Synthetic Biology Journal ›› 2020, Vol. 1 ›› Issue (3): 385-394.DOI: 10.12211/2096-8280.2020-065
• Invited Review • Previous Articles
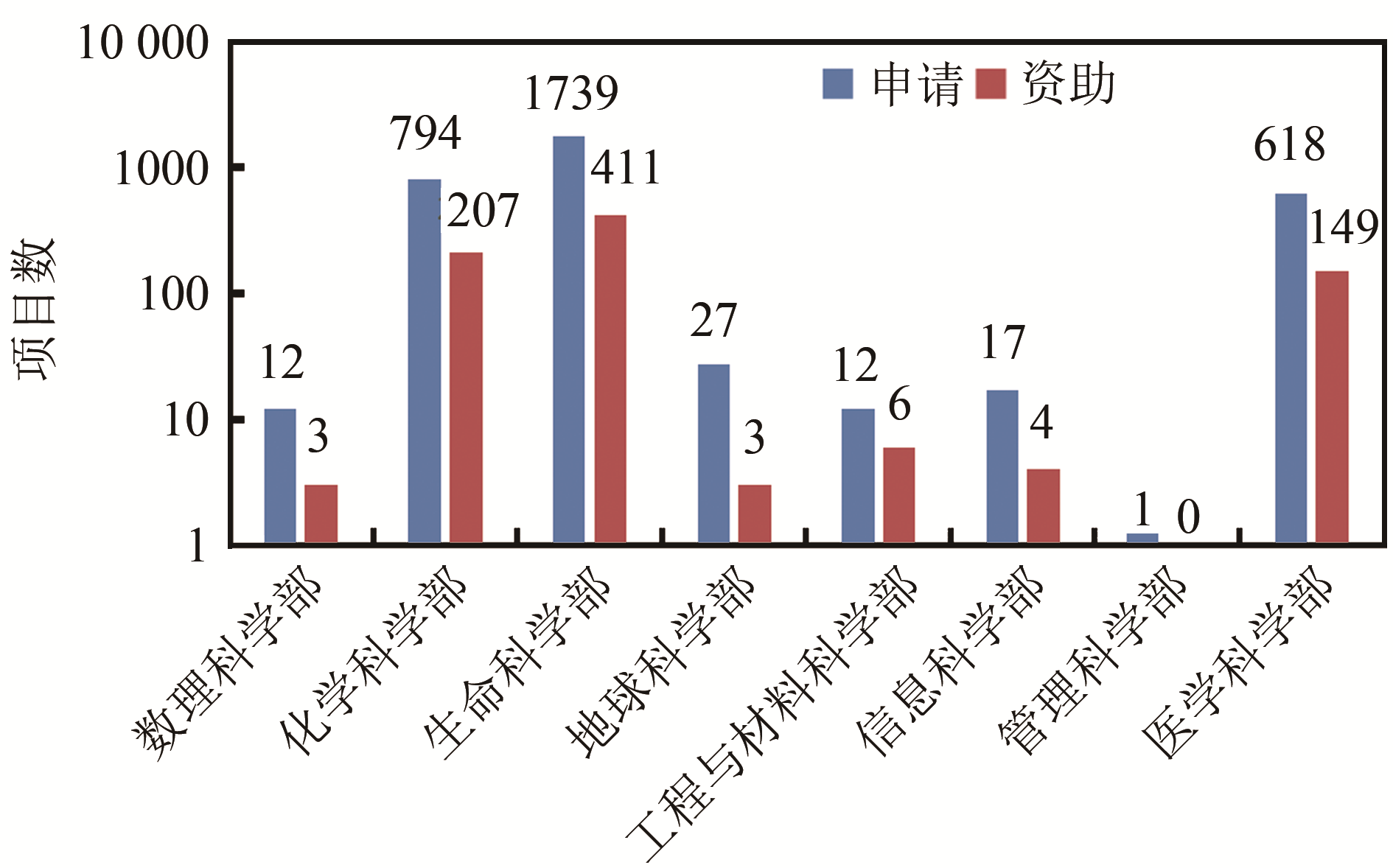
Grant and funding for synthetic biology at NSFC from 2010 to 2019
DU Quansheng1( ), HONG Wei2, ZU Yan3
), HONG Wei2, ZU Yan3
- 1.Department of Life Sciences,Natural Science Foundation of China,Beijing 100085,China
2.Key Laboratory of Endemic and Ethnic Diseases,Guizhou Medical University,Ministry of Education,Guiyang 550004,Guizhou,China
3.School of Mechanical and Material Engineering,North China University of Technology,Beijing 100144,China
-
Received:2020-05-19Revised:2020-07-13Online:2020-09-29Published:2020-06-30 -
Contact:DU Quansheng
2010—2019年国家自然科学基金资助合成生物学领域情况
- 1.国家自然科学基金委员会生命科学部,北京 100085
2.贵州医科大学地方病与少数民族疾病教育部重点实验室,贵州 贵阳 550004
3.北方工业大学机械与材料工程学院,北京 100144
-
通讯作者:杜全生 -
作者简介:杜全生,(1980—),男,博士,副研究员,研究方向为遗传学。E-mail:duqs@nsfc.gov.cn
CLC Number:
Cite this article
DU Quansheng, HONG Wei, ZU Yan. Grant and funding for synthetic biology at NSFC from 2010 to 2019[J]. Synthetic Biology Journal, 2020, 1(3): 385-394.
杜全生, 洪伟, 祖岩. 2010—2019年国家自然科学基金资助合成生物学领域情况[J]. 合成生物学, 2020, 1(3): 385-394.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2020-065
| 1 | WU Y, LI B Z, ZHAO M, et al. Bug mapping and fitness testing of chemically synthesized chromosome X[J]. Science, 2017, 355(6329): aaf4706. |
| 2 | XIE Z X, LI B Z, MITCHELL L A, et al. “Perfect” designer chromosome V and behavior of a ring derivative[J]. Science, 2017, 355(6329): eaaf4704. |
| 3 | QIN Z, STOILOV P, ZHANG X, et al. SEASTAR: systematic evaluation of alternative transcription start sites in RNA[J].Nucleic Acids Research, 2018, 46(8): e45. |
| 4 | CUI J, CUI H, YANG M, et al. Tongue coating microbiome as a potential biomarker for gastritis including precancerous cascade[J]. Protein Cell, 2019, 10(7): 496-509. |
| 5 | MIAO Z, DENG K, WANG X, et al. DEsingle for detecting three types of differential expression in single-cell RNA-seq data[J]. Bioinformatics, 2018, 34(18): 3223-3224. |
| 6 | CHEN W H, QIN Z J, WANG J, et al. The MASTER (methylation-assisted tailorable ends rational) ligation method for seamless DNA assembly[J]. Nucleic Acids Research, 2013, 41(8): e93. |
| 7 | LU AN G, LU X. Tailoring cyanobacterial cell factory for improved industrial properties[J]. Biotechnology Advances, 2018, 36(2): 430-442. |
| 8 | LUAN G, ZHANG S, LU X. Engineering Cyanobacteria chassis cells toward more efficient photosynthesis[J]. Current Opinion in Biotechnology, 2020, 62: 1-6. |
| 9 | ZHANG L, LIANG Y, WU W, et al. Microbial synthesis of propane by engineering valine pathway and aldehyde-deformylating oxygenase[J]. Biotechnology for Biofuels, 2016, 9(1): 80. |
| 10 | LIU W, LUO Z, WANG Y, et al. Rapid pathway prototyping and engineering using in vitro and in vivo synthetic genome SCRaMbLE-in methods[J]. Nature Communications, 2018, 9(1): 1936. |
| 11 | LIN Y, ZOU X, ZHENG Y, et al. Improving chromosome synthesis with a semiquantitative phenotypic assay and refined assembly strategy[J]. ACS Synthetic Biology, 2019, 8(10): 2203-2211. |
| 12 | JIAO Z, FENGHUI QIAN, FENG D, et al. De novo engineering of Corynebacterium glutamicum for l-proline production[J]. ACS Synthetic Biology, 2020, DOI: https://doi.org/10.1021/acssynbio.0c00249 . |
| 13 | SHAO J, WANG M, YU G, et al. Synthetic far-red light-mediated CRISPR-dCas9 device for inducing functional neuronal differentiation[J]. Proceedings of The National Academy of Sciences of The United States of America, 2018, 115(29): 6722-6730. |
| 14 | XIE M, YE H, WANG H, et al. beta-cell-mimetic designer cells provide closed-loop glycemic control[J]. Science, 2016, 354(6317): 1296-1301. |
| 15 | ZHANG Z B, WANG Q Y, KE Y X, et al. Design of tunable oscillatory dynamics in a synthetic NF-kappaB signaling circuit[J]. Cell Systems, 2017, 5(5): 460-470. |
| 16 | LIU R, ZHU F, LU L, et al. Metabolic engineering of fatty acyl-ACP reductase-dependent pathway to improve fatty alcohol production in Escherichia coli [J]. Metabolic Engineering, 2014, 22: 10-21. |
| 17 | GUO D, ZHU J, DENG Z, et al. Metabolic engineering of Escherichia coli for production of fatty acid short-chain esters through combination of the fatty acid and 2-keto acid pathways[J]. Metabolic Engineering, 2014, 22: 69-75. |
| 18 | WU T, YE L, ZHAO D, et al. Membrane engineering-A novel strategy to enhance the production and accumulation of beta-carotene in Escherichia coli [J]. Metabolic Engineering, 2017, 43(Pt A): 85-91. |
| 19 | LI Q, FAN F, GAO X, et al. Balanced activation of IspG and IspH to eliminate MEP intermediate accumulation and improve isoprenoids production in Escherichia coli [J]. Metabolic Engineering, 2017, 44: 13-21. |
| 20 | ZHU X, ZHAO D, QIU H, et al. The CRISPR/Cas9-facilitated multiplex pathway optimization (CFPO) technique and its application to improve the Escherichia coli xylose utilization pathway[J]. Metabolic Engineering, 2017, 43(Pt A): 37-45. |
| 21 | GU Y, XU X, WU Y, et al. Advances and prospects of Bacillus subtilis cellular factories: From rational design to industrial applications[J]. Metabolic Engineering, 2018, 50: 109-121. |
| 22 | WU Y, CHEN T, LIU Y, et al. Design of a programmable biosensor-CRISPRi genetic circuits for dynamic and autonomous dual-control of metabolic flux in Bacillus subtilis[J]. Nucleic Acids Research, 2020, 48(2):996-1009. |
| 23 | NIU T, LIU Y, LI J, et al. Engineering a glucosamine-6-phosphate responsive glmS ribozyme switch enables dynamic control of metabolic flux in Bacillus subtilis for overproduction of N-acetylglucosamine[J]. ACS Synthetic Biology 2018, 7(10): 2423-2435. |
| 24 | SHAO Y, LU N, XUE X, et al. Creating functional chromosome fusions in yeast with CRISPR-Cas9[J]. Nature Protocols, 2019, 14(8): 2521-2545. |
| 25 | ZHANG Y, WEN W H, PU J Y,et al. Extracellularly oxidative activation and inactivation of matured prodrug for cryptic self-resistance in naphthyridinomycin biosynthesis[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(44): 11232-11237. |
| 26 | LI W N, MA L, SHEN X L,et al. Targeting metabolic driving and intermediate influx in lysine catabolism for high-level glutarate production[J]. Nature Communications, 2019, 10(1): 3337. |
| 27 | XU J Y, XU Y, XU Z, et al. Protein acylation is a general regulatory mechanism in biosynthetic pathway of acyl-CoA-derived natural products[J]. Cell Chemical Biology, 2018, 25(8): 984-995. |
| 28 | LUO Y Z, LI B Z, LIU D, et al. Engineered biosynthesis of natural products in heterologous hosts[J]. Chemical Society Reviews, 2015, 44(15): 5265-5290. |
| 29 | TAN Z T, ZHU C J, FU J W, et al. Regulating cofactor balance in vivo with a synthetic flavin analogue[J]. Angewandte Chemie-International Edition, 2018, 57(50): 16464-16468. |
| 30 | CAO W, MA W, WANG X, et al. Enhanced pinocembrin production in Escherichia coli by regulating cinnamic acid metabolism.[J]. Scientific Reports, 2016, 6(1): 32640. |
| 31 | LIU Z, LI X, ZHANG J T, et al. Autism-like behaviours and germline transmission in transgenic monkeys overexpressing MeCP2[J]. Nature, 2016, 530(7588): 98-102. |
| 32 | YU X, QIU W, YANG L, et al. Defining multistep cell fate decision pathways during pancreatic development at single‐cell resolution[J]. The EMBO Journal, 2019, 38(8): e100164. |
| 33 | LU Y L, ZHOU Y P, TIAN W D. Combining Hi-C data with phylogenetic correlation to predict the target genes of distal regulatory elements in human genome[J]. Nucleic Acids Research, 2013, 41(22). |
| 34 | ZHENG X, CHENG Q, YAO F, et al. Biosynthesis of the pyrrolidine protein synthesis inhibitor anisomycin involves novel gene ensemble and cryptic biosynthetic steps[J]. Proceedings of The National Academy of Sciences of The United States of America, 2017, 114(16):4 135-4140. |
| 35 | ZHANG Y, ZOU Y, BROCK N L, et al. Characterization of 2-oxindole forming heme enzyme MarE, expanding the functional diversity of the tryptophan dioxygenase superfamily[J]. Journal of The American Chemical Society, 2017, 139(34): 11887-11894. |
| 36 | HUANG T, DUAN Y, ZOU Y, et al. NRPS protein MarQ catalyzes flexible adenylation and specific S-methylation[J].ACS Chemical Biology, 2018, 13(9): 2387-2391. |
| 37 | YANG J, JIANG S, LIU X, et al. Aptamer-binding directed DNA origami pattern for logic gates[J]. ACS Applied Materials & Interfaces, 2016, 8(49): 34054-34060. |
| 38 | YIN L, GUO X, LIU L, et al. Self-assembled multimeric-enzyme nanoreactor for robust and efficient biocatalysis[J]. ACS Biomaterials Science & Engineering, 2018, 4(6): 2095-2099. |
| 39 | HUANG C, YANG C, FANG Z, et al. Discovery of stealthin derivatives and implication of the amidotransferase FlsN3 in the biosynthesis of nitrogen-containing fluostatins[J]. Marine Drugs, 2019, 17(3): 150. |
| 40 | ZHANG D, ZHANG F, LIU W, et al. A KAS-III heterodimer in lipstatin biosynthesis nondecarboxylatively condenses C8 and C14 fatty acyl-CoA substrates by a variable mechanism during the establishment of a C22 aliphatic skeleton.[J]. Journal of the American Chemical Society, 2019, 141(9): 3993-4001. |
| 41 | WANG J, LIU Y, LIU Y, et al. Time-resolved protein activation by proximal decaging in living systems[J]. Nature, 2019, 569(7757): 509-513. |
| 42 | JI Z, NIE Q, YIN Y, et al. Activation and characterization of a cryptic gene cluster reveal two series of aromatic polyketides biosynthesized by divergent tic tailoring pathways[J]. Angewandte Chemie-International Edition,2019, 58: 18046-18054. |
| 43 | MA W, CAO W, ZHANG B, et al. Engineering a pyridoxal 5'-phosphate supply for cadaverine production by using Escherichia coli whole-cell biocatalysis[J]. Scientific Reports, 2015, 5(1): 15630-15630. |
| 44 | 钱万强, 江海燕, 朱庆平, 等. 国内外合成生物学资助体系及产业投入分析[J].中国基础科学, 2014(1): 49-52. |
| QIAN W Q, JIANG H Y, ZHU Q P, et al. Analysis of the funding systems and industry investment of synthetic biology in China and main developed countries[J]. China Basic Science, 2014(1): 49-52. | |
| 45 | 杜瑾, 刘夺, 赵广荣, 等. 合成生物学学科发展概况[J].中国科学基金, 2011, 25(3): 143-147. |
| DU J, LIU D, ZHAO G R, et al. General situation on the disciplinary development in synthetic biology[J]. Science Foundation in China, 2011, 25(3): 143-147. | |
| 46 | 赵国屏. 合成生物学——生物工程产业化发展的新时期[J].生物产业技术, 2019(1): 2. |
| Zhao G P. Synthetic biology——The new era of industrial development of bioengineering[J]. Biotechnology & Business, 2019(1): 2. | |
| 47 | SLEATOR R D. JCVI-syn3.0-A synthetic genome stripped bare![J]. Bioengineered, 2016, 7(2): 53-56. |
| 48 | 马延和, 江会锋, 娄春波, 等. 合成生物与生物安全[J].中国科学院院刊, 2016, 31(4): 432-438. |
| MA Y H, JIANG H F, LOU C B, et al. Synthetic life and biosecurity[J]. Bulletin of Chinese Academy of Sciences, 2016, 31(4): 432-438. | |
| 49 | 肖尧. 美国国家科学院发布《合成生物学时代的生物防御》报告[J].科技中国, 2018(7): 106-106. |
| XIAO R. Biodefense in the age of synthetic biology[J]. China Scitechnology Business, 2018(7): 106. | |
| 50 | 刘晓, 曾艳, 王力为, 等. 创新政策体系保障合成生物学科技与产业发展[J].中国科学院院刊, 2018, 33(11): 1260-1268. |
| LIU X, ZENG Y, WANG L W, et al. Innovative policy system to ensure the development of synthetic biology[J]. Bulletin of Chinese Academy of Sciences, 2018, 33(11): 1260-1268. | |
| 51 | 周光明, 陈大明, 熊燕, 等. 英国合成生物学规划及其影响与启示[J].中国细胞生物学学报, 2019, 41(11): 2091-2100. |
| ZHOU G M, CHEN D M, XIONG Y, et al. UK synthetic biology strategic planning and its enlightenment[J]. Chinese Journal of Cell Biology, 2019, 41(11): 2091-2100. | |
| 52 | 王璞玥, 唐鸿志, 吴震州, 等. “合成生物学”研究前沿与发展趋势[J].中国科学基金, 2018, 32(5): 545-551. |
| WANG P Y, TANG H Z, WU Z Z, et al. Frontiers and trends in synthetic biology[J]. Science Foundation in China, 2018, 32(5): 545-551. |
| [1] | GAO Ge, BIAN Qi, WANG Baojun. Synthetic genetic circuit engineering: principles, advances and prospects [J]. Synthetic Biology Journal, 2025, 6(1): 45-64. |
| [2] | LI Jiyuan, WU Guosheng. Two hypothesises for the origins of organisms from the synthetic biology perspective [J]. Synthetic Biology Journal, 2025, 6(1): 190-202. |
| [3] | JIAO Hongtao, QI Meng, SHAO Bin, JIANG Jinsong. Legal issues for the storage of DNA data [J]. Synthetic Biology Journal, 2025, 6(1): 177-189. |
| [4] | TANG Xinghua, LU Qianneng, HU Yilin. Philosophical reflections on synthetic biology in the Anthropocene [J]. Synthetic Biology Journal, 2025, 6(1): 203-212. |
| [5] | XU Huaisheng, SHI Xiaolong, LIU Xiaoguang, XU Miaomiao. Key technologies for DNA storage: encoding, error correction, random access, and security [J]. Synthetic Biology Journal, 2025, 6(1): 157-176. |
| [6] | SHI Ting, SONG Zhan, SONG Shiyi, ZHANG Yi-Heng P. Job. In vitro BioTransformation (ivBT): a new frontier of industrial biomanufacturing [J]. Synthetic Biology Journal, 2024, 5(6): 1437-1460. |
| [7] | CHAI Meng, WANG Fengqing, WEI Dongzhi. Synthesis of organic acids from lignocellulose by biotransformation [J]. Synthetic Biology Journal, 2024, 5(6): 1242-1263. |
| [8] | SHAO Mingwei, SUN Simian, YANG Shimao, CHEN Guoqiang. Bioproduction based on extremophiles [J]. Synthetic Biology Journal, 2024, 5(6): 1419-1436. |
| [9] | CHEN Yu, ZHANG Kang, QIU Yijing, CHENG Caiyun, YIN Jingjing, SONG Tianshun, XIE Jingjing. Progress of microbial electrosynthesis for conversion of CO2 [J]. Synthetic Biology Journal, 2024, 5(5): 1142-1168. |
| [10] | ZHENG Haotian, LI Chaofeng, LIU Liangxu, WANG Jiawei, LI Hengrun, NI Jun. Design, optimization and application of synthetic carbon-negative phototrophic community [J]. Synthetic Biology Journal, 2024, 5(5): 1189-1210. |
| [11] | CHEN Ziling, XIANG Yangfei. Integrated development of organoid technology and synthetic biology [J]. Synthetic Biology Journal, 2024, 5(4): 795-812. |
| [12] | CAI Bingyu, TAN Xiangtian, LI Wei. Advances in synthetic biology for engineering stem cell [J]. Synthetic Biology Journal, 2024, 5(4): 782-794. |
| [13] | XIE Huang, ZHENG Yilei, SU Yiting, RUAN Jingyi, LI Yongquan. An overview on reconstructing the biosynthetic system of actinomycetes for polyketides production [J]. Synthetic Biology Journal, 2024, 5(3): 612-630. |
| [14] | ZHA Wenlong, BU Lan, ZI Jiachen. Advances in synthetic biology for producing potent pharmaceutical ingredients of traditional Chinese medicine [J]. Synthetic Biology Journal, 2024, 5(3): 631-657. |
| [15] | HUI Zhen, TANG Xiaoyu. Applications of the CRISPR/Cas9 editing system in the study of microbial natural products [J]. Synthetic Biology Journal, 2024, 5(3): 658-671. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||