合成生物学 ›› 2020, Vol. 1 ›› Issue (4): 413-426.DOI: 10.12211/2096-8280.2020-047
基因组编辑技术及其在合成生物学中的应用
曹中正1,2, 张心怡3, 徐艺源3, 周卓3,4,5,6, 魏文胜1,3,4,5,6
- 1.北大-清华生命科学联合中心,北京 100871
2.北京大学前沿交叉学科研究院,北京 100871
3.北京大学生命科学学院,北京 100871
4.北京大学生物医学前沿创新中心,北京 100871
5.北京未来基因诊断高精尖创新中心,北京 100871
6.北京大学基因组编辑研究中心,北京 100871
-
收稿日期:2020-04-13修回日期:2020-09-24出版日期:2020-08-31发布日期:2020-11-02 -
通讯作者:魏文胜 -
作者简介:曹中正(1992—),男,博士研究生,专业方向为生物化学与分子生物学。E-mail:czz949290@aliyun.com
魏文胜(1969—),男,博士,教授,研究方向为基因组编辑与功能性基因组学。E-mail:wswei@pku.edu.cn -
基金资助:国家自然科学基金重点项目(31930016)
Genome editing technology and its applications in synthetic biology
CAO Zhongzheng1,2, ZHANG Xinyi3, XU Yiyuan3, ZHOU Zhuo3,4,5,6, WEI Wensheng1,3,4,5,6
- 1.PKU-Tsinghua Center for Life Sciences,Beijing 100871,China
2.Academy for Advanced Interdisciplinary Studies,Peking University,Beijing 100871,China
3.School of Life Science,Peking University,Beijing 100871,China
4.Biomedical Pioneering Innovation Center,Peking University,Beijing 100871,China
5.Beijing Advanced Innovation Center of Genomics,Beijing 100871,China
6.Peking University Genome Editing Research Center,Beijing 100871,China
-
Received:2020-04-13Revised:2020-09-24Online:2020-08-31Published:2020-11-02 -
Contact:WEI Wensheng
摘要:
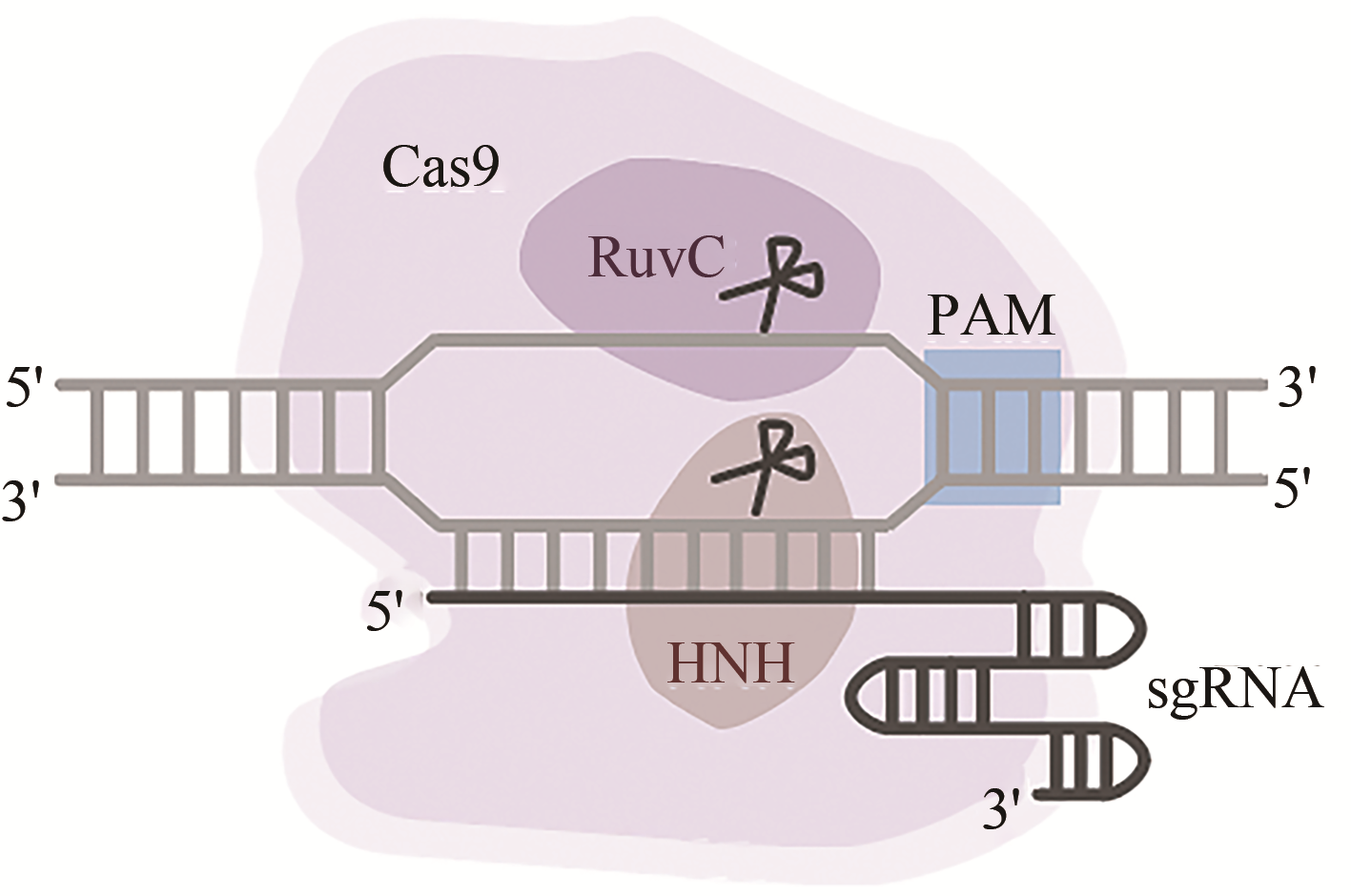
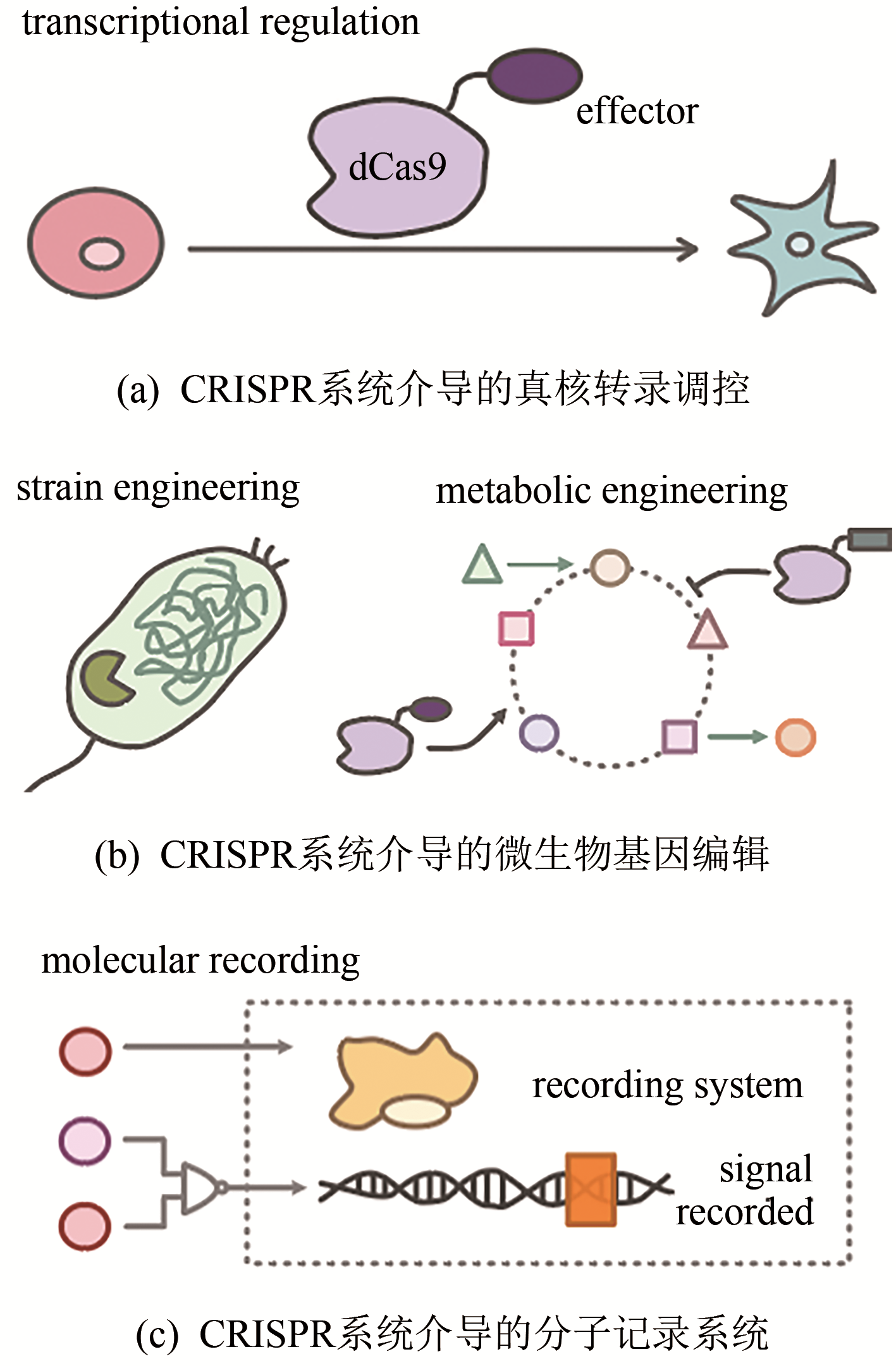
基因组编辑技术是一种能够定向修改基因组的强有力工具。近年来,CRISPR/Cas9系统因其易于构建、编辑效率高等优点逐渐成为应用最为广泛的基因组编辑工具。合成生物学作为一门整合了工程学思维以及生物学原理的新生交叉学科,在生物学、医学、化学、农业、能源和环境等领域发挥着重要的作用。合成生物学对于DNA等遗传物质的合成、组装和编辑等操作有着巨大的需求,因此基因组编辑技术在合成生物学中有着广泛的应用。本文综述了以ZFN和TALEN为代表的早期基因组编辑技术,以及新型CRISPR/Cas9基因组编辑技术的原理、发展、作用机制、系统优化、衍生技术以及应用。同时也介绍了基因组编辑技术在基因表达调控、微生物基因编辑和分子记录等合成生物学领域的应用,并展望了基因组编辑技术的前景以及在合成生物学领域的发展趋势。
中图分类号:
引用本文
曹中正, 张心怡, 徐艺源, 周卓, 魏文胜. 基因组编辑技术及其在合成生物学中的应用[J]. 合成生物学, 2020, 1(4): 413-426.
CAO Zhongzheng, ZHANG Xinyi, XU Yiyuan, ZHOU Zhuo, WEI Wensheng. Genome editing technology and its applications in synthetic biology[J]. Synthetic Biology Journal, 2020, 1(4): 413-426.
| 1 | KIM Yang Gyun, Jooyeun CHA, CHANDRASEGARAN S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain [J]. Proceedings of the National Academy of Sciences of the United States of America, 1996, 93: 1156-1160. |
| 2 | CHRISTIAN M, CERMAK T, DOYLE E L, et al. Targeting DNA double-strand breaks with TAL effector nucleases [J]. Genetics, 2010, 186: 757-761. |
| 3 | CONG Le, RAN F A, COX D, et al. Multiplex genome engineering using CRISPR/Cas systems [J]. Science, 2013, 339: 819-823. |
| 4 | JINEK M, CHYLINSKI K, FONFARA I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity [J]. Science, 2012, 337: 816-821. |
| 5 | MALI P, YANG Luhan, ESVELT K M, et al. RNA-guided human genome engineering via Cas9 [J]. Science, 2013, 339: 823-826. |
| 6 | LEE M S, GIPPERT G P, SOMAN K V, et al. Three-dimensional solution structure of a single zinc finger DNA-binding domain [J]. Science, 1989, 245: 635-637. |
| 7 | MILLER J, MCLACHLAN A D, KLUG A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes [J]. EMBO Journal, 1985, 4: 1609-1614. |
| 8 | WOLFE S A, NEKLUDOVA L, PABO C O. DNA recognition by Cys2His2 zinc finger proteins [J]. Annual Review of Biophysics and Biomolecular Structure, 2000, 29: 183-212. |
| 9 | CHOO Yen, SANCHEZ-GARCIA I, KLUG A. In vivo repression by a site-specific DNA-binding protein designed against an oncogenic sequence [J]. Nature, 1994, 372: 642-645. |
| 10 | DESJARLAIS J R, BERG J M. Use of a zinc-finger consensus sequence framework and specificity rules to design specific DNA binding proteins [J]. Proceedings of the National Academy of Sciences of the United States of America, 1993, 90: 2256-2260. |
| 11 | LI Lin, WU L P, CHANDRASEGARAN S. Functional domains in Fok I restriction endonuclease [J]. Proceedings of the National Academy of Sciences of the United States of America, 1992, 89: 4275-4279. |
| 12 | BIBIKOVA M, CARROLL D, SEGAL D J, et al. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases [J]. Molecular and Cellular Biology, 2001, 21: 289-297. |
| 13 | BITINAITE J, WAH D A, AGGARWAL A K, et al. FokI dimerization is required for DNA cleavage [J]. Proceedings of the National Academy of Sciences of the United States of America, 1998, 95: 10570-10575. |
| 14 | LIEBER M R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway [J]. Annual Review of Biochemistry, 2010, 79: 181-211. |
| 15 | FILIPPO J SAN, SUNG P, KLEIN H. Mechanism of eukaryotic homologous recombination [J]. Annual Review of Biochemistry, 2008, 77: 229-257. |
| 16 | BONAS U, STALL R E, STASKAWICZ B. Genetic and structural characterization of the avirulence gene avrBs3 from Xanthomonas campestris pv. vesicatoria [J]. Molecular and General Genetics, 1989, 218: 127-136. |
| 17 | SZUREK B, ROSSIER O, HAUSE G, et al. Type III-dependent translocation of the Xanthomonas AvrBs3 protein into the plant cell [J]. Molecular Microbiology, 2002, 46: 13-23. |
| 18 | YANG Yinong, GABRIEL D W. Xanthomonas avirulence/pathogenicity gene family encodes functional plant nuclear targeting signals [J]. Molecular Plant-Microbe Interactions, 1995, 8: 627-631. |
| 19 | ZHU Weiguang, YANG Bing, CHITTOOR J M, et al. AvrXa10 contains an acidic transcriptional activation domain in the functionally conserved C terminus [J]. Molecular Plant-Microbe Interactions, 1998, 11: 824-832. |
| 20 | BOCH J, BONAS U. Xanthomonas AvrBs3 family-type III effectors: discovery and function [J]. Annual Review of Phytopathology, 2010, 48: 419-436. |
| 21 | BOCH J, SCHOLZE H, SCHORNACK S, et al. Breaking the code of DNA binding specificity of TAL-type III effectors [J]. Science, 2009, 326: 1509-1512. |
| 22 | MOSCOU M J, BOGDANOVE A J. A simple cipher governs DNA recognition by TAL effectors [J]. Science, 2009, 326: 1501. |
| 23 | ISHINO Y, SHINAGAWA H, MAKINO K, et al. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product [J]. Journal of Bacteriology, 1987, 169: 5429-5433. |
| 24 | GRISSA I, VERGNAUD G, POURCEL C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats [J]. BMC Bioinformatics, 2007, 8: 172. |
| 25 | BARRANGOU R, FREMAUX C, DEVEAU H, et al. CRISPR provides acquired resistance against viruses in prokaryotes [J]. Science, 2007, 315: 1709-1712. |
| 26 | GARNEAU J E, DUPUIS M E, VILLION M, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA [J]. Nature, 2010, 468: 67-71. |
| 27 | SAPRANAUSKAS R, GASIUNAS G, FREMAUX C, et al. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli [J]. Nucleic Acids Research, 2011, 39: 9275-9282. |
| 28 | GASIUNAS G, BARRANGOU R, HORVATH P, et al. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria [J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109: E2579-E2586. |
| 29 | MAKAROVA K S, WOLF Y I, ALKHNBASHI O S, et al. An updated evolutionary classification of CRISPR-Cas systems [J]. Nature Reviews: Microbiology, 2015, 13: 722-736. |
| 30 | SHMAKOV S, ABUDAYYEH O O, MAKAROVA K S, et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems [J]. Molecular Cell, 2015, 60: 385-397. |
| 31 | KOONIN E V, MAKAROVA K S, ZHANG F. Diversity, classification and evolution of CRISPR-Cas systems [J]. Current Opinion in Microbiology, 2017, 37: 67-78. |
| 32 | WRIGHT A V, NUNEZ J K, DOUDNA J. A biology and applications of CRISPR systems: harnessing nature's toolbox for genome engineering [J]. Cell, 2016, 164: 29-44. |
| 33 | RATH D, AMLINGER L, RATH A, et al. The CRISPR-Cas immune system: biology, mechanisms and applications [J]. Biochimie, 2015, 117: 119-128. |
| 34 | MOJICA F J M, DIEZ-VILLASENOR C, GARCIA-MARTINEZ J, et al. Short motif sequences determine the targets of the prokaryotic CRISPR defence system [J]. Microbiology, 2009, 155: 733-740. |
| 35 | DELTCHEVA E, CHYLINSKI K, SHARMA C M, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III [J]. Nature, 2011, 471: 602-607. |
| 36 | STERNBERG S H, REDDING S, JINEK M, et al. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9 [J]. Nature, 2014, 507: 62-67. |
| 37 | NISHIMASU H, RAN F A, HSU P D, et al. Crystal structure of Cas9 in complex with guide RNA and target DNA [J]. Cell, 2014, 156: 935-949. |
| 38 | JINEK M, EAST A, CHENG A, et al. RNA-programmed genome editing in human cells [J]. eLife, 2013, 2: e00471. |
| 39 | CHEN Baohui, GILBERT L A, CIMINI B A, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system [J]. Cell, 2013, 155: 1479-1491. |
| 40 | RAN F A, CONG Le, YAN W X, et al. In vivo genome editing using Staphylococcus aureus Cas9 [J]. Nature, 2015, 520: 186-191. |
| 41 | ZETSCHE B, GOOTENBERG J S, ABUDAYYEH O O, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system [J]. Cell, 2015, 163: 759-771. |
| 42 | ABUDAYYEH O O, GOOTENBERG J S, KONERMANN S, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector [J]. Science, 2016, 353: aaf5573. |
| 43 | QI L S, LARSON M H, GILBERT L A, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression [J]. Cell, 2013, 152: 1173-1183. |
| 44 | GILBERT L A, LARSON M H, MORSUT L, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes [J]. Cell, 2013, 154: 442-451. |
| 45 | MAEDER M L, LINDER S J, CASCIO V M, et al. CRISPR RNA-guided activation of endogenous human genes [J]. Nature Methods, 2013, 10: 977-979. |
| 46 | PEREZ-PINERA P, KOCAK D D, VOCKLEY C M, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors [J]. Nature Methods, 2013, 10: 973-976. |
| 47 | KEARNS N A, PHAM H, TABAK B, et al. Functional annotation of native enhancers with a Cas9-histone demethylase fusion [J]. Nature Methods, 2015, 12: 401-403. |
| 48 | HILTON I B, D'IPPOLITO A M, VOCKLEY C M, et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers [J]. Nature Biotechnology, 2015, 33: 510-517. |
| 49 | KOMOR A C, KIM Y B, PACKER M S, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage [J]. Nature, 2016, 533: 420-424. |
| 50 | MA Yunqing, ZHANG Jiayuan, YIN t, et al. Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells [J]. Nature Methods, 2016, 13: 1029-1035. |
| 51 | NISHIDA K, ARAZOE T, YACHIE N, et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems [J]. Science, 2016: 353. |
| 52 | REES H A, LIU D R. Base editing: precision chemistry on the genome and transcriptome of living cells [J]. Nature Reviews Genetics, 2018, 19: 770-788. |
| 53 | GAUDELLI N M, KOMOR A C, REES H A, et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage [J]. Nature, 2017, 551: 464-471. |
| 54 | ANZALONE A V, RANDOLPH P B, DAVIS J R, et al. Search-and-replace genome editing without double-strand breaks or donor DNA [J]. Nature, 2019, 576: 149-157. |
| 55 | DICARLO J E, NORVILLE J E, MALI P, et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems [J]. Nucleic Acids Research, 2013, 41: 4336-4343. |
| 56 | FRIEDLAND A E, TZUR Y B, ESVELT K M, et al. Heritable genome editing in C. elegans via a CRISPR-Cas9 system [J]. Nature Methods, 2013, 10: 741-743. |
| 57 | YU Zhongsheng, REN Mengda, WANG Zhanxiang, et al. Highly efficient genome modifications mediated by CRISPR/Cas9 in Drosophila [J]. Genetics, 2013, 195: 289-291. |
| 58 | HWANG W Y, FU Yanfang, REYON D, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system [J]. Nature Biotechnology, 2013, 31: 227-229. |
| 59 | LI Jianfeng, NORVILLE J E, AACH J, et al. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9 [J]. Nature Biotechnology, 2013, 31: 688-691. |
| 60 | JIANG Wenzhi, ZHOU Huanbin, BI Honghao, et al. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice [J]. Nucleic Acids Research, 2013, 41: e188. |
| 61 | SHEN Bin, ZHANG Jun, WU Hongya, et al. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting [J]. Cell Research, 2013, 23: 720-723. |
| 62 | LI Dali, QIU Zhongwei, SHAO Yanjiao, et al. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system [J]. Nature Biotechnology, 2013, 31: 681-683. |
| 63 | WANG T, WEI J J, SABATINI D M, et al. Genetic screens in human cells using the CRISPR-Cas9 system [J]. Science, 2014, 343: 80-84. |
| 64 | SHALEM O, SANJANA N E, HARTENIAN E, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells [J]. Science, 2014, 343: 84-87. |
| 65 | ZHOU Yuexin, ZHU Shiyou, CAI Changzu, et al. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells [J]. Nature, 2014, 509: 487-491. |
| 66 | KOIKE-YUSA H, LI Yilong, TAN E-Pien, et al. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library [J]. Nature Biotechnology, 2014, 32: 267-273. |
| 67 | HART T, CHANDRASHEKHAR M, AREGGER M, et al. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities [J]. Cell, 2015, 163: 1515-1526. |
| 68 | CANVER M C, SMITH E C, SHER F, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis [J]. Nature, 2015, 527: 192-197. |
| 69 | FULCO C P, MUNSCHAUER M, ANYOHA R, et al. Systematic mapping of functional enhancer-promoter connections with CRISPR interference [J]. Science, 2016, 354: 769-773. |
| 70 | ZHU Shiyou, LI Wei, LIU Jingze, et al. Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR-Cas9 library [J]. Nature Biotechnology, 2016, 34: 1279-1286. |
| 71 | LIU S J, HORLBECK M A, Seung Woo CHO, et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells [J]. Science, 2017: 355. |
| 72 | LIU Ying, CAO Zhongzheng, WANG Yinan, et al. Genome-wide screening for functional long noncoding RNAs in human cells by Cas9 targeting of splice sites [J]. Nature Biotechnology, 2018. |
| 73 | ADAMSON B, NORMAN T M, JOST M, et al. A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response [J]. Cell, 2016, 167: 1867-1882. |
| 74 | DATLINGER P, RENDEIRO A F, SCHMIDL C, et al. Pooled CRISPR screening with single-cell transcriptome readout [J]. Nature Methods, 2017, 14: 297-301. |
| 75 | DIXIT A, PARNAS O, LI Biyu, et al. Perturb-seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens [J]. Cell, 2016, 167: 1853-1866. |
| 76 | JAITIN D A, WEINER A, YOFE I, et al. Dissecting immune circuits by linking CRISPR-pooled screens with single-cell RNA-seq [J]. Cell, 2016, 167: 1883-1896. |
| 77 | XIE Shiqi, DUAN Jialei, LI Boxun, et al. Multiplexed engineering and analysis of combinatorial enhancer activity in single cells [J]. Molecular Cell, 2017, 66: 285-299. |
| 78 | GOOTENBERG J S, ABUDAYYEH O O, KELLNER M J, et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6 [J]. Science, 2018, 360: 439-444. |
| 79 | GOOTENBERG J S, ABUDAYYEH O O, Jeong Wook LEE, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2 [J]. Science, 2017, 356: 438-442. |
| 80 | CHEN J S, MA Enbo, HARRINGTON L B, et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity [J]. Science, 2018, 360: 436-439. |
| 81 | LI Shiyuan, CHENG Qiuxiang, WANG Jingman, et al. CRISPR-Cas12a-assisted nucleic acid detection [J]. Cell Discovery, 2018, 4: 20. |
| 82 | LI Linxian, LI Shiyuan, WU Na, et al. HOLMESv2: a CRISPR-Cas12b-assisted platform for nucleic acid detection and DNA methylation quantitation [J]. ACS Synthetic Biology, 2019, 8: 2228-2237. |
| 83 | GILPATRICK T, LEE I, GRAHAM J E, et al. Targeted nanopore sequencing with Cas9-guided adapter ligation [J]. Nature Biotechnology, 2020, 38(4):433-438. |
| 84 | PORTEUS M H. A new class of medicines through DNA editing [J]. New England Journal of Medicine, 2019, 380: 947-959. |
| 85 | DEVER D P, BAK R O, REINISCH A, et al. CRISPR/Cas9 beta-globin gene targeting in human haematopoietic stem cells [J]. Nature, 2016, 539: 384-389. |
| 86 | XU Lei, WANG Jun, LIU Yulin, et al. CRISPR-edited stem cells in a patient with HIV and acute lymphocytic leukemia [J]. New England Journal of Medicine, 2019, 381: 1240-1247. |
| 87 | EYQUEM J, MANSILLA-SOTO J, GIAVRIDIS T, et al. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection [J]. Nature, 2017, 543: 113-117. |
| 88 | REN Jiangtao, ZHANG Xuhua, LIU Xiaojun, et al. A versatile system for rapid multiplex genome-edited CAR T cell generation [J]. Oncotarget, 2017, 8: 17002-17011. |
| 89 | LIU Xiaojuan, ZHANG Yongping, CHENG Chen, et al. CRISPR-Cas9-mediated multiplex gene editing in CAR-T cells [J]. Cell Research, 2017, 27: 154-157. |
| 90 | LI Meiru, LI Xiaoxia, ZHOU Zejiao, et al. Reassessment of the four yield-related Genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system [J]. Frontiers in Plant Science, 2016, 7: 377. |
| 91 | XU Rongfang, YANG Yachun, QIN Ruiying, et al. Rapid improvement of grain weight via highly efficient CRISPR/Cas9-mediated multiplex genome editing in rice [J]. J Genet Genomics, 2016, 43: 529-532. |
| 92 | SOYK S, MULLER N A, PARK Soon Ju, et al. Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato [J]. Nature Genetics, 2017, 49: 162-168. |
| 93 | SUN Yongwei, JIAO Guiai, LIU Zupei, et al. Generation of high-amylose rice through CRISPR/Cas9-mediated targeted mutagenesis of starch branching enzymes [J]. Frontiers in Plant Science, 2017: 8, 298. |
| 94 | MORINEAU C, BELLEC Y, TELLIER F, et al. Selective gene dosage by CRISPR-Cas9 genome editing in hexaploid Camelina sativa [J]. Plant Biotechnology Journal, 2017, 15: 729-739. |
| 95 | DONG O X, YU Shu, JAIN R, et al. Marker-free carotenoid-enriched rice generated through targeted gene insertion using CRISPR-Cas9 [J]. Nat Commun, 2020, 11: 1178. |
| 96 | WANG Fujun, WANG Chunlian, LIU Piqing, et al. Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF Transcription Factor Gene OsERF922 [J]. PloS One, 2016, 11: e0154027. |
| 97 | MCLEOD C, SMITH P, MCGUINNESS S L, et al. Human case of Balantidium infection in Australia [J]. Pathology, 2015, 47: 603-604. |
| 98 | ALI Z, ALI S, TASHKANDI M, et al. CRISPR/Cas9-mediated immunity to Geminiviruses: differential interference and evasion [J]. Scientific Reports, 2016, 6: 26912. |
| 99 | ANDRIANANTOANDRO E, BASU S, KARIG D K, et al. Synthetic biology: new engineering rules for an emerging discipline [J]. Molecular Systems Biology, 2006, 2: 2006.0028. |
| 100 | RIVENBARK A G, STOLZENBURG S, BELTRAN A S, et al. Epigenetic reprogramming of cancer cells via targeted DNA methylation [J]. Epigenetics, 2012, 7: 350-360. |
| 101 | KHALIL A S, LU T K, BASHOR C J, et al. A synthetic biology framework for programming eukaryotic transcription functions [J]. Cell, 2012, 150: 647-658. |
| 102 | KEUNG A J, BASHOR C J, KIRIAKOV S, et al. Using targeted chromatin regulators to engineer combinatorial and spatial transcriptional regulation [J]. Cell, 2014, 158: 110-120. |
| 103 | MAEDER M L, ANGSTMAN J F, RICHARDSON M E, et al. Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins [J]. Nature Biotechnology, 2013, 31: 1137-1142. |
| 104 | MENDENHALL E M, WILLIAMSON K E, REYON D, et al. Locus-specific editing of histone modifications at endogenous enhancers [J]. Nature Biotechnology, 2013, 31: 1133-1136. |
| 105 | KONERMANN S, BRIGHAM M D, TREVINO A, et al. Optical control of mammalian endogenous transcription and epigenetic states [J]. Nature, 2013, 500: 472-476. |
| 106 | ZHANG Feng, CONG Le, LODATO S, et al. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription [J]. Nature Biotechnology, 2011, 29: 149-153. |
| 107 | CHAVEZ A, SCHEIMAN J, VORA S, et al. Highly efficient Cas9-mediated transcriptional programming [J]. Nature Methods, 2015, 12: 326-328. |
| 108 | TANENBAUM M E, GILBERT L A, QI L S, et al. A protein-tagging system for signal amplification in gene expression and fluorescence imaging [J]. Cell, 2014, 159: 635-646. |
| 109 | KONERMANN S, BRIGHAM M D, TREVINO A E, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex [J]. Nature, 2015, 517: 583-588. |
| 110 | CHAKRABORTY S, JI HaYeun, KABADI A M, et al. A CRISPR/Cas9-based system for reprogramming cell lineage specification [J]. Stem Cell Reports, 2014, 3: 940-947. |
| 111 | KEARNS N A, GENGA R M, ENUAMEH M S, et al. Cas9 effector-mediated regulation of transcription and differentiation in human pluripotent stem cells [J]. Development, 2014, 141: 219-223. |
| 112 | LIU X S, WU Hao, JI Xiong, et al. Editing DNA methylation in the mammalian genome [J]. Cell, 2016, 167: 233-247. |
| 113 | KWON D Y, ZHAO Yingtao, LAMONICA J M, et al. Locus-specific histone deacetylation using a synthetic CRISPR-Cas9-based HDAC [J]. Nature Communications, 2017, 8: 15315. |
| 114 | ZETSCHE B, VOLZ S E, ZHANG Feng. A split-Cas9 architecture for inducible genome editing and transcription modulation [J]. Nature Biotechnology, 2015, 33: 139-142. |
| 115 | POLSTEIN L R, GERSBACH C A. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation [J]. Nature Chemical Biology, 2015, 11: 198-200. |
| 116 | NIHONGAKI Y, YAMAMOTO S, KAWANO F, et al. CRISPR-Cas9-based photoactivatable transcription system [J]. Chemistry and Biology, 2015, 22: 169-174. |
| 117 | GAO Yuchen, XIONG Xin, WONG S, et al. Complex transcriptional modulation with orthogonal and inducible dCas9 regulators [J]. Nature Methods, 2016, 13: 1043-1049. |
| 118 | LIAO Hsin-Kai, HATANAKA F, ARAOKA T, et al. In vivo target gene activation via CRISPR/Cas9-mediated trans-epigenetic modulation [J]. Cell, 2017, 171: 1495-1507. |
| 119 | LIU X S, WU Hao, KRZISCH M, et al. Rescue of fragile X syndrome neurons by DNA methylation editing of the FMR1 gene [J]. Cell, 2018, 172: 979-992 e976. |
| 120 | MORENO A M, FU Xin, ZHU Jie, et al. In situ gene therapy via AAV-CRISPR-Cas9-mediated targeted gene regulation [J]. Molecular Therapy, 2018, 26: 1818-1827. |
| 121 | YE Wei, ZHANG Weimin, LIU Taomei, et al. Improvement of ethanol production in Saccharomyces cerevisiae by high-efficient disruption of the ADH2 gene using a novel recombinant TALEN vector [J]. Frontiers in Microbiology, 2016, 7: 1067. |
| 122 | YE Wei, LIU Taomei, ZHU Muzi, et al. An easy and efficient strategy for the enhancement of epothilone production mediated by TALE-TF and CRISPR/dcas9 systems in Sorangium cellulosum [J]. Frontiers in Bioengineering and Biotechnology, 2019, 7: 334. |
| 123 | LINDNER S N, PETROV D P, HAGMANN C T, et al. Phosphotransferase system-mediated glucose uptake is repressed in phosphoglucoisomerase-deficient Corynebacterium glutamicum strains [J]. Applied and Environmental Microbiology, 2013, 79: 2588-2595. |
| 124 | SAWADA K, ZEN-IN S, WADA M, et al. Metabolic changes in a pyruvate kinase gene deletion mutant of Corynebacterium glutamicum ATCC 13032 [J]. Metabolic Engineering, 2010, 12: 401-407. |
| 125 | WU Junjun, DU Guocheng, CHEN Jian, et al. Enhancing flavonoid production by systematically tuning the central metabolic pathways based on a CRISPR interference system in Escherichia coli [J]. Scientific Reports, 2015, 5: 13477. |
| 126 | SCHWARTZ C, CURTIS N, LOBS A K, et al. Multiplexed CRISPR activation of cryptic sugar metabolism enables yarrowia lipolytica growth on cellobiose [J]. Biotechnology Journal, 2018, 13: e1700584. |
| 127 | COBB R E, WANG Yajie, ZHAO Huimin. High-efficiency multiplex genome editing of Streptomyces species using an engineered CRISPR/Cas system [J]. ACS Synthetic Biology, 2015, 4: 723-728. |
| 128 | JIA Haiyan, ZHANG Longmei, WANG Tongtong, et al. Development of a CRISPR/Cas9-mediated gene-editing tool in Streptomyces rimosus [J]. Microbiology, 2017, 163: 1148-1155. |
| 129 | ZHANG Yueping, WANG Juan, WANG Zibai, et al. A gRNA-tRNA array for CRISPR-Cas9 based rapid multiplexed genome editing in Saccharomyces cerevisiae [J]. Nature Communications, 2019, 10: 1053. |
| 130 | LI Lei, WEI Keke, ZHENG Guosong, et al. CRISPR-Cpf1-assisted multiplex genome editing and transcriptional repression in Streptomyces [J]. Applied and Environmental Microbiology, 2018, 84. |
| 131 | TONG Y J, WHITFORD C M, ROBERTSEN H L, et al. Highly efficient DSB-free base editing for streptomycetes with CRISPR-BEST [J]. Proceedings of the National Academy of Sciences of the United States of America, 2019, 116: 20366-20375. |
| 132 | COHEN D R, TOWNSEND C A. A dual role for a polyketide synthase in dynemicin enediyne and anthraquinone biosynthesis [J]. Nature Chemistry, 2018, 10: 231-236. |
| 133 | ZHANG M M, WONG Fongtian, WANG Yajie, et al. CRISPR-Cas9 strategy for activation of silent Streptomyces biosynthetic gene clusters [J]. Nature Chemical Biology, 2017. doi:10.1038/nchembio.2341 . |
| 134 | FARZADFARD F, LU T K. Emerging applications for DNA writers and molecular recorders [J]. Science, 2018, 361: 870-875. |
| 135 | TANG Weixin, LIU D R. Rewritable multi-event analog recording in bacterial and mammalian cells [J]. Science, 2018, 360: eaap8992. |
| 136 | FARZADFARD F, GHARAEI N, HIGASHIKUNI Y, et al. Single-nucleotide-resolution computing and memory in living cells [J]. Molecular Cell, 2019, 75: 769-780. |
| 137 | MCKENNA A, FINDLAY G M, GAGNON J A, et al. Whole-organism lineage tracing by combinatorial and cumulative genome editing [J]. Science, 2016, 353: aaf7907. |
| 138 | FRIEDA K L, LINTON J M, HORMOZ S, et al. Synthetic recording and in situ readout of lineage information in single cells [J]. Nature, 2017, 541: 107-111. |
| 139 | KALHOR R, MALI P, CHURCH G M. Rapidly evolving homing CRISPR barcodes [J]. Nature Methods, 2017, 14: 195-200. |
| 140 | PERLI S D, CUI C H, LU T K. Continuous genetic recording with self-targeting CRISPR-Cas in human cells [J]. Science, 2016: 353. |
| 141 | SHIPMAN S L, NIVALA J, MACKLIS J D, et al. Molecular recordings by directed CRISPR spacer acquisition [J]. Science, 2016, 353: aaf1175. |
| 142 | SHIPMAN S L, NIVALA J, MACKLIS J D, et al. CRISPR-Cas encoding of a digital movie into the genomes of a population of living bacteria [J]. Nature, 2017, 547: 345-349. |
| 143 | SHETH R U, Sung Sun YIM, WU F L, et al. Multiplex recording of cellular events over time on CRISPR biological tape [J]. Science, 2017, 358: 1457-1461. |
| 144 | HU J H, MILLER S M, GEURTS M H, et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity [J]. Nature, 2018, 556: 57-63. |
| 145 | KLEINSTIVER B P, PREW M S, TSAI S Q, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities [J]. Nature, 2015, 523: 481-485. |
| 146 | MILLER S M, WANG T, RANDOLPH P B, et al. Continuous evolution of SpCas9 variants compatible with non-G PAMs [J]. Nature Biotechnology, 2020, 38: 471-481. |
| 147 | NISHIMASU H, SHI Xi, ISHIGURO S, et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space [J]. Science, 2018, 361: 1259-1262. |
| 148 | WALTON R T, CHRISTIE K A, WHITTAKER M N, et al. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants [J]. Science, 2020. |
| 149 | CHEN J S, DAGDAS Y S, KLEINSTIVER B P, et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy [J]. Nature, 2017, 550: 407-410. |
| 150 | KLEINSTIVER B P, PATTANAYAK V, PREW M S, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects [J]. Nature, 2016, 529: 490-495. |
| 151 | SLAYMAKER I M, GAO Linyi, ZETSCHE B, et al. Rationally engineered Cas9 nucleases with improved specificity [J]. Science, 2016, 351: 84-88. |
| 152 | BURSTEIN D, HARRINGTON L B, STRUTT S C, et al. New CRISPR-Cas systems from uncultivated microbes [J]. Nature, 2017, 542: 237-241. |
| 153 | KIM Eunji, Taeyoung KOO, PARK Sung Wook, et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni [J]. Nature Communications, 2017, 8: 14500. |
| 154 | MA Dacheng, PENG Shuguang, HUANG Weiren, et al. Rational design of mini-Cas9 for transcriptional activation [J]. ACS Synthetic Biology, 2018, 7: 978-985. |
| 155 | LUO M L, LEENAY R T, BEISEL C L. Current and future prospects for CRISPR-based tools in bacteria [J]. Biotechnology and Bioengineering, 2016, 113: 930-943. |
| [1] | 高歌, 边旗, 王宝俊. 合成基因线路的工程化设计研究进展与展望[J]. 合成生物学, 2025, 6(1): 45-64. |
| [2] | 李冀渊, 吴国盛. 合成生物学视域下有机体的两种隐喻[J]. 合成生物学, 2025, 6(1): 190-202. |
| [3] | 焦洪涛, 齐蒙, 邵滨, 蒋劲松. DNA数据存储技术的法律治理议题[J]. 合成生物学, 2025, 6(1): 177-189. |
| [4] | 任家卫, 张金鹏, 徐国强, 张晓梅, 许正宏, 张晓娟. 大肠杆菌中终止子对下游转录单元基因表达的影响[J]. 合成生物学, 2025, 6(1): 213-227. |
| [5] | 唐兴华, 陆钱能, 胡翌霖. 人类世中对合成生物学的哲学反思[J]. 合成生物学, 2025, 6(1): 203-212. |
| [6] | 徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176. |
| [7] | 石婷, 宋展, 宋世怡, 张以恒. 体外生物转化(ivBT):生物制造的新前沿[J]. 合成生物学, 2024, 5(6): 1437-1460. |
| [8] | 柴猛, 王风清, 魏东芝. 综合利用木质纤维素生物转化合成有机酸[J]. 合成生物学, 2024, 5(6): 1242-1263. |
| [9] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [10] | 陈雨, 张康, 邱以婧, 程彩云, 殷晶晶, 宋天顺, 谢婧婧. 微生物电合成技术转化二氧化碳研究进展[J]. 合成生物学, 2024, 5(5): 1142-1168. |
| [11] | 郑皓天, 李朝风, 刘良叙, 王嘉伟, 李恒润, 倪俊. 负碳人工光合群落的设计、优化与应用[J]. 合成生物学, 2024, 5(5): 1189-1210. |
| [12] | 夏孔晨, 徐维华, 吴起. 光酶催化混乱性反应的研究进展[J]. 合成生物学, 2024, 5(5): 997-1020. |
| [13] | 陈子苓, 向阳飞. 类器官技术与合成生物学协同研究进展[J]. 合成生物学, 2024, 5(4): 795-812. |
| [14] | 蔡冰玉, 谭象天, 李伟. 合成生物学在干细胞工程化改造中的研究进展[J]. 合成生物学, 2024, 5(4): 782-794. |
| [15] | 谢皇, 郑义蕾, 苏依婷, 阮静怡, 李永泉. 放线菌聚酮类化合物生物合成体系重构研究进展[J]. 合成生物学, 2024, 5(3): 612-630. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||