 | 肌醇 | 2倍[ |
| 1 | SUN X, SHEN X, JAIN R, et al. Synthesis of chemicals by metabolic engineering of microbes[J]. Chemical Society Reviews, 2015, 44(11): 3760-3785. |
| 2 | SHEN X, WANG J, WANG J, et al. High-level de novo biosynthesis of arbutin in engineered Escherichia coli [J]. Metabolic Engineering, 2017, 42: 52-58. |
| 3 | WANG J, SHEN X, YUAN Q, et al. Microbial synthesis of pyrogallol using genetically engineered Escherichia coli [J]. Metabolic Engineering, 2018, 45: 134-141. |
| 4 | ATSUMI S, HANAI T, LIAO J C. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels[J]. Nature, 2008, 451(7174): 86-89. |
| 5 | CHEN X, GAO C, GUO L, et al. DCEO biotechnology: tools to design, construct, evaluate, and optimize the metabolic pathway for biosynthesis of chemicals[J]. Chemical Reviews, 2018, 118(1): 4-72. |
| 6 | SHEN X, WANG J, LI C, et al. Dynamic gene expression engineering as a tool in pathway engineering[J]. Current Opinion in Biotechnology, 2019, 59: 122-129. |
| 7 | TAN S Z, PRATHER K L. Dynamic pathway regulation: recent advances and methods of construction[J]. Current Opinion in Chemical Biology, 2017, 41: 28-35. |
| 8 | XU P, GU Q, WANG W, et al. Modular optimization of multi-gene pathways for fatty acids production in E. coli [J]. Nature Communications, 2013, 4: 1409. |
| 9 | FARMER W R, LIAO J C. Improving lycopene production in Escherichia coli by engineering metabolic control[J]. Nature Biotechnology, 2000, 18(5): 533-537. |
| 10 | DOONG S J, GUPTA A, PRATHER K L J. Layered dynamic regulation for improving metabolic pathway productivity in Escherichia coli [J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(12): 2964-2969. |
| 11 | WU J, LIU Y, ZHAO S, et al. Application of dynamic regulation to increase L-phenylalanine production in Escherichia coli [J]. Journal of Microbiology and Biotechnology, 2019, 29(6): 923-932. |
| 12 | SPITZ F, FURLONG E E. Transcription factors: from enhancer binding to developmental control[J]. Nature Reviews Genetics, 2012, 13(9): 613-626. |
| 13 | ZHANG F, KEASLING J. Biosensors and their applications in microbial metabolic engineering[J]. Trends in Microbiology, 2011, 19(7): 323-329. |
| 14 | XU P. Production of chemicals using dynamic control of metabolic fluxes[J]. Current Opinion in Biotechnology, 2018, 53: 12-19. |
| 15 | WU J, YU O, DU G, et al. Fine-tuning of the fatty acid pathway by synthetic antisense RNA for enhanced (2S)-naringenin production from L-tyrosine in Escherichia coli [J]. Applied and Environmental Microbiology, 2014, 80(23): 7283-7292. |
| 16 | CHEN Z, HUANG J, WU Y, et al. Metabolic engineering of Corynebacterium glutamicum for the production of 3-hydroxypropionic acid from glucose and xylose[J]. Metabolic Engineering, 2017, 39: 151-158. |
| 17 | SHEN X, MAHAJANI M, WANG J, et al. Elevating 4-hydroxycoumarin production through alleviating thioesterase-mediated salicoyl-CoA degradation[J]. Metabolic Engineering, 2017, 42: 59-65. |
| 18 | XU P, LI L, ZHANG F, et al. Improving fatty acids production by engineering dynamic pathway regulation and metabolic control[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(31): 11299-11304. |
| 19 | LIU D, XIAO Y, EVANS B S, et al. Negative feedback regulation of fatty acid production based on a malonyl-CoA sensor-actuator[J]. ACS Synthetic Biology, 2015, 4(2): 132-140. |
| 20 | HANKO E K R, MINTON N P, MALYS N. A transcription factor-based biosensor for detection of itaconic acid[J]. ACS Synthetic Biology, 2018, 7(5): 1436-1446. |
| 21 | KOCH M, PANDI A, BORKOWSKI O, et al. Custom-made transcriptional biosensors for metabolic engineering[J]. Current Opinion in Biotechnology, 2019, 59: 78-84. |
| 22 | LIANG C, ZHANG X, WU J, et al. Dynamic control of toxic natural product biosynthesis by an artificial regulatory circuit[J]. Metabolic Engineering, 2020, 57: 239-246. |
| 23 | DABIRIAN Y, LI X, CHEN Y, et al. Expanding the dynamic range of a transcription factor-based biosensor in Saccharomyces cerevisiae [J]. ACS Synthetic Biology, 2019, 8(9): 1968-1975. |
| 24 | CHEN Y, HO J M L, SHIS D L, et al. Tuning the dynamic range of bacterial promoters regulated by ligand-inducible transcription factors[J]. Nature Communications, 2018, 9(1): 64. |
| 25 | MANNAN A A, LIU D, ZHANG F, et al. Fundamental design principles for transcription-factor-based metabolite biosensors[J]. ACS Synthetic Biology, 2017, 6(10): 1851-1859. |
| 26 | TIAN J, YANG G, GU Y, et al. Developing an endogenous quorum-sensing based CRISPRi circuit for autonomous and tunable dynamic regulation of multiple targets in industrial Streptomyces [J]. Nucleic Acids Research, 2020. DOI: 10.1093/nar/gkaa602 . |
| 27 | CHO S, SHIN J, CHO B K. Applications of CRISPR/Cas system to bacterial metabolic engineering[J]. International Journal of Molecular Sciences, 2018, 19(4): 1089. |
| 28 | WU Y, CHEN T, LIU Y, et al. Design of a programmable biosensor-CRISPRi genetic circuits for dynamic and autonomous dual-control of metabolic flux in Bacillus subtilis [J]. Nucleic Acids Research, 2020, 48(2): 996-1009. |
| 29 | YANG Y, LIN Y, WANG J, et al. Sensor-regulator and RNAi based bifunctional dynamic control network for engineered microbial synthesis[J]. Nature Communications, 2018, 9(1): 3043. |
| 30 | GALIZI R, JARAMILLO A. Engineering CRISPR guide RNA riboswitches for in vivo applications[J]. Current Opinion in Biotechnology, 2019, 55: 103-113. |
| 31 | LIU D, EVANS T, ZHANG F. Applications and advances of metabolite biosensors for metabolic engineering[J]. Metabolic Engineering, 2015, 31: 35-43. |
| 32 | JANG S, JANG S, XIU Y, et al. Development of artificial riboswitches for monitoring of naringenin in vivo [J]. ACS Synthetic Biology, 2017, 6(11): 2077-2085. |
| 33 | BREAKER R R. Riboswitches and translation control[J]. Cold Spring Harbor Perspectives in Biology, 2018, 10(11): a032797. |
| 34 | LOTZ T S, SUESS B. Small-molecule-binding riboswitches[J]. Microbiology Spectrum, 2018, 6(4): 75-88. |
| 35 | WACHSMUTH M, FINDEISS S, WEISSHEIMER N, et al. De novo design of a synthetic riboswitch that regulates transcription termination[J]. Nucleic Acids Research, 2013, 41(4): 2541-2551. |
| 36 | MASUDA S, LZAWA S. Applied RNA bioscience[M]. Singapore: Springer, 2018: 33-46. |
| 37 | ZHOU L B, ZENG A P. Exploring lysine riboswitch for metabolic flux control and improvement of L-lysine synthesis in Corynebacterium glutamicum [J]. ACS Synthetic Biology, 2015, 4(6): 729-734. |
| 38 | ZHOU L, REN J, LI Z, et al. Characterization and engineering of a Clostridium glycine riboswitch and its use to control a novel metabolic pathway for 5-aminolevulinic acid production in Escherichia coli [J]. ACS Synthetic Biology, 2019, 8(10): 2327-2335. |
| 39 | PANG Q, HAN H, LIU X, et al. In vivo evolutionary engineering of riboswitch with high-threshold for N-acetylneuraminic acid production[J]. Metabolic Engineering, 2020, 59: 36-43. |
| 40 | HAN L, HAN D, LI L, et al. Discovery and identification of medium‐chain fatty acid responsive promoters in Saccharomyces cerevisiae [J]. Engineering in Life Sciences, 2020, 5: 186-196. |
| 41 | MAURY J, KANNAN S, JENSEN N B, et al. Glucose-dependent promoters for dynamic regulation of metabolic pathways[J]. Frontiers in Bioengineering and Biotechnology, 2018, 6: 63. |
| 42 | ABISADO R G, BENOMAR S, KLAUS J R, et al. Bacterial quorum sensing and microbial community interactions[J]. mBio, 2018, 9(3): e02331-17. |
| 43 | DANG H T, KOMATSU S, MASUDA H, et al. Characterization of LuxI and LuxR protein homologs of N-Acylhomoserine lactone-dependent quorum sensing system in Pseudoalteromonas sp. 520P1[J]. Marine Biotechnology, 2017, 19(1): 1-10. |
| 44 | WHITELEY M, DIGGLE S P, GREENBERG E P. Progress in and promise of bacterial quorum sensing research[J]. Nature, 2017, 551(7680): 313-320. |
| 45 | KIM E M, WOO H M, TIAN T, et al. Autonomous control of metabolic state by a quorum sensing (QS)-mediated regulator for bisabolene production in engineered E. coli [J]. Metabolic Engineering, 2017, 44: 325-336. |
| 46 | BAO S H, LI W Y, LIU C J, et al. Quorum-sensing based small RNA regulation for dynamic and tuneable gene expression[J]. Biotechnology Letters, 2019, 41(10): 1147-1154. |
| 47 | KIMATU J N. Advances in plant pathology[M]. London: IntechOpen, 2018: 68-89. |
| 48 | PAPENFORT K, BASSLER B L. Quorum sensing signal-response systems in Gram-negative bacteria[J]. Nature Reviews Microbiology, 2016, 14(9): 576-588. |
| 49 | SHEN Y P, FONG L S, YAN Z B, et al. Combining directed evolution of pathway enzymes and dynamic pathway regulation using a quorum-sensing circuit to improve the production of 4-hydroxyphenylacetic acid in Escherichia coli [J]. Biotechnology for Biofuels, 2019, 12: 94. |
| 50 | GU F, JIANG W, MU Y, et al. Quorum sensing-based dual-function switch and its application in solving two key metabolic engineering problems[J]. ACS Synthetic Biology, 2020, 9(2): 209-217. |
| 51 | GUPTA A, REIZMAN I M, REISCH C R, et al. Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit[J]. Nature Biotechnology, 2017, 35(3): 273-279. |
| 52 | DOONG S J, GUPTA A, PRATHER K L J. Layered dynamic regulation for improving metabolic pathway productivity in Escherichia coli [J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(12): 2964-2969. |
| 53 | DINH C V, PRATHER K L J. Development of an autonomous and bifunctional quorum-sensing circuit for metabolic flux control in engineered Escherichia coli [J]. Proceedings of the National Academy of Sciences of the United States of America, 2019, 116(51): 25562-25568. |
| 54 | ZENG W, DU P, LOU Q, et al. Rational design of an ultrasensitive quorum-sensing switch[J]. ACS Synthetic Biology, 2017, 6(8): 1445-1452. |
| 55 | DAER R, BARRETT C M, MELENDEZ E L, et al. Characterization of diverse homoserine lactone synthases in Escherichia coli [J]. Plos One, 2018, 13(8): e0202294. |
| 56 | TEKEL S J, SMITH C L, LOPEZ B, et al. Engineered orthogonal quorum sensing systems for synthetic gene regulation in Escherichia coli [J]. Frontiers in Bioengineering and Biotechnology, 2019, 7: 80. |
| 57 | SCOTT S R, HASTY J. Quorum sensing communication modules for microbial consortia[J]. ACS Synthetic Biology, 2016, 5(9): 969-977. |
| 58 | HARDER B J, BETTENBROCK K, KLAMT S. Temperature-dependent dynamic control of the TCA cycle increases volumetric productivity of itaconic acid production by Escherichia coli [J]. Biotechnology and Bioengineering, 2018, 115(1): 156-164. |
| 59 | ZHOU P, XIE W, YAO Z, et al. Development of a temperature-responsive yeast cell factory using engineered Gal4 as a protein switch[J]. Biotechnology and Bioengineering, 2018, 115(5): 1321-1330. |
| 60 | HWANG H J, KIM J W, JU S Y, et al. Application of an oxygen-inducible nar promoter system in metabolic engineering for production of biochemicals in Escherichia coli [J]. Biotechnology and Bioengineering, 2017, 114(2): 468-473. |
| 61 | YIN X, SHIN H D, LI J, et al. Pgas, a low-pH-induced promoter, as a tool for dynamic control of gene expression for metabolic engineering of Aspergillus niger [J]. Applied and Environmental Microbiology, 2017, 83(6): e03222-16. |
| 62 | BANARES A B, VALDEHUESA K N G, RAMOS K R M, et al. A pH-responsive genetic sensor for the dynamic regulation of D-xylonic acid accumulation in Escherichia coli [J]. Applied Microbiology and Biotechnology, 2020, 104(5): 2097-2108. |
| 63 | LIU Z, ZHANG J, JIN J, et al. Programming bacteria with light-sensors and applications in synthetic biology[J]. Frontiers in Microbiology, 2018, 9: 2692. |
| 64 | TANDAR S T, SENOO S, TOYA Y, et al. Optogenetic switch for controlling the central metabolic flux of Escherichia coli [J]. Metabolic Engineering, 2019, 55: 68-75. |
| 65 | RAMAKRISHNAN P, TABOR J J. Repurposing Synechocystis PCC6803 UirS-UirR as a UV-violet/Green photoreversible transcriptional regulatory tool in E. coli [J]. ACS Synthetic Biology, 2016, 5(7): 733-740. |
| 66 | BAUMSCHLAGER A, AOKI S K, KHAMMASH M. Dynamic blue light-inducible T7 RNA polymerases (Opto-T7RNAPs) for precise spatiotemporal gene expression control[J]. ACS Synthetic Biology, 2017, 6(11): 2157-2167. |
| 67 | ZHAO E M, ZHANG Y, MEHL J, et al. Optogenetic regulation of engineered cellular metabolism for microbial chemical production[J]. Nature, 2018, 555(7698): 683-687. |
| 68 | SHEETS M B, WONG W W, DUNLOP M J. Light-inducible recombinases for bacterial optogenetics[J]. ACS Synthetic Biology, 2020, 9(2): 227-235. |
| 69 | BOTHFELD W, KAPOV G, TYO K E J. A glucose-sensing toggle switch for autonomous, high productivity genetic control[J]. ACS Synthetic Biology, 2017, 6(7): 1296-1304. |
| 70 | DAVID F, NIELSEN J, SIEWERS V. Flux control at the malonyl-CoA node through hierarchical dynamic pathway regulation in Saccharomyces cerevisiae [J]. ACS Synthetic Biology, 2016, 5(3): 224-233. |
| 71 | MOTLAGH H N, WRABL J O, LI J, et al. The ensemble nature of allostery[J]. Nature, 2014, 508(7496): 331-339. |
| 72 | DOKHOLYAN N V. Controlling allosteric networks in proteins[J]. Chemical Reviews, 2016, 116(11): 6463-6487. |
| 73 | CHEN Z, RAPPERT S, ZENG A P. Rational design of allosteric regulation of homoserine dehydrogenase by a nonnatural inhibitor L-lysine[J]. ACS Synthetic Biology, 2015, 4(2): 126-131. |
| 74 | GAO C, HOU J, XU P, et al. Programmable biomolecular switches for rewiring flux in Escherichia coli [J]. Nature Communications, 2019, 10(1): 3751. |
| 75 | BROCKMAN I M, PRATHER K L J. Dynamic knockdown of E. coli central metabolism for redirecting fluxes of primary metabolites[J]. Metabolic Engineering, 2015, 28: 104-113. |
| 76 | HE F, STUMPF M P H. Quantifying dynamic regulation in metabolic pathways with nonparametric flux inference[J]. Biophysical Journal, 2019, 116(10): 2035-2046. |
| 77 | JABARIVELISDEH B, WALDHERR S. Optimization of bioprocess productivity based on metabolic-genetic network models with bilevel dynamic programming[J]. Biotechnology and Bioengineering, 2018, 115(7): 1829-1841. |
| [1] | GUO Shuyuan, ZHANG Qiannan, Gulikezi· MAIMAITIREXIATI, YANG Yiqun, YU Tao. Advances in microbial production of liquid biofuels [J]. Synthetic Biology Journal, 2025, 6(1): 18-44. |
| [2] | SHAO Mingwei, SUN Simian, YANG Shimao, CHEN Guoqiang. Bioproduction based on extremophiles [J]. Synthetic Biology Journal, 2024, 5(6): 1419-1436. |
| [3] | ZHAO Liang, LI Zhenshuai, FU Liping, LYU Ming, WANG Shi’an, ZHANG Quan, LIU Licheng, LI Fuli, LIU Ziyong. Progress in biomanufacturing of lipids and single cell protein from one-carbon compounds [J]. Synthetic Biology Journal, 2024, 5(6): 1300-1318. |
| [4] | ZHU Fanghuan, CEN Xuecong, CHEN Zhen. Research progress of diols production by microbes [J]. Synthetic Biology Journal, 2024, 5(6): 1367-1385. |
| [5] | ZHENG Haotian, LI Chaofeng, LIU Liangxu, WANG Jiawei, LI Hengrun, NI Jun. Design, optimization and application of synthetic carbon-negative phototrophic community [J]. Synthetic Biology Journal, 2024, 5(5): 1189-1210. |
| [6] | YU Wei, GAO Jiaoqi, ZHOU Yongjin. Bioconversion of one carbon feedstocks for producing organic acids [J]. Synthetic Biology Journal, 2024, 5(5): 1169-1188. |
| [7] | CHEN Xiwei, ZHANG Huaran, ZOU Yi. Biosynthesis and metabolic engineering of fungal non-ribosomal peptides [J]. Synthetic Biology Journal, 2024, 5(3): 571-592. |
| [8] | XIE Huang, ZHENG Yilei, SU Yiting, RUAN Jingyi, LI Yongquan. An overview on reconstructing the biosynthetic system of actinomycetes for polyketides production [J]. Synthetic Biology Journal, 2024, 5(3): 612-630. |
| [9] | ZHAO Jingyu, ZHANG Jian, QI Qingsheng, WANG Qian. Research progress in biosensors based on bacterial two-component systems [J]. Synthetic Biology Journal, 2024, 5(1): 38-52. |
| [10] | SUN Huili, CUI Jinyu, LUAN Guodong, LYU Xuefeng. Progress of cyanobacterial synthetic biotechnology for efficient light-driven carbon fixation and ethanol production [J]. Synthetic Biology Journal, 2023, 4(6): 1161-1177. |
| [11] | YAN Xiongying, WANG Zhen, LOU Jiyun, ZHANG Haoyu, HUANG Xingyu, WANG Xia, YANG Shihui. Progress in the construction of microbial cell factories for efficient biofuel production [J]. Synthetic Biology Journal, 2023, 4(6): 1082-1121. |
| [12] | CHENG Zhenzhen, ZHANG Jian, GAO Cong, LIU Liming, CHEN Xiulai. Progress in metabolic engineering of microorganisms for the utilization of formate [J]. Synthetic Biology Journal, 2023, 4(4): 756-778. |
| [13] | LIU Jiayu, YANG Zhihan, YANG Lei, ZHU Liying, ZHU Zhengming, JIANG Ling. Advances in the development of Clostridium tyrobutyricum cell factories driven by synthetic biotechnology [J]. Synthetic Biology Journal, 2022, 3(6): 1174-1200. |
| [14] | GUO Shuyuan, WU Lianghuan, LIU Xiangjian, WANG Bo, YU Tao. Developing C1-based metabolic network in methylotrophy for biotransformation [J]. Synthetic Biology Journal, 2022, 3(1): 116-137. |
| [15] | CHEN Jiuzhou, WANG Yu, PU Wei, ZHENG Ping, SUN Jibin. Advances and perspective on bioproduction of 5-aminolevulinic acid [J]. Synthetic Biology Journal, 2021, 2(6): 1000-1016. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||