• 特约评述 •
N-糖链合成的研究进展
丁怡1,2, 苏敏2, 高晓冬1,2, 王宁1,2
- 1.江南大学,糖化学与生物技术教育部重点实验室,生物工程学院,江苏 无锡 214122
2.中国科学院 过程工程研究所,生物药制备与递送全国重点实验室,北京 100190
-
收稿日期:2025-06-04修回日期:2025-07-22出版日期:2025-08-01 -
通讯作者:高晓冬,王宁 -
作者简介:丁怡 (1995—),女,博士研究生。研究方向为糖生物学,糖疫苗。E-mail:814469449@qq.com高晓冬 (1965—),男,研究员,博士生导师,研究方向为糖生物学;生物医药工程;生物化学与分子生物学;细胞生物学等。E-mail:xdgao@ipe.ac.cn王宁 (1984—),女,研究员,博士生导师,研究方向为糖化学生物学;糖疫苗;糖基化相关疾病;糖化学等。E-mail:wangning@ipe.ac.cn -
基金资助:国家自然科学基金(22077053);国家自然科学基金(32271342);北京市自然科学基金(2242022);中国科学院关键核心技术攻坚先导专项(C类先导专项)(XDC0290302)
Advances in Research of N-Glycan Synthesis
Ding Yi1,2, Su Min2, Gao Xiaodong1,2, Wang Ning1,2
- 1.Key Laboratory of Carbohydrate Chemistry and Biotechnology,Ministry of Education,School of Biotechnology,Jiangnan University,Wuxi 214122,China
2.State Key Laboratory of Biopharmaceutical Preparation and Delivery,Institute of Process Engineering,Chinese Academy of Sciences,Beijing,100190,China
-
Received:2025-06-04Revised:2025-07-22Online:2025-08-01 -
Contact:Gao Xiaodong, Wang Ning
摘要:
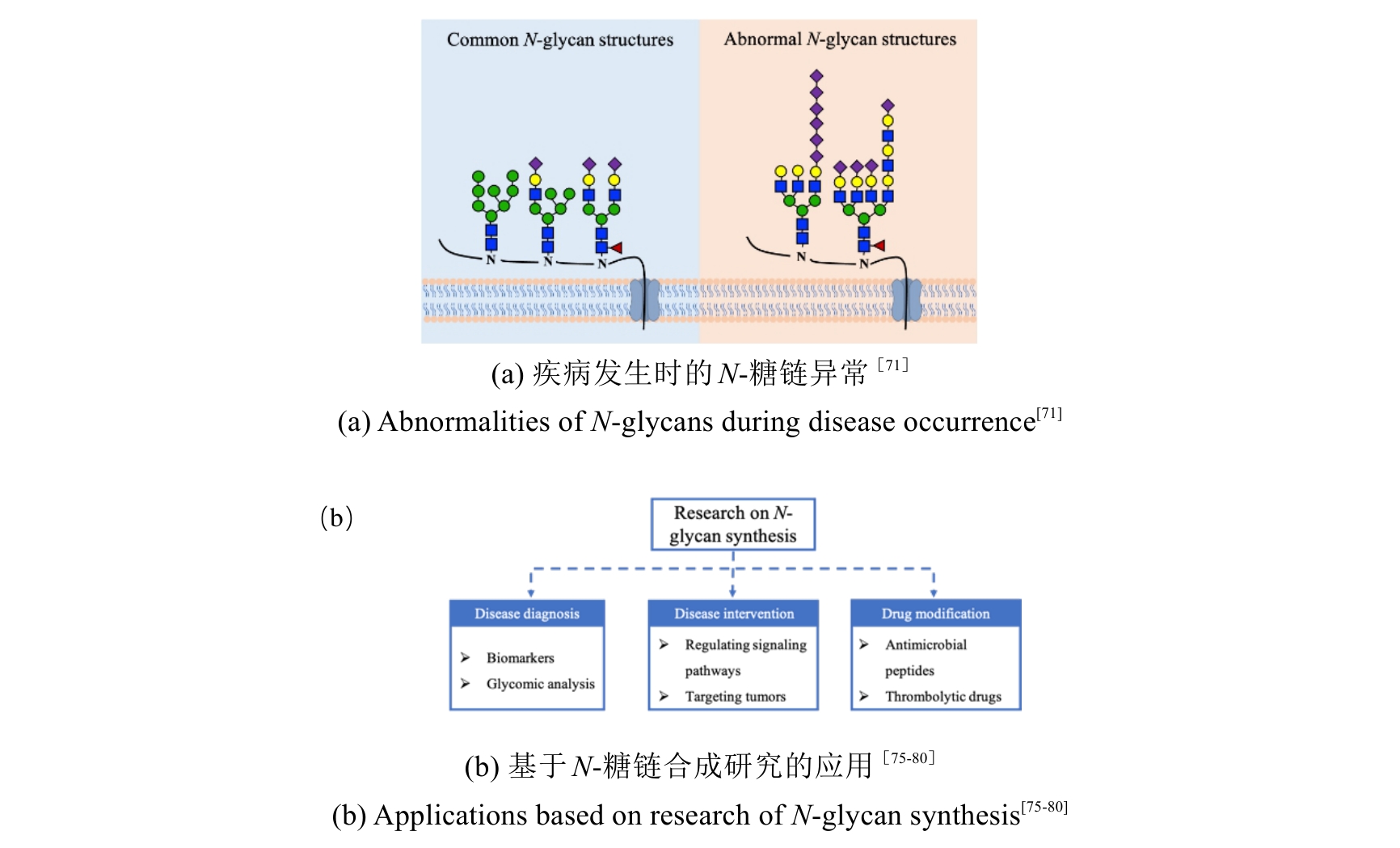
糖基化是一种常见的蛋白质翻译后修饰,N-糖链的合成起始于内质网并在高尔基体中完成。糖链结构的精确合成与调控不仅促进蛋白质结构与功能的成熟,还与多种疾病的发生发展密切相关。近年来,N-糖链生物合成机制陆续阐明,使其合成通路逐渐清晰,进而推动了合成方法的发展和应用转化。本综述系统梳理了近年来N-糖链合成领域的重要工作和进展,旨在为糖合成生物学研究提供可行的策略。在解析N-糖链体内合成的过程中,活体成像与单细胞分析、多组学整合等前沿技术的涌现,使糖链合成动态网络相关研究取得了突破性进展;在N-糖链体外合成的研究中,酶法催化、化学酶法结合、细胞工厂等技术的发展和优化实现了复杂糖链的可控合成,为其功能研究和应用提供了重要工具;在疾病诊断与干预方面,神经退行性疾病、肿瘤和心血管疾病等多种疾病中存在异常合成的N-糖链,为诊断标志物的发现和干预策略的设计提供了新思路。
中图分类号:
引用本文
丁怡, 苏敏, 高晓冬, 王宁. N-糖链合成的研究进展[J]. 合成生物学, DOI: 10.12211/2096-8280.2025-053.
Ding Yi, Su Min, Gao Xiaodong, Wang Ning. Advances in Research of N-Glycan Synthesis[J]. Synthetic Biology Journal, DOI: 10.12211/2096-8280.2025-053.
| [1] | SCHWARZ F, AEBI M. Mechanisms and principles of N-linked protein glycosylation[J]. Current Opinion in Structural Biology, 2011, 21(5): 576-582. |
| [2] | HELENIUS A, AEBI M. Roles of N-Linked Glycans in the Endoplasmic Reticulum[J]. Annual Review of Biochemistry, 2004, 73(1): 1019-1049. |
| [3] | DWEK R A. Glycobiology: Toward Understanding the Function of Sugars[J]. Chemical Reviews, 1996, 96(2): 683-720. |
| [4] | SCHJOLDAGER K T, NARIMATSU Y, JOSHI H J, et al. Global view of human protein glycosylation pathways and functions[J]. Nature Reviews Molecular Cell Biology, 2020, 21(12): 729-749. |
| [5] | VARKI A. Biological roles of glycans[J]. Glycobiology, 2017, 27(1): 3-49. |
| [6] | MOREMEN K W, TIEMEYER M, NAIRN A V. Vertebrate protein glycosylation: diversity, synthesis and function[J]. Nature Reviews Molecular Cell Biology, 2012, 13(7): 448-462. |
| [7] | AEBI M, BERNASCONI R, CLERC S, et al. N-glycan structures: recognition and processing in the ER[J]. Trends in Biochemical Sciences, 2010, 35(2): 74-82. |
| [8] | PETRESCU A, WORMALD M, DWEK R. Structural aspects of glycomes with a focus on N-glycosylation and glycoprotein folding[J]. Current Opinion in Structural Biology, 2006, 16(5): 600-607. |
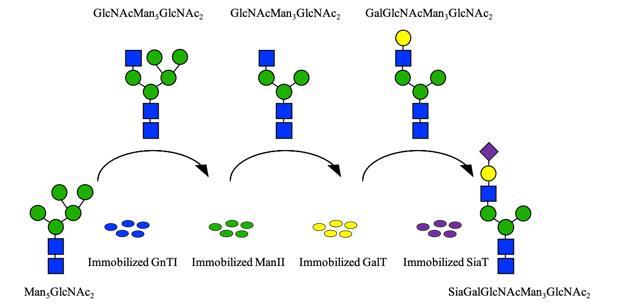
| [9] | OHTSUBO K, MARTH J D. Glycosylation in Cellular Mechanisms of Health and Disease[J]. Cell, 2006, 126(5): 855-867. |
| [10] | HE M, ZHOU X, WANG X. Glycosylation: mechanisms, biological functions and clinical implications[J]. Signal Transduction and Targeted Therapy, 2024, 9(1): 194. |
| [11] | LI C W, LIM S O, XIA W, et al. Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity[J]. Nature Communications, 2016, 7(1): 12632. |
| [12] | STANLEY P. Genetics of glycosylation in mammalian development and disease[J]. Nature Reviews Genetics, 2024, 25(10). |
| [13] | HU F, GAO J, ZHENG J, et al. N-GlycoPred: A hybrid deep learning model for accurate identification of N-glycosylation sites[J]. Methods, 2024, 227: 48-57 |
| [14] | HAO Z, GUO Q, FENG Y, et al. Investigation of the Catalytic Mechanism of a Soluble N-glycosyltransferase Allows Synthesis of N-glycans at Noncanonical Sequons[J]. JACS Au, 2023, 3(8): 2144-2155. |
| [15] | ZHOU X, LIU G, YANG M, et al. Glycosyl N-phenyl pentafluorobenzimidates as a new generation of imidate donors for catalytic glycosylation[J]. Chinese Chemical Letters, 2024: 110734. |
| [16] | POWELL W C, JING R, HERLORY M, et al. Chemical Synthesis Reveals Pathogenic Role of N-Glycosylation in Microtubule-Associated Protein Tau[J]. Journal of the American Chemical Society, 2025, 147(8): 6995-7007. |
| [17] | YU B. Gold(I)-Catalyzed Glycosylation with Glycosyl o-Alkynylbenzoates as Donors[J]. Accounts of Chemical Research, 2018, 51(2): 507-516. |
| [18] | BAO B, KELLMAN B P, CHIANG A W T, et al. Correcting for sparsity and interdependence in glycomics by accounting for glycan biosynthesis[J]. Nature Communications, 2021, 12(1): 4988. |
| [19] | GREEN R S, STONE E L, TENNO M, et al. Mammalian N-Glycan Branching Protects against Innate Immune Self-Recognition and Inflammation in Autoimmune Disease Pathogenesis[J]. Immunity, 2007, 27(2): 308-320. |
| [20] | RABINOVICH G A, CROCI D O. Regulatory Circuits Mediated by Lectin-Glycan Interactions in Autoimmunity and Cancer[J]. Immunity, 2012, 36(3): 322-335. |
| [21] | LIU H, LI X, REN Y, et al. In Situ Visualization of RNA-Specific Sialylation on Living Cell Membranes to Explore N-Glycosylation Sites[J]. Journal of the American Chemical Society, 2024, 146(12): 8780-8786. |
| [22] | TIAN Y, WANG Y, ZHANG Y, et al. One-Step Labeling Strategy for the Profiling of Multiple Types of Protein Glycosylation[J]. Analytical Chemistry, 2025, 97(14): 7833-7841. |
| [23] | KAJI H, SAITO H, YAMAUCHI Y, et al. Lectin affinity capture, isotope-coded tagging and mass spectrometry to identify N-linked glycoproteins[J]. Nature Biotechnology, 2003, 21(6): 667-672. |
| [24] | SONG X, JU H, LASANAJAK Y, et al. Oxidative release of natural glycans for functional glycomics[J]. Nature Methods, 2016, 13(6): 528-534. |
| [25] | GEMMER M, CHAILLET M L, VAN LOENHOUT J, et al. Visualization of translation and protein biogenesis at the ER membrane[J]. Nature, 2023, 614(7946): 160-167. |
| [26] | Tsui C K, Twells N, Durieux J,et al.CRISPR screens and lectin microarrays identify high mannose N-glycan regulators[J].Nature Communications, 15(1):1-13. |
| [27] | IVES C M, SINGH O, D'ANDREA S, et al. Restoring protein glycosylation with GlycoShape[J]. Nature Methods, 2024, 21(11): 2117-2127. |
| [28] | YANG J. Sequential genome-wide CRISPR-Cas9 screens identify genes regulating cell-surface expression of tetraspanins[J]. Cell Report, 2023:1-25. |
| [29] | TIMOSHCHUK A, SHARAPOV S, AULCHENKO Y S. Twelve Years of Genome-Wide Association Studies of Human Protein N-Glycosylation[J]. Engineering, 2023, 26: 17-31. |
| [30] | Klari T S, IvanGudelj,GabrielSantpere,et al.Human-specific features and developmental dynamics of the brain N-glycome[J].Science Advances, 2023, 9(49):21. |
| [31] | MA W, XU Z, JIANG Y, et al. Divergent Enzymatic Assembly of a Comprehensive 64‐Membered IgG N ‐Glycan Library for Functional Glycomics[J]. Advanced Science, 2023, 10(30): 2303832. |
| [32] | WIDMALM G. Glycan Shape, Motions, and Interactions Explored by NMR Spectroscopy[J]. JACS Au, 2024, 4(1): 20-39. |
| [33] | Carnielli C M, De L M T M, de Sá Patroni F.M.Ribeiro A.C.P.Bro T.B.Sobroza E.Matos L.L.Kowalski L.P.Leme A.F.P.Kawahara R.Thaysen-Andersen M.Comprehensive Glycoprofiling of Oral Tumors Associates N-Glycosylation With Lymph Node Metastasis and Patient Survival[J].Molecular & cellular proteomics: MCP, 2023, 22(7):100586-100586. |
| [34] | ISENRICH F N, LOSFELD M E, AEBI M, et al. Microfluidic mimicry of the Golgi-linked N-glycosylation machinery[J]. Lab on a Chip, 2025, 25(8): 1907-1917. |
| [35] | CINDRIĆ A, PRIBIĆ T, LAUC G. High-throughput N-glycan analysis in aging and inflammaging: State of the art and future directions[J]. Seminars in Immunology, 2024, 73: 101890. |
| [36] | SCHEPER A F, SCHOFIELD J, BOHARA R, et al. Understanding glycosylation: Regulation through the metabolic flux of precursor pathways[J]. Biotechnology Advances, 2023, 67: 108184. |
| [37] | WILSON M P, KENTACHE T, ALTHOFF C R, et al. A pseudoautosomal glycosylation disorder prompts the revision of dolichol biosynthesis[J]. Cell, 2024, 187(14): 3585-3601.e22. |
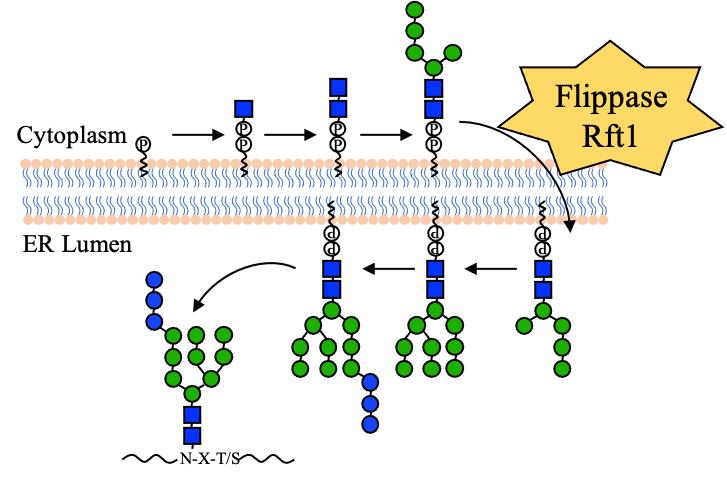
| [38] | CHEN S, PEI C X, XU S, et al. Rft1 catalyzes lipid-linked oligosaccharide translocation across the ER membrane[J]. Nature Communications, 2024, 15(1): 5157. |
| [39] | MA M, DUBEY R, JEN A, et al. Regulated N-glycosylation controls chaperone function and receptor trafficking[J]. Science, 2024, 386(6722): 667-672. |
| [40] | DURAN R, HOUBEN B, VLEESCHOUWER M D, et al. N-glycosylation as a eukaryotic protective mechanism against protein aggregation[J]. Science Advances, 2024, 10: 1-16. |
| [41] | SUN R, KIM A M J, MURRAY A A, et al. N-Glycosylation Facilitates 4-1BB Membrane Localization by Avoiding Its Multimerization[J]. Cells, 2022, 11(1): 162. |
| [42] | CHENG Q, HU X, ZHANG X, et al. N-glycosylation at N57/100/110 affects CD44s localization, function and stability in hepatocellular carcinoma[J]. European Journal of Cell Biology, 2023, 102(4): 151360. |
| [43] | VAN KOOYK Y, RABINOVICH G A. Protein-glycan interactions in the control of innate and adaptive immune responses[J]. Nature Immunology, 2008, 9(6): 593-601. |
| [44] | ITSKANOV S, PARK E. Mechanism of Protein Translocation by the Sec61 Translocon Complex[J]. Cold Spring Harbor Perspectives in Biology, 2023, 15(1): a041250. |
| [45] | PHOOMAK C, RINIS N, BARO M, et al. Signal recognition particle receptor-β (SR-β) coordinates cotranslational N-glycosylation[J]. Science Advances, 2023, 9(11): eade8079. |
| [46] | GREGORCZYK P, PORĘBSKA N, ŻUKOWSKA D, et al. N-glycosylation acts as a switch for FGFR1 trafficking between the plasma membrane and nuclear envelope[J]. Cell Communication and Signaling, 2023, 21(1): 177. |
| [47] | FANG J, ZHANG Y, HUANG C, et al. Glucose-mediated N-glycosylation of RPTPα affects its subcellular localization and Src activation[J]. Oncogene, 2023, 42(14): 1058-1071. |
| [48] | HU Z F, ZHONG K, CAO H. Recent advances in enzymatic and chemoenzymatic synthesis of N- and O-glycans[J]. Current Opinion in Chemical Biology, 2024, 78: 102417. |
| [49] | RØNNE KRISTENSEN B, PEDERSEN C M. Self‐Promoted N-Glycosylation: Extended Substrate Scope and Substituent Effects[J]. European Journal of Organic Chemistry, 2023, 26(16): e202300213. |
| [50] | XIE X, ZHAO S, HAN Y, et al. Direct construction of aryl amide N-glycosides from glycosyl oxamic acids via photoredox palladium-catalyzed aminocarbonylations[J]. Chem Catalysis, 2024, 4(10): 101109. |
| [51] | ZHONG L, WANG Q, WANG Y, et al. Facile and stereospecific synthesis of diverse β-N-glycosyl sulfonamide scaffolds via palladium catalysis[J]. Chemical Communications, 2023, 59(86): 12907-12910. |
| [52] | DONG W, YANG X, LI X, et al. Investigation of N-Glycan Functions in Receptor for Advanced Glycation End Products V Domain through Chemical Glycoprotein Synthesis[J]. Journal of the American Chemical Society, 2024, 146(27): 18270-18280. |
| [53] | ZHANG J, LUO Z X, WU X, et al. Photosensitizer-free visible-light-promoted glycosylation enabled by 2-glycosyloxy tropone donors[J]. Nature Communications, 2023, 14(1): 8025. |
| [54] | SUN Q, WANG Q, QIN W, et al. N-glycoside synthesis through combined copper- and photoredox-catalysed N-glycosylation of N-nucleophiles[J]. Nature Synthesis, 2024, 3(5): 623-632. |
| [55] | LIU D P, ZHANG X S, LIU S, et al. Dehydroxylative radical N-glycosylation of heterocycles with 1-hydroxycarbohydrates enabled by copper metallaphotoredox catalysis[J]. Nature Communications, 2024, 15(1): 3401. |
| [56] | HUANG W, LI C, LI B, et al. Glycosynthases Enable a Highly Efficient Chemoenzymatic Synthesis of N-Glycoproteins Carrying Intact Natural N-Glycans[J]. Journal of the American Chemical Society, 2009, 131(6): 2214-2223. |
| [57] | LI C, WANG L X. Chemoenzymatic Methods for the Synthesis of Glycoproteins[J]. Chemical Reviews, 2018, 118(17): 8359-8413. |
| [58] | DING W, CHEN X, SUN Z, et al. A Radical Activation Strategy for Versatile and Stereoselective N‐Glycosylation[J]. Angewandte Chemie International Edition, 2024, 63(36): e202409004. |
| [59] | LI D, LI C, CHEN Q, et al. Generalizing a Ligation Site at the N-Glycosylation Sequon for Chemical Synthesis of N-Linked Glycopeptides and Glycoproteins[J]. Journal of the American Chemical Society, 2024, 146(42): 29017-29027. |
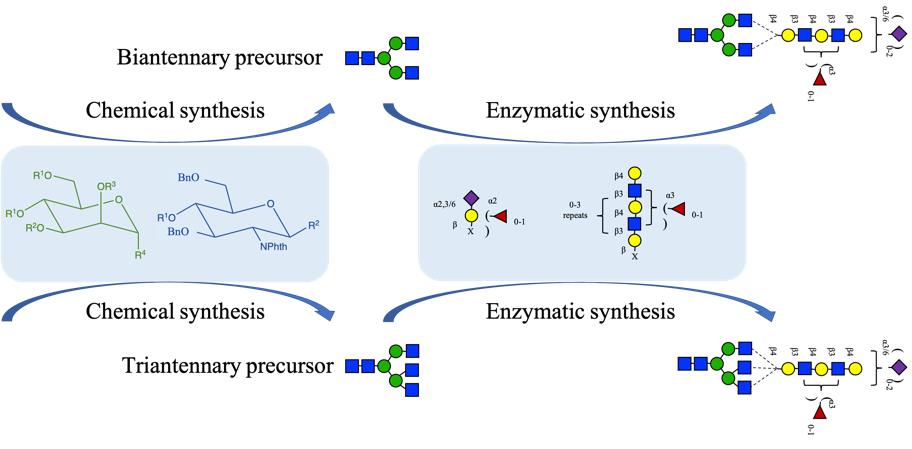
| [60] | Li R, Chen P, Zeng Y-F, et al. Expedient Assembly of Multiantennary N Glycans from Common N-Glycan Cores with Orthogonal Protection for the Profiling of Glycan-Binding Proteins[J]. Journal of the American Chemical Society, 2025,147(34): 12937-12948. |
| [61] | SCHWARZ F, HUANG W, LI C, et al. A combined method for producing homogeneous glycoproteins with eukaryotic N-glycosylation[J]. Nature Chemical Biology, 2010, 6(4): 264-266. |
| [62] | Liu C C, Ye J, Cao H. Chemical Evolution of Enzyme-Catalyzed Glycosylation[J]. Accounts of Chemical Research, 2024, 57(4):636-647. |
| [63] | Wei F, Zang L, Zhang P, et al.Concise chemoenzymatic synthesis of N-glycans[J].Chem, 2024(9):10. |
| [64] | Dubey K K, Kumar A, Baldia A, et al.Biomanufacturing of glycosylated antibodies: Challenges, solutions, and future prospects[J].Biotechnology Advances, 2023, 69(000):16. |
| [65] | Lin C-L, Sojitra M, Carpenter E J, et al. Chemoenzymatic synthesis of genetically- encoded multivalent liquid N-glycan arrays[J]. Nature Communications, 2023,14(1): 5237. |
| [66] | PINHO S S, REIS C A. Glycosylation in cancer: mechanisms and clinical implications[J]. Nature Reviews Cancer, 2015, 15(9): 540-555. |
| [67] | REILY C, STEWART T J, RENFROW M B, et al. Publisher Correction: Glycosylation in health and disease[J]. Nature Reviews Nephrology, 2025, 21(3): 216-216. |
| [68] | PONGRACZ T, MAYBORODA O A, WUHRER M. The Human Blood N-Glycome: Unraveling Disease Glycosylation Patterns[J]. JACS Au, 2024, 4(5): 1696-1708. |
| [69] | HENNET T, CABALZAR J. Congenital disorders of glycosylation: a concise chart of glycocalyx dysfunction[J]. Trends in Biochemical Sciences, 2015, 40(7): 377-384. |
| [70] | ZHU Z, SUN J, XU W, et al. MGAT4A/Galectin9‐Driven N‐Glycosylation Aberration as a Promoting Mechanism for Poor Prognosis of Endometrial Cancer with TP53 Mutation[J]. Advanced Science, 2024, 11(48): 2409764 (1-19. |
| [71] | ZHOU X, MOTTA F, SELMI C, et al. Antibody glycosylation in autoimmune diseases[J]. Autoimmunity Reviews, 2021, 20(5): 102804. |
| [72] | MENDELSOHN R, CHEUNG P, BERGER L, et al. Complex N-Glycan and Metabolic Control in Tumor Cells[J]. Cancer Research, 2007, 67(20): 9771-9780. |
| [73] | REBELO A L, DRAKE R R, MARCHETTI-DESCHMANN M, et al. Changes in tissue protein N-glycosylation and associated molecular signature occur in the human Parkinsonian brain in a region-specific manner[J]. PNAS Nexus, 2023, 3(1): 439. |
| [74] | LIN Y, LUBMAN D M. The role of N-glycosylation in cancer[J]. Acta Pharmaceutica Sinica B, 2024, 14(3): 1098-1110. |
| [75] | FENG X, ZHENG X, LIN A, et al. FBN1 knockout promotes cervical artery dissection by inducing N-glycosylation alternation of extracellular matrix proteins in rat VSMCs[J]. Cellular Signalling, 2023, 110: 110834. |
| [76] | ANNIBALINI G, SALTARELLI R, FERRI L, et al. Impaired myoblast differentiation and muscle IGF‐1 receptor signaling pathway activation after N‐glycosylation inhibition[J]. FASEB Journal, 2024, 38(13). |
| [77] | IWAMOTO S, KOBAYASHI T, HANAMATSU H, et al. Tolerable glycometabolic stress boosts cancer cell resilience through altered N-glycosylation and Notch signaling activation[J]. Cell Death & Disease, 2024, 15(1): 53. |
| [78] | CHANG X, OBIANWUNA U E, WANG J, et al. Glycosylated proteins with abnormal glycosylation changes are potential biomarkers for early diagnosis of breast cancer[J]. International Journal of Biological Macromolecules, 2023, 236: 123855. |
| [79] | BANGARH R, KHATANA C, KAUR S, et al. Aberrant protein glycosylation: Implications on diagnosis and Immunotherapy[J]. Biotechnology Advances, 2023, 66: 108149. |
| [80] | SENRA R L, PEREIRA H S, SCHITTINO L M P, et al. Co-expression of human sialyltransferase improves N-glycosylation in Leishmania tarentolae and optimizes the production of humanized therapeutic glycoprotein IFN-beta[J]. Journal of Biotechnology, 2024, 394: 24-33. |
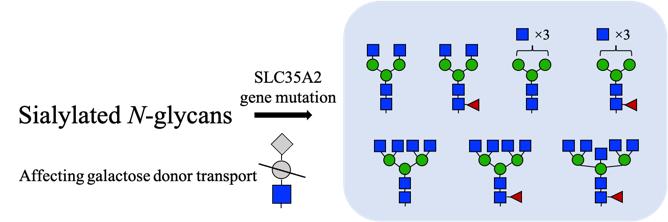
| [81] | LIU X, TANG Q, XIA X, et al. Somatic variants in SLC35A2 leading to defects in N-glycosylation in mild malformation of cortical development with oligodendroglial hyperplasia in epilepsy(MOGHE)[J]. Acta Neuropathologica, 2025, 149(1): 13. |
| [82] | HUNG C H, WU S Y, YAO C I D, et al. Defective N-glycosylation of IL6 induces metastasis and tyrosine kinase inhibitor resistance in lung cancer[J]. Nature Communications, 2024, 15(1): 7885. |
| [83] | ZHANG M, LI M, LI H, et al. Dysregulation of N-glycosylation by Rpn1 knockout in spermatocytes induces male infertility via endoplasmic reticulum stress in mice[J]. International Journal of Biological Sciences, 2025, 21(5): 2360-2379. |
| [84] | Geng J., Ma S., Tang H., & Zhang C. (2025). Pathogenesis and Therapeutic Perspectives of Tubular Injury in Diabetic Kidney Disease: An Update[J]. Biomedicines, 13(6), 1424. |
| [85] | Lee S., Choi J., Mohanty J., Sousa L. P., Tome F., Pardon E., Steyaert J., Lemmon M. A., Lax I., & Schlessinger J. (2018). Structures of β-Klotho reveal a 'zip code'-like mechanism for endocrine FGF signaling[J]. Nature, 553(7689), 501-505. |
| [86] | LI Q, LIU W, ZHANG Y, et al. ALG5 downregulation inhibits osteogenesis and promotes adipogenesis by regulating the N-glycosylation of SLC6A9 in osteoporosis[J]. Cellular and Molecular Life Sciences, 2025, 82(1): 35. |
| [87] | TRZOS S, LINK-LENCZOWSKI P, POCHEĆ E. The role of N-glycosylation in B-cell biology and IgG activity. The aspects of autoimmunity and anti-inflammatory therapy[J]. Frontiers in Immunology, 2023, 14: 1188838. |
| [88] | LIU M, PENG T, HU L, et al. N-glycosylation-mediated CD147 accumulation induces cardiac fibrosis in the diabetic heart through ALK5 activation[J]. International Journal of Biological Sciences, 2023, 19(1): 137-155. |
| [89] | LI R, GAO J, WANG L, et al. Multivalent interactions between fully glycosylated influenza virus hemagglutinins mediated by glycans at distinct N-glycosylation sites[J]. npj Viruses, 2024, 2(1): 48. |
| [90] | GENG L, YI X, LIN Y, et al. Site-specific analysis and functional characterization of N-linked glycosylation for β-Klotho protein[J]. International Journal of Biological Macromolecules, 2025, 289: 138846. |
| [91] | Tyurenkov I. N., Perfilova V. N., Nesterova A. A., & Glinka Y. (2021). Klotho Protein and Cardio-Vascular System[J]. Biochemistry (Moscow), 86(2), 132-145. |
| [92] | MIAO Z, CAO Q, LIAO R, et al. Elevated transcription and glycosylation of B3GNT5 promotes breast cancer aggressiveness[J]. Journal of Experimental & Clinical Cancer Research, 2022, 41(1): 169. |
| [93] | CHATHAM J C, PATEL R P. Protein glycosylation in cardiovascular health and disease[J]. Nature Reviews Cardiology, 2024, 21(8): 525-544. |
| [94] | LIU R, WANG G, QIAN Y, et al. Hexosamine biosynthesis dysfunction-induced LIFR N-glycosylation deficiency exacerbates steatotic liver ischemia/reperfusion injury[J]. Metabolism, 2025, 168: 156258. |
| [95] | PARK J S, CHOI H J, JUNG K M, et al. Production of recombinant human IgG1 Fc with beneficial N-glycosylation pattern for anti-inflammatory activity using genome-edited chickens[J]. Communications Biology, 2023, 6(1): 589. |
| [96] | ZHAO T, LIU S, WANG P, et al. Protective RBD-dimer vaccines against SARS-CoV-2 and its variants produced in glycoengineered Pichia pastoris[J]. PLOS Pathogens, 2024, 20(8): e1012487. |
| [97] | Li Y, Fu B, Wang M,et al.Urinary extracellular vesicle N-glycomics identifies diagnostic glycosignatures for bladder cancer[J].Nature Communications,(2025)16:2292]. |
| [98] | TRBOJEVIĆ-AKMAČIĆ I, VUČKOVIĆ F, PRIBIĆ T, et al. Comparative analysis of transferrin and IgG N-glycosylation in two human populations[J]. Communications Biology, 2023, 6(1): 312. |
| [99] | KRANJC J, KRAMER L, MIKELJ M, et al. Modulating antibody N-glycosylation through feed additives using a multi-tiered approach[J]. Frontiers in Bioengineering and Biotechnology, 2024, 12: 1448925. |
| [100] | CONG W, SHEN H, JIANG Y, et al. Design, Synthesis, and Anti-Osteoporotic Characterization of Arginine N-Glycosylated Teriparatide Analogs via the Silver-catalyzed Solid-Phase Glycosylation Strategy[J]. Journal of Medicinal Chemistry, 2024, 67(2): 1360-1369. |
| [101] | LI X, DING Y, XUE J, et al. Peptide Double-Stapling and Arginine N-Glycosylation Triggered the Development of Therapeutic Antimicrobial Peptides Capable of Killing Drug-Resistant Bacteria in Mice[J]. Journal of Medicinal Chemistry, 2025, 68(4): 4511-4526. |
| [102] | PU Y, ZHOU B, BING J, et al. Ultrasound-triggered and glycosylation inhibition-enhanced tumor piezocatalytic immunotherapy[J]. Nature Communications, 2024, 15(1): 9023. |
| [103] | Chi, Jack Junlong, et al. "MGAT1-Guided Complex N-Glycans on CD73 Regulate Immune Evasion in Triple-Negative Breast Cancer[J]. Nature Communications, 2025, 16: 3225. |
| [104] | VELIČKOVIĆ D, SHAPIRO J P, PARIKH S V, et al. Protein N-Glycans in Healthy and Sclerotic Glomeruli in Diabetic Kidney Disease[J]. Journal of the American Society of Nephrology, 2024, 35(9): 1198-1207. |
| [105] | RUDMAN N, KAUR S, SIMUNOVIĆ V, et al. Integrated glycomics and genetics analyses reveal a potential role for N-glycosylation of plasma proteins and IgGs, as well as the complement system, in the development of type 1 diabetes[J]. Diabetologia, 2023, 66(6): 1071-1083. |
| [106] | Hajare A. D., Dagar N., & Gaikwad A. B. (2025). Klotho antiaging protein: molecular mechanisms and therapeutic potential in diseases[J]. Molecular Biomedicine, 6(19), 1-24. |
| [107] | WANG J, ZHANG H min, ZHU G hua, et al. STT3-mediated aberrant N-glycosylation of CD24 inhibits paclitaxel sensitivity in triple-negative breast cancer[J]. Acta Pharmacologica Sinica, 2025, 46(4): 1097-1110. |
| [108] | MA H, CHEN X, MO S, et al. Targeting N-glycosylation of 4F2hc mediated by glycosyltransferase B3GNT3 sensitizes ferroptosis of pancreatic ductal adenocarcinoma[J]. Cell Death & Differentiation, 2023, 30(8): 1988-2004. |
| [109] | LIU Z, MENG X, ZHANG Y, et al. FUT8-mediated aberrant N-glycosylation of SEMA7A promotes head and neck squamous cell carcinoma progression[J]. International Journal of Oral Science, 2024, 16(1): 26. |
| [110] | WEI Q, ZHANG J, DAO Y, et al. Exploiting Complex-Type N-Glycan to Improve the in Vivo Stability of Bioactive Peptides[J]. CCS Chemistry, 2023, 5(7): 1623-1634. |
| [111] | LIU Z, MENG X, TANG X, et al. A novel allosteric driver mutation of β-glucuronidase promotes head and neck squamous cell carcinoma progression through STT3B-mediated PD‐L1 N‐glycosylation[J]. MedComm, 2025, 6(2): e70062. |
| [112] | KOGELMANN B, MELNIK S, BOGNER M, et al. A genome‐edited N. benthamiana line for industrial‐scale production of recombinant glycoproteins with targeted N‐glycosylation[J]. Biotechnology Journal, 2024, 19(1): 2300323. |
| [113] | SHI C, WANG Y, WU M, et al. Promoting anti-tumor immunity by targeting TMUB1 to modulate PD-L1 polyubiquitination and glycosylation[J]. Nature Communications, 2022, 13(1): 6951. |
| [114] | NIE H, SAINI P, MIYAMOTO T, et al. Targeting branched N-glycans and fucosylation sensitizes ovarian tumors to immune checkpoint blockade[J]. Nature Communications, 2024, 15(1): 2853. |
| [115] | SHU Z, DWIVEDI B, SWITCHENKO J M, et al. PD-L1 deglycosylation promotes its nuclear translocation and accelerates DNA double-strand-break repair in cancer[J]. Nature Communications, 2024, 15(1): 6830. |
| [116] | PINHO S S, ALVES I, GAIFEM J, et al. Immune regulatory networks coordinated by glycans and glycan-binding proteins in autoimmunity and infection[J]. Cellular & Molecular Immunology, 2023, 20(10): 1101-1113. |
| [117] | WANI W Y, ZUNKE F, BELUR N R, et al. The hexosamine biosynthetic pathway rescues lysosomal dysfunction in Parkinson's disease patient iPSC derived midbrain neurons[J]. Nature Communications, 2024, 15(1): 5206. |
| [118] | VARADHARAJ V, PETERSEN W, BATRA S K, et al. Sugar symphony: glycosylation in cancer metabolism and stemness[J]. Trends in Cell Biology, 2024: S096289242400206X. |
| [119] | DU X, QI Z, CHEN S, et al. Synthetic Retinoid Sulfarotene Selectively Inhibits Tumor‐Repopulating Cells of Intrahepatic Cholangiocarcinoma via Disrupting Cytoskeleton by P-Selectin/PSGL1 N-Glycosylation Blockage[J]. Advanced Science, 2025, 12(3): 2407519. |
| [120] | MA B, CHEN H, GONG J, et al. Enhancing Protein Solubility via Glycosylation: From Chemical Synthesis to Machine Learning Predictions[J]. Biomacromolecules, 2024, 25(5): 3001-3010. |
| [1] | 黄扬, 李一叶, 聂广军. 合成生物学赋能细胞膜纳米颗粒的精准诊疗[J]. 合成生物学, 2025, (): 1-24. |
| [2] | 李全飞, 陈乾, 杨凯, 刘浩, 贺坤东, 潘亮, 李莎, 雷鹏, 谷益安, 孙良, 邱益彬, 张琦, 徐虹, 王瑞. 高黏性蛋白材料的生物合成及应用[J]. 合成生物学, 2025, (): 1-20. |
| [3] | 肖和, 姚明菊, 乔雪. 长糖链皂苷的生物合成研究进展[J]. 合成生物学, 2025, (): 1-7. |
| [4] | 李明辰, 钟博子韬, 余元玺, 姜帆, 张良, 谭扬, 虞慧群, 范贵生, 洪亮. DeepSeek模型分析及其在AI辅助蛋白质工程中的应用[J]. 合成生物学, 2025, 6(3): 636-650. |
| [5] | 宋成治, 林一瀚. AI+定向进化赋能蛋白改造及优化[J]. 合成生物学, 2025, 6(3): 617-635. |
| [6] | 田晓军, 张日新. 合成基因回路面临的细胞“经济学窘境”[J]. 合成生物学, 2025, 6(3): 532-546. |
| [7] | 姜百翼, 钱珑. 活细胞记录器在细胞谱系追踪中的应用和前景[J]. 合成生物学, 2025, 6(3): 651-668. |
| [8] | 李倩, FERRELL JR.James E., 陈于平. 细胞质浓度:细胞生物学的老问题、新参数[J]. 合成生物学, 2025, 6(3): 497-515. |
| [9] | 张成辛. 基于文本数据挖掘的蛋白功能预测:机遇与挑战[J]. 合成生物学, 2025, 6(3): 603-616. |
| [10] | 王珂, 陈文慧, 雷春阳, 聂舟. CRISPR/Cas系统在分子诊断领域的应用研究进展[J]. 合成生物学, 2025, (): 1-27. |
| [11] | 甘牡丹, 左静蕊, 曹友志. 工程菌的生物安全防控策略[J]. 合成生物学, 2025, (): 1-14. |
| [12] | 王倩, 果士婷, 辛波, 钟成, 王钰. L-精氨酸的微生物合成研究进展[J]. 合成生物学, 2025, 6(2): 290-305. |
| [13] | 高琪, 肖文海. 酵母合成单萜类化合物的研究进展[J]. 合成生物学, 2025, 6(2): 357-372. |
| [14] | 张梦瑶, 蔡鹏, 周雍进. 合成生物学助力萜类香精香料可持续生产[J]. 合成生物学, 2025, 6(2): 334-356. |
| [15] | 伊进行, 唐宇琳, 李春雨, 吴鹤云, 马倩, 谢希贤. 氨基酸衍生物在化妆品中的应用及其生物合成研究进展[J]. 合成生物学, 2025, 6(2): 254-289. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||