• •
遗传编码荧光探针在疾病诊断中的最新进展
李睿1,2,3, 左方婷1,2,4, 杨弋1,2
- 1.华东理工大学光遗传学与合成生物学跨学科研究中心,生物反应器工程国家重点实验室,中国 上海 200237
2.华东理工大学药学院,上海市细胞代谢光遗传学技术前沿科学研究基地,中国 上海 200237
3.南方医科大学深圳医院,中国 深圳 518110
4.同济大学附属杨浦医院,中国 上海 200090
-
收稿日期:2025-05-13修回日期:2025-08-28出版日期:2025-08-29 -
通讯作者:杨弋 -
作者简介:李睿 (1996—),女,博士研究生。研究方向合成生物技术与生物传感技术结合在监测细胞内实时动态代谢过程。E-mail:y20180020@mail.ecust.edu.cn杨弋 (1973—),男,博士生导师,主要研究对象为利用合成生物技术与光遗传学技术控制与监测细胞内分子过程的前沿技术;癌症及代谢类疾病药理及药物筛选技术;蛋白质特异性标记、翻译后修饰的鉴定、与细胞内原位成像;蛋白质药物生产技术等。E-mail:yiyang@ecust.edu.cn -
基金资助:国家重点研发计划“基因表达时空精准操控技术研究”(2022YFC3400100);国家自然科学基金-创新研究群体项目“细胞代谢监测与调控”(32121005);上海市青年科技英才扬帆计划“近红外荧光RNA的开发与应用研究”(24YF2709300);博士后创新人才支持计划“基于新型胆汁酸生物传感器在肠道菌群中时空动态监测与调控”
Recent advances in genetically encoded fluorescent sensors for disease diagnosis
Li Rui1,2,3, Zuo Fangting1,2,4, Yang Yi1,2
- 1.Interdisciplinary Research Center of Optogenetics and Synthetic Biology,State Key Laboratory of Bioreactor Engineering,East China University of Science and Technology,Shanghai 200237,China
2.Shanghai Advanced Research Base of Cell Metabolism Genetics,School of Pharmacy,East China University of Science and Technology,Shanghai 200237,China
3.Shenzhen Hospital,Southern Medical University,Shenzhen 518110,China
4.Yangpu Hospital,Tongji University,shanghai 200090,China.
-
Received:2025-05-13Revised:2025-08-28Online:2025-08-29 -
Contact:Yang Yi
摘要:
近年来,遗传编码荧光探针在结构优化与疾病诊断应用中取得了快速发展。通过蛋白质工程,荧光蛋白在光稳定性、灵敏度和光谱范围方面显著提升,并涌现出多种新型传感机制,实现了离子、代谢物及神经递质等生理信号的实时可视化。与此同时,荧光RNA在折叠稳定性、激活效率和亮度上不断突破,多色工具箱的建立使 RNA 动态成像成为可能。这两类探针已广泛应用于肿瘤代谢、糖尿病及神经疾病研究,在代谢监测、病理状态识别和早期诊断等方面展现出独特优势,推动了疾病机制解析与诊断技术进步。未来,随着探针性能持续优化和设计创新,遗传编码荧光探针有望在基础研究和临床转化中发挥更大作用,为精准诊断和个性化医疗提供有力支持。
中图分类号:
引用本文
李睿, 左方婷, 杨弋. 遗传编码荧光探针在疾病诊断中的最新进展[J]. 合成生物学, DOI: 10.12211/2096-8280.2025-045.
Li Rui, Zuo Fangting, Yang Yi. Recent advances in genetically encoded fluorescent sensors for disease diagnosis[J]. Synthetic Biology Journal, DOI: 10.12211/2096-8280.2025-045.
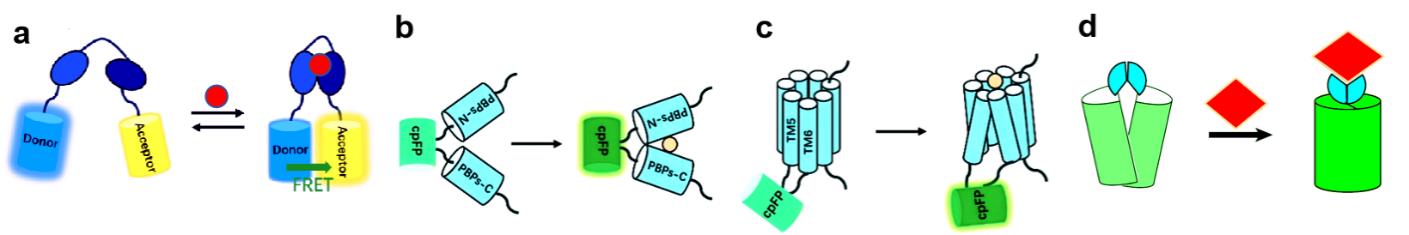
图1 遗传编码荧光探针设计构建图(a)基于 FRET 原理构建遗传编码荧光探针的示意图;(b)基于周质结合蛋白 构建的遗传编码荧光探针,结合底物后其构象变化将会改变环化荧光蛋白(Circularly Permuted Fluorescent Protein,cpFP)的荧光;(c)基于G蛋白偶联受体 构建的遗传编码荧光探针,cpFP 通常插入在G蛋白偶联受体位于胞内的第五和第六跨膜结构域之间,结合底物后G蛋白偶联受体的构象变化将会改变 cpFP 的荧光[6];(d)基于ATOM原理构建遗传编码荧光探针,配体结合诱导荧光蛋白结构域折叠,恢复其天然构象,使发色团成熟[5]。
Fig.1 Design and construction of genetically encoded fluorescent sensors(a) Diagram of genetically encoded fluorescent sensor based on FRET principle. (b) PBP-based genetically encoded indicators, following substrate binding, conformational changes of PBPs will change fluorescence of cpFP. (c) GPCRs-based genetically encoded indicators, cpFP is inserted into the intracellular loop of GPCRs between transmembrane domains 5 and 6, following substrate binding, conformational changes of GPCRs will change fluorescence of cpFP[6]. (d) ATOM-based genetically encoded fluorescence indicators, ligand is binded to fluorescence protein domain, restored its natural conformation, and matured the chromophore[5].
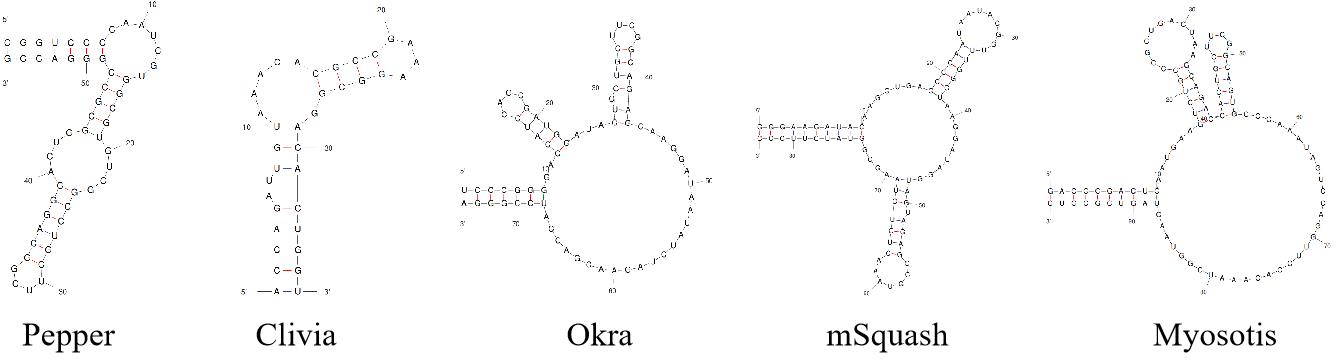
图2 Pepper[14]、Clivia[19]、Okra[20]、mSquash[23]和Myosotis[21]适配体结构示意图
Fig.2 Schematic diagram of Pepper[14], Clivia[19], Okra[20], mSquash[23] and Myosotis[21] aptamer structures.
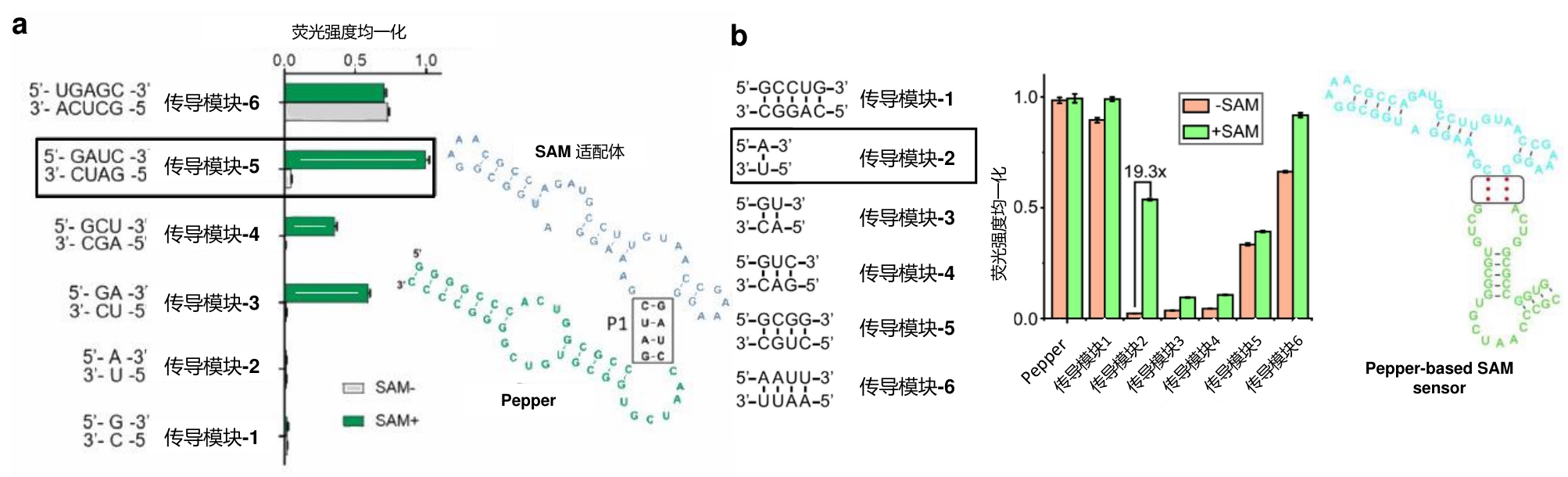
图3 检测SAM的RNA传感器(a)Pepper-SAM-C14U探针设计原理及其对SAM的响应[17];(b)基于Pepper和RhoBAST的比率型SAM传感器设计原理及响应[18]。
Fig.3 RNA sensors for detecting SAM(a) Design principle of Pepper-SAM-C14U sensor and its response to SAM[17]. (b) Design principle of ratiometric SAM sensor based on Pepper and RhoBAST and its response to SAM[18].
| [1] | WANG X, DING Q, GROLEAU R R, et al. Fluorescent Probes for Disease Diagnosis [J]. Chemical Reviews, 2024, 124(11): 7106-64. |
| [2] | GHOUNEIMY A, MAHAS A, MARSIC T, et al. CRISPR-Based Diagnostics: Challenges and Potential Solutions toward Point-of-Care Applications [J]. ACS Synth Biol, 2023, 12(1): 1-16. |
| [3] | HAN Y, YANG J, LI Y, et al. Bright and sensitive red voltage indicators for imaging action potentials in brain slices and pancreatic islets [J]. Sci Adv, 2023, 9(47): eadi4208. |
| [4] | ZHANG Y, RóZSA M, LIANG Y, et al. Fast and sensitive GCaMP calcium indicators for imaging neural populations [J]. Nature, 2023, 615(7954): 884-91. |
| [5] | SEKHON H, HA J H, PRESTI M F, et al. Adaptable, turn-on maturation (ATOM) fluorescent biosensors for multiplexed detection in cells [J]. Nature Methods, 2023, 20(12): 1920-9. |
| [6] | WU Z, LIN D, LI Y. Pushing the frontiers: tools for monitoring neurotransmitters and neuromodulators [J]. Nat Rev Neurosci, 2022, 23(5): 257-74. |
| [7] | CHANG H, CLEMENS S, GAO P, et al. Fluorogenic Rhodamine-Based Chemigenetic Biosensor for Monitoring Cellular NADPH Dynamics [J]. Journal of the American Chemical Society, 2024, 146(30): 20569-76. |
| [8] | HU L, CAO W, JIANG Y, et al. Designing artificial fluorescent proteins and biosensors by genetically encoding molecular rotor-based amino acids [J]. Nature Chemistry, 2024, 16(12): 1960-71. |
| [9] | LU S, HOU Y, ZHANG X E, et al. Live cell imaging of DNA and RNA with fluorescent signal amplification and background reduction techniques [J]. Front Cell Dev Biol, 2023, 11: 1216232. |
| [10] | SONG Q, TAI X, REN Q, et al. Structure-based insights into fluorogenic RNA aptamers [J]. Acta Biochim Biophys Sin (Shanghai), 2024, 57(1): 108-18. |
| [11] | GAO W K C Z Y, RONG X X, ET AL. . Progress in RNA dynamic imaging technology in live cells [J]. SCIENTIA SINICA Vitae, 2024, 54(4): 651-67. |
| [12] | ZUO F T, ZHANG Y Q, YANG H M, et al. Progress on fluorescent RNA and fluorescent RNA-based biosensing technology [J]. Yi Chuan, 2024, 46(2): 92-108. |
| [13] | LU X, KONG K Y S, UNRAU P J. Harmonizing the growing fluorogenic RNA aptamer toolbox for RNA detection and imaging [J]. Chemical Society Reviews, 2023, 52(12): 4071-98. |
| [14] | HUANG K, CHEN X, LI C, et al. Structure-based investigation of fluorogenic Pepper aptamer [J]. Nature Chemical Biology, 2021, 17(12): 1289-95. |
| [15] | WANG Q, XIAO F, SU H, et al. Inert Pepper aptamer-mediated endogenous mRNA recognition and imaging in living cells [J]. Nucleic Acids Res, 2022, 50(14): e84. |
| [16] | ZHENG H, LIU X, LIU L, et al. Imaging of endogenous RNA in live cells using sequence-activated fluorescent RNA probes [J]. Nucleic Acids Res, 2024. |
| [17] | FANG M, LI H, XIE X, et al. Imaging intracellular metabolite and protein changes in live mammalian cells with bright fluorescent RNA-based genetically encoded sensors [J]. Biosens Bioelectron, 2023, 235: 115411. |
| [18] | CHEN Z, CHEN W, REHEMAN Z, et al. Genetically encoded RNA-based sensors with Pepper fluorogenic aptamer [J]. Nucleic Acids Research, 2023, 51(16): 8322-36. |
| [19] | JIANG L, XIE X, SU N, et al. Large Stokes shift fluorescent RNAs for dual-emission fluorescence and bioluminescence imaging in live cells [J]. Nature Methods, 2023, 20(10): 1563-72. |
| [20] | ZUO F, JIANG L, SU N, et al. Imaging the dynamics of messenger RNA with a bright and stable green fluorescent RNA [J]. Nature Chemical Biology, 2024, 20(10): 1272-81. |
| [21] | JIANG L, ZUO F, PAN Y, et al. Bright and Stable Cyan Fluorescent RNA Enables Multicolor RNA Imaging in Live Escherichia coli [J]. Small, 2025, 21(9): e2405165. |
| [22] | CHEN Z, CHEN W, XU C, et al. Near-infrared fluorogenic RNA for in vivo imaging and sensing [J]. Nature Communications, 2025, 16(1): 518. |
| [23] | YIN P, HUANG C, ZHANG L, et al. Developing Orthogonal Fluorescent RNAs for Photoactive Dual-Color Imaging of RNAs in Live Cells [J]. Angewandte Chemie International Edition, 2025, n/a(n/a): e202424060. |
| [24] | HOU J, GUO P, WANG J, et al. Artificial dynamic structure ensemble-guided rational design of a universal RNA aptamer-based sensing tag [J]. Proceedings of the National Academy of Sciences of the United States of America, 2024, 121(52): e2414793121. |
| [25] | WONG F, HE D, KRISHNAN A, et al. Deep generative design of RNA aptamers using structural predictions [J]. Nature Computational Science, 2024, 4(11): 829-39. |
| [26] | WU Z, CUI Y, WANG H, et al. Neuronal activity-induced, equilibrative nucleoside transporter-dependent, somatodendritic adenosine release revealed by a GRAB sensor [J]. Proceedings of the National Academy of Sciences of the United States of America, 2023, 120(14): e2212387120. |
| [27] | ZHUO Y, LUO B, YI X, et al. Improved green and red GRAB sensors for monitoring dopaminergic activity in vivo [J]. Nature Methods, 2024, 21(4): 680-91. |
| [28] | ZENG J, LI X, ZHANG R, et al. Local 5-HT signaling bi-directionally regulates the coincidence time window for associative learning [J]. Neuron, 2023, 111(7): 1118-35.e5. |
| [29] | DENG F, WAN J, LI G, et al. Improved green and red GRAB sensors for monitoring spatiotemporal serotonin release in vivo [J]. Nature Methods, 2024, 21(4): 692-702. |
| [30] | TOUHARA K K, ROSSEN N D, DENG F, et al. Topological segregation of stress sensors along the gut crypt–villus axis [J]. Nature, 2025. |
| [31] | QIAN T, WANG H, WANG P, et al. A genetically encoded sensor measures temporal oxytocin release from different neuronal compartments [J]. Nature Biotechnology, 2023, 41(7): 944-57. |
| [32] | FENG J, DONG H, LISCHINSKY J E, et al. Monitoring norepinephrine release in vivo using next-generation GRAB(NE) sensors [J]. Neuron, 2024, 112(12): 1930-42.e6. |
| [33] | WANG H, QIAN T, ZHAO Y, et al. A tool kit of highly selective and sensitive genetically encoded neuropeptide sensors [J]. Science, 2023, 382(6672): eabq8173. |
| [34] | XIA X, LI Y. A high-performance GRAB sensor reveals differences in the dynamics and molecular regulation between neuropeptide and neurotransmitter release [J]. Nature Communications, 2025, 16(1): 819. |
| [35] | WU T, KUMAR M, ZHANG J, et al. A genetically encoded far-red fluorescent indicator for imaging synaptically released Zn2+ [J]. Science Advances, 9(9): eadd2058. |
| [36] | YANG L, PATHIRANAGE V, ZHOU S, et al. Genetically Encoded Red Fluorescent Indicators for Imaging Intracellular and Extracellular Potassium Ions [J]. bioRxiv, 2024: 2024.12.20.629597. |
| [37] | LAI C, YANG L, PATHIRANAGE V, et al. Genetically encoded green fluorescent sensor for probing sulfate transport activity of solute carrier family 26 member a2 (Slc26a2) protein [J]. Communications Biology, 2024, 7(1): 1375. |
| [38] | RAHMAN T, PATEL S. Recent developments in probing the levels and flux of selected organellar cations as well as organellar mechanosensitivity [J]. Current Opinion in Chemical Biology, 2025, 87: 102600. |
| [39] | MARVIN J S, KOKOTOS A C, KUMAR M, et al. iATPSnFR2: A high-dynamic-range fluorescent sensor for monitoring intracellular ATP [J]. Proceedings of the National Academy of Sciences, 2024, 121(21): e2314604121. |
| [40] | WANG K, CHEN T-L, ZHANG X-X, et al. Unveiling tryptophan dynamics and functions across model organisms via quantitative imaging [J]. BMC Biology, 2024, 22(1): 258. |
| [41] | LIU B, ZHAO Z, WANG P, et al. GlutaR: A High‐Performance Fluorescent Protein-Based Sensor for Spatiotemporal Monitoring of Glutamine Dynamics In Vivo [J]. Angewandte Chemie International Edition, 2024. |
| [42] | LI X, ZHANG Y, XU L, et al. Ultrasensitive sensors reveal the spatiotemporal landscape of lactate metabolism in physiology and disease [J]. Cell Metab, 2023, 35(1): 200-11 e9. |
| [43] | LI R, LI Y, JIANG K, et al. Lighting up arginine metabolism reveals its functional diversity in physiology and pathology [J]. Cell Metabolism, 2025, 37(1): 291-304.e9. |
| [44] | HUANG D, ZHANG C, XIAO M, et al. Redox metabolism maintains the leukemogenic capacity and drug resistance of AML cells [J]. Proceedings of the National Academy of Sciences of the United States of America, 2023, 120(13): e2210796120. |
| [45] | CHANDRASEKHARAN A, TIWARI S K, MUNIRPASHA H A, et al. Genetically encoded caspase sensor and RFP-LC3 for temporal analysis of apoptosis-autophagy [J]. International Journal of Biological Macromolecules, 2024, 257(Pt 2): 128807. |
| [46] | XU B, WANG Y, BAHRIZ S M F M, et al. Probing spatiotemporal PKA activity at the ryanodine receptor and SERCA2a nanodomains in cardomyocytes [J]. Cell Communication and Signaling, 2022, 20(1): 143. |
| [47] | ROMERO-SUAREZ D, WULFF T, RONG Y, et al. A Reporter System for Cytosolic Protein Aggregates in Yeast [J]. ACS Synth Biol, 2021, 10(3): 466-77. |
| [48] | LIU S, LIU J, FOOTE A, et al. Digital and Tunable Genetically Encoded Tension Sensors Based on Engineered Coiled-Coils [J]. Angewandte Chemie International Edition, 2025, 64(8): e202407359. |
| [49] | MOLNAR K, J-B MANNEVILLE. Emerging mechanobiology techniques to probe intracellular mechanics [J]. npj Biological Physics and Mechanics, 2025, 2(1): 12. |
| [50] | FREI M S, MEHTA S, ZHANG J. Next-Generation Genetically Encoded Fluorescent Biosensors Illuminate Cell Signaling and Metabolism [J]. Annual Review of Biophysics, 2024, 53(Volume 53, 2024): 275-97. |
| [51] | ZHOU L, YANG R, LI X, et al. COF-Coated Microelectrode for Space-Confined Electrochemical Sensing of Dopamine in Parkinson's Disease Model Mouse Brain [J]. Journal of the American Chemical Society, 2023, 145(43): 23727-38. |
| [52] | AGGARWAL A, LIU R, CHEN Y, et al. Glutamate indicators with improved activation kinetics and localization for imaging synaptic transmission [J]. Nature Methods, 2023, 20(6): 925-34. |
| [53] | JI D, WANG B, LO K W, et al. Pre-Defined Stem-Loop Structure Library for the Discovery of L-RNA Aptamers that Target RNA G-Quadruplexes [J]. Angewandte Chemie International Edition in English, 2025, 64(5): e202417247. |
| [54] | GUO S K, LIU C X, XU Y F, et al. Therapeutic application of circular RNA aptamers in a mouse model of psoriasis [J]. Nature Biotechnology, 2025, 43(2): 236-46. |
| [55] | DING D, ZHAO H, WEI D, et al. The First-in-Human Whole-Body Dynamic Pharmacokinetics Study of Aptamer [J]. Research (Wash D C), 2023, 6: 0126. |
| [1] | 陈涛, 赖锦涛, 胡美林, 马显才. 蛋白质优化设计与从头合成引领的疫苗研发革命[J]. 合成生物学, 2025, (): 1-25. |
| [2] | 丁怡, 苏敏, 高晓冬, 王宁. N-糖链合成的研究进展[J]. 合成生物学, 2025, (): 1-16. |
| [3] | 黄扬, 李一叶, 聂广军. 合成生物学赋能细胞膜纳米颗粒的精准诊疗[J]. 合成生物学, 2025, (): 1-24. |
| [4] | 李全飞, 陈乾, 杨凯, 刘浩, 贺坤东, 潘亮, 李莎, 雷鹏, 谷益安, 孙良, 邱益彬, 张琦, 徐虹, 王瑞. 高黏性蛋白材料的生物合成及应用[J]. 合成生物学, 2025, (): 1-20. |
| [5] | 肖和, 姚明菊, 乔雪. 长糖链皂苷的生物合成研究进展[J]. 合成生物学, 2025, (): 1-7. |
| [6] | 李明辰, 钟博子韬, 余元玺, 姜帆, 张良, 谭扬, 虞慧群, 范贵生, 洪亮. DeepSeek模型分析及其在AI辅助蛋白质工程中的应用[J]. 合成生物学, 2025, 6(3): 636-650. |
| [7] | 宋成治, 林一瀚. AI+定向进化赋能蛋白改造及优化[J]. 合成生物学, 2025, 6(3): 617-635. |
| [8] | 田晓军, 张日新. 合成基因回路面临的细胞“经济学窘境”[J]. 合成生物学, 2025, 6(3): 532-546. |
| [9] | 姜百翼, 钱珑. 活细胞记录器在细胞谱系追踪中的应用和前景[J]. 合成生物学, 2025, 6(3): 651-668. |
| [10] | 李倩, FERRELL JR.James E., 陈于平. 细胞质浓度:细胞生物学的老问题、新参数[J]. 合成生物学, 2025, 6(3): 497-515. |
| [11] | 张成辛. 基于文本数据挖掘的蛋白功能预测:机遇与挑战[J]. 合成生物学, 2025, 6(3): 603-616. |
| [12] | 王珂, 陈文慧, 雷春阳, 聂舟. CRISPR/Cas系统在分子诊断领域的应用研究进展[J]. 合成生物学, 2025, (): 1-27. |
| [13] | 甘牡丹, 左静蕊, 曹友志. 工程菌的生物安全防控策略[J]. 合成生物学, 2025, (): 1-14. |
| [14] | 王倩, 果士婷, 辛波, 钟成, 王钰. L-精氨酸的微生物合成研究进展[J]. 合成生物学, 2025, 6(2): 290-305. |
| [15] | 高琪, 肖文海. 酵母合成单萜类化合物的研究进展[J]. 合成生物学, 2025, 6(2): 357-372. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
