合成生物学 ›› 2020, Vol. 1 ›› Issue (6): 635-655.DOI: 10.12211/2096-8280.2020-027
噬菌体合成生物学研究进展和应用
袁盛建1,2, 马迎飞1
- 1.中国科学院深圳先进技术研究院,深圳合成生物学创新研究院,中国科学院定量工程生物学重点实验室,广东省合成基因组学重点实验室,深圳市合成基因组学重点实验室,广东 深圳 518055
2.中国科学院大学,北京 100049
-
收稿日期:2020-03-16修回日期:2020-11-27出版日期:2020-12-31发布日期:2021-01-15 -
通讯作者:马迎飞 -
作者简介:袁盛建(1988—),男,博士研究生。研究方向为噬菌体人工合成。E-mail:sj.yuan@siat.ac.cn
马迎飞(1978—),男,研究员,博士生导师。主要研究方向:①噬菌体组学,应用生物信息用手段,鉴定各生态系统中噬菌体宏基因组及噬菌体在各生态位中的功能等;②人工噬菌体,对噬菌体进行必需基因功能鉴定,模块化设计与合成;③噬菌体的应用,噬菌体防治耐药菌感染。E-mail:yingfei.ma@siat.ac.cn -
基金资助:广东省合成基因组学重点实验室(2019B030301006);深圳市合成基因组学重点实验室(ZDSYS201802061806209);深圳市海外高层次人才创新团队(KQTD2016112915000294)
Advances and applications of phage synthetic biology
YUAN Shengjian1,2, MA Yingfei1
- 1.Shenzhen Key Laboratory of Synthetic Genomics,Guangdong Provincial Key Laboratory of Synthetic Genomics,CAS Key Laboratory of Quantitative Engineering Biology,Shenzhen Institute of Synthetic Biology,Shenzhen Institutes of Advanced Technology,Chinese Academy of Sciences,Shenzhen 518055,Guangdong,China
2.University of Chinese Academy of Sciences,Beijing 100049,China
-
Received:2020-03-16Revised:2020-11-27Online:2020-12-31Published:2021-01-15 -
Contact:MA Yingfei
摘要:
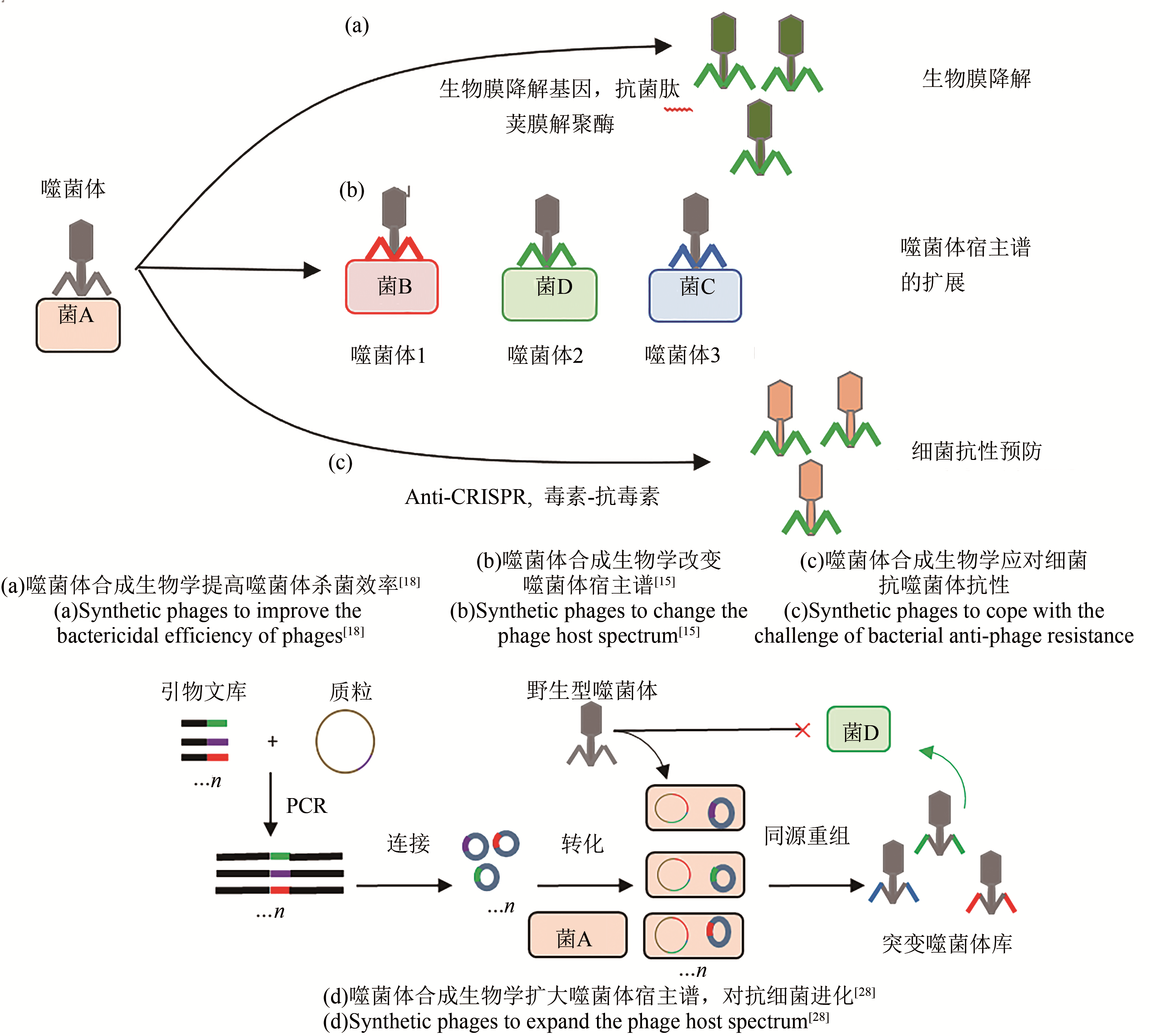
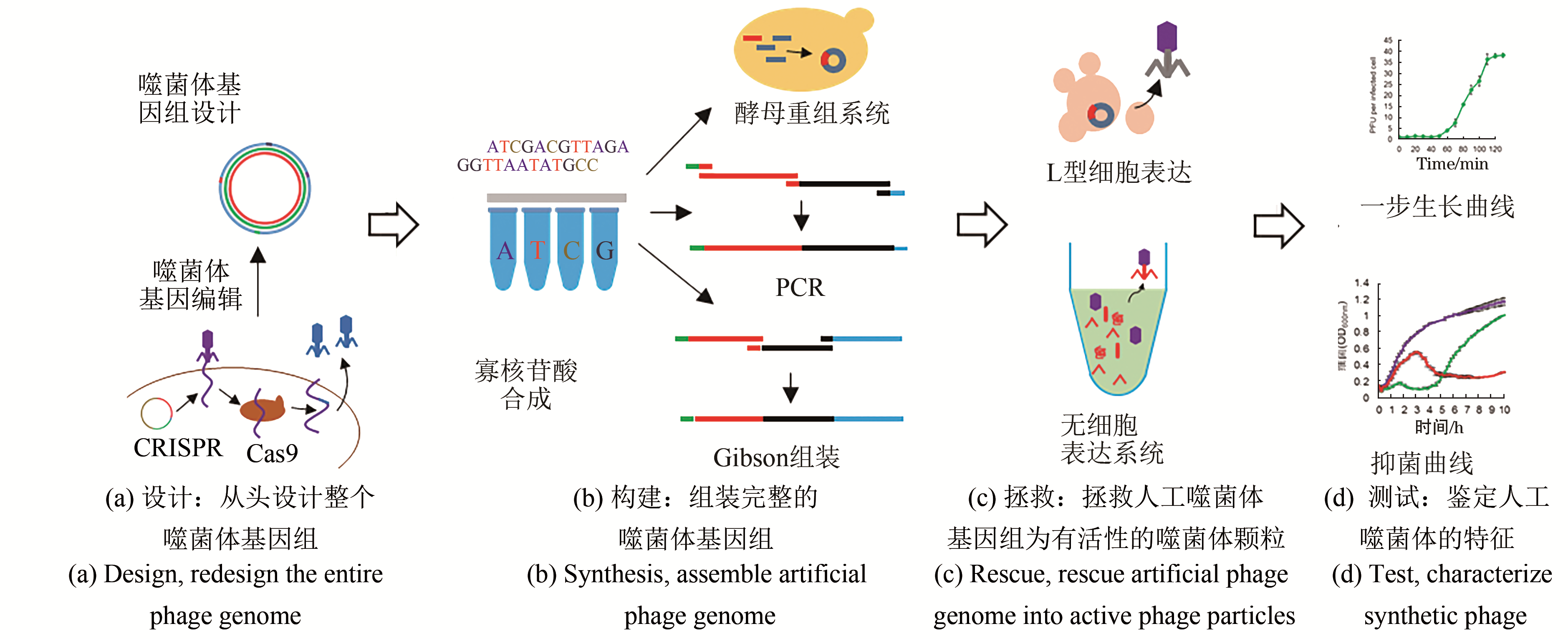
噬菌体是地球上多样性最高和最丰富的生物体,也是合成生物学研究中重要的模式生物。噬菌体基因组相对较小,结构简单,是研究基本生命过程最简单的生物系统。通过对噬菌体基因组进行编辑,乃至重新设计、合成噬菌体基因组,获得具有新的功能的噬菌体,是当前噬菌体合成生物学研究的重要内容。本文综述了当前合成生物学在解决天然噬菌体的基础研究和应用研究中的主要进展,如通过人工改造的噬菌体已成功用于提高噬菌体的侵染效率,调节噬菌体宿主范围,降低噬菌体毒性和免疫原性,提高给药后噬菌体存活周期,提高噬菌体对生物膜的降解等;此外,噬菌体展示、噬菌体辅助的持续进化和噬菌体介导的DNA转导等也成为合成生物学研究中强大的工具。总之,合成生物学的发展将为模块化设计噬菌体作为多功能生物制剂、控制多重耐药细菌、病原体检测、药物开发、菌群的调控、药物递送,甚至噬菌体纳米材料等铺平道路。
中图分类号:
引用本文
袁盛建, 马迎飞. 噬菌体合成生物学研究进展和应用[J]. 合成生物学, 2020, 1(6): 635-655.
YUAN Shengjian, MA Yingfei. Advances and applications of phage synthetic biology[J]. Synthetic Biology Journal, 2020, 1(6): 635-655.
| 146 | HSU B B, GIBSON TE, YELISEYEV V, et al. Dynamic modulation of the gut microbiota and metabolome by bacteriophages in a mouse model [J]. Cell Host and Microbe, 2019, 25(6): 803-814. |
| 147 | CITORIK R J, MIMEE M, LU T K. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases [J]. Nature Biotechnology, 2014, 32(11): 1141-1145. |
| 148 | KATO T, YUI M, DEO V K, et al. Development of Rous sarcoma Virus-like particles displaying hCC49 scFv for specific targeted drug delivery to human colon carcinoma cells [J]. Pharmaceutical Research, 2015, 32(11): 3699-3707. |
| 149 | DEO V K, KATO T, PARK E Y. Virus-like particles displaying recombinant short-chain fragment region and interleukin 2 for targeting colon cancer tumors and attracting macrophages [J]. Journal of Pharmaceutical Sciences, 2016, 105(5): 1614-1622. |
| 150 | LINO C A, CALDEIRA J C, PEABODY D S. Display of single-chain variable fragments on bacteriophage MS2 virus-like particles [J]. Journal of Nanobiotechnology, 2017, 15(1): 13. |
| 151 | VAKS L, BENHAR I. Antibacterial application of engineered bacteriophage nanomedicines: antibody-targeted, chloramphenicol prodrug loaded bacteriophages for inhibiting the growth of Staphylococcus aureus bacteria [J]. Methods in Molecular Biology, 2011, 726: 187-206. |
| 152 | ANAND P, O'NEIL A, LIN E, et al. Tailored delivery of analgesic ziconotide across a blood brain barrier model using viral nanocontainers [J]. Scientific Reports, 2015, 5: 12497. |
| 153 | QAZI S, MIETTINEN H M, WILKINSON R A, et al. Programmed self-assembly of an active P22-Cas9 nanocarrier system [J]. Molecular Pharmaceutics, 2016, 13(3): 1191-1196. |
| 154 | HUME H K C, VIDIGAL J. Synthetic biology for bioengineering virus-like particle vaccines [J]. Cellular & Molecular Immunology, 2019, 116(4): 919-35. |
| 155 | LUA L H, CONNORS N K, SAINSBURY F, et al. Bioengineering virus-like particles as vaccines [J]. Biotechnology and Bioengineering, 2014, 111(3): 425-40. |
| 156 | DENG L, ROOSE K, JOB E R, et al. Oral delivery of Escherichia coli persistently infected with M2e-displaying bacteriophages partially protects against influenza A virus [J]. Journal of Controlled Release, 2017, 264: 55-65. |
| 157 | XU Hai, BAO Xi, LU Yu, et al. Immunogenicity of T7 bacteriophage nanoparticles displaying G-H loop of foot-and-mouth disease virus (FMDV) [J]. Vet Microbiol, 2017, 205: 46-52. |
| 158 | BAHADIR A O, BALCIOGLU B K, UZYOL K S, et al. Phage displayed HBV core antigen with immunogenic activity [J]. Applied Biochemistry and Biotechnology, 2011, 165(7/8): 1437-1447. |
| 159 | GAO Jianming, LIU Z, HUANG M, et al. T7 phage displaying latent membrane protein 1 of Epstein-Barr virus elicits humoral and cellular immune responses in rats [J]. Acta Virologica, 2011, 55(2): 117-121. |
| 160 | BUTTERFIELD G L, LAJOIE M J, GUSTAFSON H H, et al. Evolution of a designed protein assembly encapsulating its own RNA genome [J]. Nature, 2017, 552(7685): 415-420. |
| 161 | HAN Lei, SHAO Changxu, LIANG Bo, et al. Genetically engineered phage-templated MnO2 nanowires: synthesis and their application in electrochemical glucose biosensor operated at neutral pH condition [J]. ACS Applied Materials & Interfaces, 2016, 8(22): 13768-13776. |
| 162 | MANIVANNAN S, KANG Inhak, Yeji SEO, et al. M13 virus-incorporated biotemplates on electrode surfaces to nucleate metal nanostructures by electrodeposition [J]. ACS Applied Materials & Interfaces, 2017, 9(38): 32965-32976. |
| 163 | Dahyun OH, QI Jifa, HAN Binghong, et al. M13 virus-directed synthesis of nanostructured metal oxides for lithium-oxygen batteries [J]. Nano Letters, 2014, 14(8): 4837-4845. |
| 164 | YI Hyunjung, GHOSH D, Moon Ho HAM, et al. M13 phage-functionalized single-walled carbon nanotubes as nanoprobes for second near-infrared window fluorescence imaging of targeted tumors [J]. Nano Letters, 2012, 12(3): 1176-1183. |
| 165 | SZOT-KARPIŃSKA K, GOLEC P, LEŚNIEWSKI A, et al. Modified filamentous bacteriophage as a scaffold for carbon nanofiber [J]. Bioconjugate Chemistry, 2016, 27(12): 2900-2910. |
| 166 | Hwa Kyoung LEE, Yujean LEE, KIM Hyori, et al. Screening of Pro-Asp sequences exposed on bacteriophage M13 as an ideal anchor for gold nanocubes [J]. ACS Synthetic Biology, 2017, 6(9): 1635-1641. |
| 167 | GIESSEN T W, SILVER P A. A catalytic nanoreactor based on in vivo encapsulation of multiple enzymes in an engineered protein nanocompartment [J]. ChemBioChem, 2016, 17(20): 1931-1935. |
| 168 | MYHRVOLD C, POLKA J K, SILVER P A. Synthetic lipid-containing scaffolds enhance production by colocalizing enzymes [J]. ACS Synthetic Biology, 2016, 5(12): 1396-4103. |
| 169 | DION M B, OECHSLIN F, MOINEAU S. Phage diversity, genomics and phylogeny [J]. Nature Reviews Microbiology, 2020, 18(3): 125-138. |
| 1 | TWORT F W. An investigation on the nature of ultra-microscopic viruses [J]. The Lancet, 1915, 186(4814): 1241-1243. |
| 2 | HENDRIX R W. Bacteriophage genomics [J]. Current Opinion in Microbiology, 2003, 6(5): 506-511. |
| 3 | MA Yingfei, YOU Xiaoyan, Guoqin MAI, et al. A human gut phage catalog correlates the gut phageome with type 2 diabetes [J]. Microbiome, 2018, 6(1): 24. |
| 4 | HERSHEY A D, CHASE M. Independent functions of viral protein and nucleic acid in growth of bacteriophage [J]. Journal of General Physiology, 1952, 36(1): 39-56. |
| 5 | CRICK F H C, BARNETT L, BRENNER S, et al. General nature of the genetic code for proteins [J]. Nature, 1961, 192(4809): 1227-1232. |
| 6 | CHAMBERLIN M, RING J. Characterization of T7-specific ribonucleic acid polymerase (I): general properties of the enzymatic reaction and the template specificity of the enzyme [J]. The Journal of Biological Chemistry, 1973, 248(6): 2235-2244. |
| 7 | SMITH H O, C A 3rd HUTCHISON, PFANNKOCH C, et al. Generating a synthetic genome by whole genome assembly: phiX174 bacteriophage from synthetic oligonucleotides [J]. Proceedings of the National Academy of Sciences of the United States of America, 2003, 100(26): 15440-15445. |
| 8 | BARRANGOU R, FREMAUX C, DEVEAU H, et al. CRISPR provides acquired resistance against viruses in prokaryotes [J]. Science, 2007, 315(5819): 1709-1712. |
| 9 | DONOHOUE P D, BARRANGOU R, MAY A P. Advances in industrial biotechnology using CRISPR-Cas systems [J]. Trends in Biotechnology, 2018, 36(2): 134-146. |
| 10 | POTERA C. Phage renaissance: new hope against antibiotic resistance [J]. Environmental Health Perspectives, 2013, 121(2): a48-53. |
| 11 | BARBU E M, CADY K C, HUBBY B. Phage therapy in the era of synthetic biology [J]. Cold Spring Harbor Perspectives in Biology, 2016, 8(10): a023879. |
| 12 | LU T K, KOERIS M S. The next generation of bacteriophage therapy [J]. Current Opinion in Microbiology, 2011, 14(5): 524-531. |
| 170 | SHARMA U, PAUL V D. Bacteriophage lysins as antibacterials [J]. Critical Care, 2017, 21(1): 99. |
| 13 | BORN Y, FIESELER L, THONY V, et al. Engineering of bacteriophages Y2:: dpoL1-C and Y2:: luxAB for efficient control and rapid detection of the fire blight pathogen, Erwinia amylovora [J]. Applied and Environmental Microbiology, 2017, 83(12): e00341. |
| 14 | ROSTOL J T, MARRAFFINI L. (Ph)ighting phages: how bacteria resist their parasites [J]. Cell Host and Microbe, 2019, 25(2): 184-194. |
| 15 | ANDO H, LEMIRE S, PIRES D P, et al. Engineering modular viral scaffolds for targeted bacterial population editing [J]. Cell Systems, 2015, 1(3): 187-196. |
| 16 | C A 3rd HUTCHISON, CHUANG Ray-Yuan, NOSKOV V N, et al. Design and synthesis of a minimal bacterial genome [J]. Science, 2016, 351(6280): aad6253. |
| 17 | BHATTARAI S R, So Young YOO, Seung-Wuk LEE, et al. Engineered phage-based therapeutic materials inhibit Chlamydia trachomatis intracellular infection [J]. Biomaterials, 2012, 33(20): 5166-5174. |
| 18 | LU T K, COLLINS J J. Dispersing biofilms with engineered enzymatic bacteriophage [J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(27): 11197-11202. |
| 19 | LIN Han, PAFF M L, MOLINEUX I J, et al. Therapeutic application of phage capsule depolymerases against K1, K5, and K30 capsulated E. coli in mice [J]. Frontiers in Microbiology, 2017, 8: 2257. |
| 20 | FLEMING D, RUMBAUGH K P. Approaches to dispersing medical biofilms [J]. Microorganisms, 2017, 5(2): 5020015. |
| 21 | CHUNG Yeon-Bo, HINKLE D C. Bacteriophage T7 DNA packaging (I): plasmids containing a T7 replication origin and the T7 concatemer junction are packaged into transducing particles during phage infection [J]. Journal of Molecular Biology, 1990, 216(4): 911-926. |
| 22 | HASHIMOTO C, FUJISAWA H. Packaging and transduction of non-T3 DNA by bacteriophage T3 [J]. Virology, 1988, 166(2): 432-439. |
| 23 | KROM RJ, BHARGAVA P, LOBRITZ M A, et al. Engineered phagemids for nonlytic, targeted antibacterial therapies [J]. Nano Letters, 2015, 15(7): 4808-4813. |
| 24 | BERTOZZI SILVA J, STORMS Z, SAUVAGEAU D. Host receptors for bacteriophage adsorption [J]. FEMS Microbiology Letters, 2016, 363(4): fnw002. |
| 25 | HAMDI S, ROUSSEAU G M, LABRIE S J, et al. Characterization of two polyvalent phages infecting Enterobacteriaceae [J]. Scientific Reports, 2017, 7: 40349. |
| 26 | STEPHEN T. ABEDON R L C. The Bacteriophages [M]. Oxford, UK: Oxford University Press, 2006. |
| 27 | YOSEF I, GOREN M G, GLOBUS R, et al. Extending the host range of bacteriophage particles for DNA transduction [J]. Molecular Cell, 2017, 66(5): 721-728. |
| 28 | YEHL K, LEMIRE S, YANG A C, et al. Engineering phage host-range and suppressing bacterial resistance through phage tail fiber mutagenesis [J]. Cell, 2019, 179(2): 459-469. |
| 29 | BRÜSSOW H. What is needed for phage therapy to become a reality in Western medicine? [J]. Virology, 2012, 434(2): 138-142. |
| 30 | HARGREAVES K R, CLOKIE M R. Clostridium difficile phages: still difficult? [J]. Frontiers in Microbiology, 2014, 5: 184. |
| 31 | CANCHAYA C, FOURNOUS G, BRÜSSOW H. The impact of prophages on bacterial chromosomes [J]. Molecular Microbiology, 2004, 53(1): 9-18. |
| 32 | HOWARD-VARONA C, HARGREAVES K R, ABEDON S T, et al. Lysogeny in nature: mechanisms, impact and ecology of temperate phages [J]. ISME Journal, 2017, 11(7): 1511-1520. |
| 33 | KILCHER S, STUDER P, MUESSNER C, et al. Cross-genus rebooting of custom-made, synthetic bacteriophage genomes in L-form bacteria [J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(3): 567-572. |
| 34 | ZHANG Hongming, FOUTS D E, DEPEW J, et al. Genetic modifications to temperate Enterococcus faecalis phage Ef11 that abolish the establishment of lysogeny and sensitivity to repressor, and increase host range and productivity of lytic infection [J]. Microbiology, 2013, 159(Pt 6): 1023-1035. |
| 35 | PARK Joo Youn, MOON Bo Youn, PARK Juw Won, et al. Genetic engineering of a temperate phage-based delivery system for CRISPR/Cas9 antimicrobials against Staphylococcus aureus [J]. Scientific Reports, 2017, 7: 44929. |
| 36 | YOSEF I, MANOR M, KIRO R, et al. Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria [J]. Proceedings of the National Academy of Sciences of the United States of America,2015, 112(23): 7267-7272. |
| 37 | MARINELLI LJ, PIURI M, SWIGONOV Z, et al. BRED: a simple and powerful tool for constructing mutant and recombinant bacteriophage genomes [J]. PLoS One, 2008, 3(12): e3957. |
| 38 | DEDRICK R M, GUERRERO-BUSTAMANTE C A, GARLENA R A, et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus [J]. Nature Medicine, 2019, 25(5): 730-733. |
| 39 | FISCHETTI V A. Phage therapy: a practical approach [M]. Cham: Springer, 2019: 317-334. |
| 40 | PASTAGIA M, SCHUCH R, FISCHETTI V A, et al. Lysins: the arrival of pathogen-directed anti-infectives [J]. Journal of Medical Microbiology, 2013, 62(10): 1506-1516. |
| 41 | LOEFFLER J M, NELSON D, FISCHETTI V A. Rapid killing of Streptococcus pneumoniae with a bacteriophage cell wall hydrolase [J]. Science, 2001, 294(5549): 2170-2172. |
| 42 | LETRADO P, CORSINI B, DÍEZ-MARTÍNEZ R, et al. Bactericidal synergism between antibiotics and phage endolysin Cpl-711 to kill multidrug-resistant pneumococcus [J]. Future Microbiology, 2018, 13(11): 1215-1223. |
| 43 | SHEN Yang, BARROS M, VENNEMANN T, et al. A bacteriophage endolysin that eliminates intracellular streptococci [J]. eLife, 2016, 5: 13152. |
| 44 | SHEN Yang, KÖLLER T, KREIKEMEYER B, et al. Rapid degradation of Streptococcus pyogenes biofilms by PlyC, a bacteriophage-encoded endolysin [J]. Journal of Antimicrobial Chemotherapy, 2013, 68(8): 1818-1824. |
| 45 | YANG Hang, ZHANG Huaidong, WANG Jing, et al. A novel chimeric lysin with robust antibacterial activity against planktonic and biofilm methicillin-resistant Staphylococcus aureus [J]. Scientific Reports, 2017, 7: 40182. |
| 46 | SCHUCH R, KHAN B K, RAZ A, et al. Bacteriophage lysin CF-301, a potent antistaphylococcal biofilm agent [J]. Antimicrobial Agents and Chemotherapy, 2017, 61(7): e02666. |
| 47 | ZHANG Yufeng, CHENG Mengjun, ZHANG Hao, et al. Antibacterial effects of phage lysin LysGH15 on planktonic cells and biofilms of diverse Staphylococci [J]. Applied and Environmental Microbiology, 2018, 84(15): 00886. |
| 48 | LAI Meng-Jiun, LIU Chih-Chin, JIANG Shinn-Jong, et al. Antimycobacterial activities of endolysins derived from a mycobacteriophage, BTCU-1 [J]. Molecules, 2015, 20(10): 19277-19290. |
| 49 | KIM Shukho, Da-Won LEE, JIN Jong-Sook, et al. Antimicrobial activity of LysSS, a novel phage endolysin, against Acinetobacter baumannii and Pseudomonas aeruginosa [J]. Journal of Global Antimicrobial Resistance, 2020, 22: 32-39. |
| 50 | BRIERS Y, WALMAGH M, PUYENBROECK V VAN, et al. Engineered endolysin-based "Artilysins" to combat multidrug-resistant gram-negative pathogens [J]. mBio, 2014, 5(4): e01379. |
| 51 | DEFRAINE V, SCHUERMANS J, GRYMONPREZ B, et al. Efficacy of Artilysin Art-175 against resistant and persistent Acinetobacter baumannii [J]. Antimicrobial Agents and Chemotherapy, 2016, 60(6): 3480-3488. |
| 52 | FERNÁNDEZ-RUIZ I, COUTINHO F H, RODRIGUEZ-VALERA F. Thousands of novel endolysins discovered in uncultured phage genomes [J]. Frontiers in Microbiology, 2018, 9: 1033. |
| 53 | SMITH G P. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface [J]. Science, 1985, 228(4705): 1315-1317. |
| 54 | LIU I-Ju, HSUEH Po-Ren, LIN Chin-Tarng, et al. Disease-specific B cell epitopes for serum antibodies from patients with severe acute respiratory syndrome (SARS) and serologic detection of SARS antibodies by epitope-based peptide antigens [J]. The Journal of Infectious Diseases, 2004, 190(4): 797-809. |
| 55 | SANTAMARIA H, MANOUTCHARIAN K, ROCHA L, et al. Identification of peptide sequences specific for serum antibodies from human papillomavirus-infected patients using phage display libraries [J]. Clinical Immunology, 2001, 101(3): 296-302. |
| 56 | KHURANA S, SUGUITAN A L JR, RIVERA Y, et al. Antigenic fingerprinting of H5N1 avian influenza using convalescent sera and monoclonal antibodies reveals potential vaccine and diagnostic targets [J]. PLoS Medicine, 2009, 6(4): e1000049. |
| 57 | WU Chien-Hsun, LIU I-Ju, LU Ruei-Min, et al. Advancement and applications of peptide phage display technology in biomedical science [J]. Journal of Biomedical Science, 2016, 23: 8. |
| 58 | HESS K L, JEWELL C M. Phage display as a tool for vaccine and immunotherapy development [J]. Bioengineering & Translational Medicine, 2020, 5(1): e10142. |
| 59 | FRENZEL A, SCHIRRMANN T, HUST M. Phage display-derived human antibodies in clinical development and therapy [J]. MAbs, 2016, 8(7): 1177-1194. |
| 60 | ESVELT KM, CARLSON JC, LIU D R. A system for the continuous directed evolution of biomolecules [J]. Nature, 2011, 472(7344): 499-503. |
| 61 | CARLSON J C, BADRAN A H, GUGGIANA-NILO D A, et al. Negative selection and stringency modulation in phage-assisted continuous evolution [J]. Nature Chemical Biology, 2014, 10(3): 216-222. |
| 62 | BRYSON D I, FAN Chenguang. Continuous directed evolution of aminoacyl-tRNA synthetases [J]. Nature Chemical Biology, 2017, 13(12): 1253-1260. |
| 63 | PACKER M S, REES H A, LIU D R. Phage-assisted continuous evolution of proteases with altered substrate specificity [J]. Nature Communications, 2017, 8(1): 956. |
| 64 | HUBBARD B P, BADRAN A H. Continuous directed evolution of DNA-binding proteins to improve TALEN specificity [J]. Nature Methods, 2015, 12(10): 939-942. |
| 65 | BADRAN A H, GUZOV V M, HUAI Qing, et al. Continuous evolution of Bacillus thuringiensis toxins overcomes insect resistance [J]. Nature, 2016, 533(7601): 58-63. |
| 66 | JOHNSTON C W, BADRAN A H, COLLINS J J. Continuous bioactivity-dependent evolution of an antibiotic biosynthetic pathway [J]. Nature Communications, 2020, 11(1): 4202. |
| 67 | SOUSA R, MUKHERJEE S. T7 RNA polymerase [J] Progress in Nucleic Acid Research and Molecular Biology, 2003, 73: 1-41. |
| 68 | LENNEMAN B R, ROTHMAN-DENES L B. Structural and biochemical investigation of bacteriophage N4-encoded RNA polymerases [J]. Biomolecules, 2015, 5(2): 647-667. |
| 69 | TEMME K, HILL R, SEGALL-SHAPIRO T H, et al. Modular control of multiple pathways using engineered orthogonal T7 polymerases [J]. Nucleic Acids Research, 2012, 40(17): 8773-8781. |
| 70 | MEYER A J, ELLEFSON J W, ELLINGTON A D. Directed evolution of a panel of orthogonal T7 RNA polymerase variants for in vivo or in vitro synthetic circuitry [J]. ACS Synthetic Biology, 2015, 4(10): 1070-1076. |
| 71 | ELOWITZ M B, LEIBLER S. A synthetic oscillatory network of transcriptional regulators [J]. Nature, 2000, 403(6767): 335-338. |
| 72 | STIRLING F, BITZAN L, O'KEEFE S, et al. Rational design of evolutionarily stable microbial kill switches [J]. Molecular Cell, 2017, 68(4): 686-697. |
| 73 | NOMAN N, INNISS M, IBA H, et al. Pulse detecting genetic circuit - a new design approach [J]. PLoS One, 2016, 11(12): e0167162. |
| 74 | TAO Pan, WU Xiaorong, TANG Weichun, et al. Engineering of bacteriophage T4 genome using CRISPR-Cas9 [J]. ACS Synthetic Biology, 2017, 6(10): 1952-1961. |
| 75 | PAEZ-ESPINO D, ELOE-FADROSH E A, PAVLOPOULOS G A, et al. Uncovering Earth's virome [J]. Nature, 2016, 536(7617): 425-430. |
| 76 | FEH R T, KARCAGI I, BLATTNER F R, et al. Bacteriophage recombineering in the lytic state using the lambda red recombinases [J]. Microbial Biotechnology, 2012, 5(4): 466-476. |
| 77 | SHIN Jonghyeon, JARDINE P, NOIREAUX V. Genome replication, synthesis, and assembly of the bacteriophage T7 in a single cell-free reaction [J]. ACS Synthetic Biology, 2012, 1(9): 408-413. |
| 78 | HORVATH P, BARRANGOU R. CRISPR/Cas, the immune system of bacteria and archaea [J]. Science, 2010, 327(5962): 167-170. |
| 79 | MAKAROVA K S, HAFT D H, BARRANGOU R, et al. Evolution and classification of the CRISPR-Cas systems [J]. Nature Reviews Microbiology 2011, 9(6): 467-477. |
| 80 | MARTEL B, MOINEAU S. CRISPR-Cas: an efficient tool for genome engineering of virulent bacteriophages [J]. Nucleic Acids Research, 2014, 42(14): 9504-9513. |
| 81 | KIRO R, SHITRIT D, QIMRON U. Efficient engineering of a bacteriophage genome using the type I-E CRISPR-Cas system [J]. RNA Biology, 2014, 11(1): 42-44. |
| 82 | LEMAY M L, TREMBLAY D M, MOINEAU S. Genome engineering of virulent lactococcal phages using CRISPR-Cas9 [J]. ACS Synthetic Biology, 2017, 6(7): 1351-1358. |
| 83 | SHEN Juntao, ZHOU Jinjie, CHEN Guoqiang, et al. Efficient genome engineering of a virulent Klebsiella bacteriophage using CRISPR-Cas9 [J]. Journal of Virology, 2018, 92(17): e00534. |
| 84 | SCHILLING T, DIETRICH S, HOPPERT M, et al. A CRISPR-Cas9-based toolkit for fast and precise in vivo genetic engineering of Bacillus subtilis phages [J]. Viruses, 2018, 10(5): 241. |
| 85 | HUPFELD M, TRASANIDOU D, RAMAZZINI L, et al. A functional type II-A CRISPR-Cas system from Listeria enables efficient genome editing of large non-integrating bacteriophage [J]. Nucleic Acids Research, 2018, 46(13): 6920-6933. |
| 86 | YUAN Shengjian, CHEN Ling, LIU Quan, et al. Characterization and genomic analyses of Aeromonas hydrophila phages AhSzq-1 and AhSzw-1, isolates representing new species within the T5virus genus [J]. Archives of Virology, 2018, 163(7): 1985-1988. |
| 87 | YANG Zhenlong, YUAN Shengjian, CHEN Ling, et al. Complete genome analysis of bacteriophage AsXd-1, a new member of the genus Hk97virus, family Siphoviridae [J]. Archives of Virology, 2018, 163(11): 3195-3197. |
| 88 | LI Erna, YIN Zhe, MA Yanyan, et al. Identification and molecular characterization of bacteriophage phiAxp-2 of Achromobacter xylosoxidans [J]. Scientific Reports, 2016, 6: 34300. |
| 89 | POPE W H, BOWMAN C A, RUSSELL D A, et al. Whole genome comparison of a large collection of mycobacteriophages reveals a continuum of phage genetic diversity [J]. eLife, 2015, 4: e06416. |
| 90 | CHAN L Y, KOSURI S, ENDY D. Refactoring bacteriophage T7 [J]. Molecular Systems Biology, 2005, 1: 2005.0018. |
| 91 | GIBSON D G, YOUNG L, CHUANG Ray-Yuan, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases [J]. Nature Methods, 2009, 6(5): 343-345. |
| 92 | LEMIRE S, YEHL K M, LU T K. Phage-based applications in synthetic biology [J]. Annual Review of Virology, 2018, 5(1): 453-476. |
| 93 | GIBSON D G. Synthesis of DNA fragments in yeast by one-step assembly of overlapping oligonucleotides [J]. Nucleic Acids Research, 2009, 37(20): 6984-6990. |
| 94 | JASCHKE P R, LIEBERMAN E K, RODRIGUEZ J, et al. A fully decompressed synthetic bacteriophage øX174 genome assembled and archived in yeast [J]. Virology, 2012, 434(2): 278-284. |
| 95 | OLDFIELD L M, GRZESIK P, VOORHIES A A, et al. Genome-wide engineering of an infectious clone of herpes simplex virus type 1 using synthetic genomics assembly methods [J]. Proceedings of the National Academy of Sciences of the United States Of America, 2017, 114(42): E8885. |
| 96 | ALLAN E J, HOISCHEN C, GUMPERT J. Bacterial L-Forms [J]. Advances in Applied Microbiology, 2009, 68: 1-39. |
| 97 | SWARTZ J R. Universal cell-free protein synthesis [J]. Nature Biotechnology, 2009, 27(8): 731-732. |
| 98 | CHAPPELL J, JENSEN K, FREEMONT P S. Validation of an entirely in vitro approach for rapid prototyping of DNA regulatory elements for synthetic biology [J]. Nucleic Acids Research, 2013, 41(5): 3471-3481. |
| 99 | RUSTAD M, EASTLUND A, MARSHALL R, et al. Synthesis of infectious bacteriophages in an E. coli-based cell-free expression system [J]. Journal of Visualized Experiments, 2017, 126: 56144. |
| 100 | RUSTAD M, NOIREAUX V, EASTLUND A, et al. Cell-free TXTL synthesis of infectious bacteriophage T4 in a single test tube reaction [J]. Synthetic Biology, 2018, 3(1): ysy002. |
| 101 | MESYANZHINOV V V, LEIMAN P G, KOSTYUCHENKO V A, et al. Molecular architecture of bacteriophage T4 [J]. Biochemistry (Moscow), 2004, 69(11): 1190-1202. |
| 102 | BROWN R, LENGELING A, WANG Baojun. Phage engineering: how advances in molecular biology and synthetic biology are being utilized to enhance the therapeutic potential of bacteriophages [J]. Quantitative Biology, 2017, 5(1): 42-54. |
| 103 | PASIN F, MENZEL W, DAR S J A. Harnessed viruses in the age of metagenomics and synthetic biology: an update on infectious clone assembly and biotechnologies of plant viruses [J]. Plant Biotechnology Journal, 2019, 17(6): 1010-1026. |
| 104 | HEMMINGA M A, VOS W L, NAZAROV P V, et al. Viruses: incredible nanomachines. New advances with filamentous phages [J]. European Biophysics Journal, 2010, 39(4): 541-550. |
| 105 | KUPFERSCHMIDT K. Resistance fighters [J]. Science, 2016, 352(6287): 758-761. |
| 106 | REARDON S. Phage therapy gets revitalized [J]. Nature, 2014, 510(7503): 15-16. |
| 107 | SUMMERS W C. The strange history of phage therapy [J]. Bacteriophage, 2012, 2(2): 130-133. |
| 108 | KUTTER E, DE VOS D, GVASALIA G, et al. Phage therapy in clinical practice: treatment of human infections [J]. Current Pharmaceutical Biotechnology 2010, 11(1): 69-86. |
| 109 | ROHWER F, SEGALL A M. In retrospect: a century of phage lessons [J]. Nature, 2015, 528(7580): 46-48. |
| 110 | LUONG T, SALABARRIA AC, ROACH DR. Phage therapy in the resistance era: where do we stand and where are we going? [J]. Clinical Therapeutics, 2020, 42(9): 1659-1680. |
| 111 | BAO Juan, WU Nannan, ZENG Yigang, et al. Non-active antibiotic and bacteriophage synergism to successfully treat recurrent urinary tract infection caused by extensively drug-resistant Klebsiella pneumoniae [J]. Emerging Microbes & Infections, 2020, 9(1): 771-774. |
| 112 | MIĘDZYBRODZKI R, BORYSOWSKI J, WEBER-DĄBROWSKA B, et al. Clinical aspects of phage therapy [J] Advances in Virus Research, 2012, 83: 73-121. |
| 113 | HEILMANN S, SNEPPEN K, KRISHNA S. Sustainability of virulence in a phage-bacterial ecosystem [J]. Journal of Virology, 2010, 84(6): 3016-22. |
| 114 | CHEN Ling, YUAN Shengjian, LIU Quan, et al. In vitro design and evaluation of phage cocktails against Aeromonas salmonicida [J]. Frontiers in Microbiology, 2018, 9: 1476. |
| 115 | CANCHAYA C, PROUX C, FOURNOUS G, et al. Prophage genomics [J]. Microbiology and Molecular Biology Reviews, 2003, 67(2): 238-276. |
| 116 | GÓRSKI A, MIĘDZYBRODZKI R, BORYSOWSKI J, et al. Phage as a modulator of immune responses: practical implications for phage therapy[J]. Advances in Virus Research, 2012, 83: 41-71. |
| 117 | COOPER C J, MIRZAEI M K, NILSSON A S. Adapting drug approval pathways for bacteriophage-based therapeutics[J]. Frontiers in Microbiology, 2016, 7: 1209. |
| 118 | SAMSON J E, MAGADAN A H, SABRI M, et al. Revenge of the phages: defeating bacterial defences [J]. Nature Reviews Microbiology, 2013, 11(10): 675-687. |
| 119 | LABRIE S J, SAMSON J E, MOINEAU S. Bacteriophage resistance mechanisms [J]. Nature Reviews Microbiology, 2010, 8(5): 317-327. |
| 120 | OJALA V, LAITALAINEN J, JALASVUORI M. Fight evolution with evolution: plasmid-dependent phages with a wide host range prevent the spread of antibiotic resistance [J]. Evolutionary Applications, 2013, 6(6): 925-932. |
| 121 | MATSUDA T, FREEMAN T A, HILBERT D W, et al. Lysis-deficient bacteriophage therapy decreases endotoxin and inflammatory mediator release and improves survival in a murine peritonitis model [J]. Surgery, 2005, 137(6): 639-646. |
| 122 | MUROOKA Y, TAKIZAWA N, HARADA T. Introduction of bacteriophage Mu into bacteria of various genera and intergeneric gene transfer by RP4:: Mu [J]. Journal of Bacteriology, 1981, 145(1): 358-368. |
| 123 | MUROOKA Y, HARADA T. Expansion of the host range of coliphage P1 and gene transfer from enteric bacteria to other gram-negative bacteria [J]. Applied and Environmental Microbiology, 1979, 38(4): 754-757. |
| 124 | NAMURA M, HIJIKATA T, MIYANAGA K, et al. Detection of Escherichia coli with fluorescent labeled phages that have a broad host range to E. coli in sewage water [J]. Biotechnology Progress, 2008, 24(2): 481-486. |
| 125 | EDGAR R, MCKINSTRY M, HWANG Jeeseong, et al. High-sensitivity bacterial detection using biotin-tagged phage and quantum-dot nanocomplexes [J]. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(13): 4841-4845. |
| 126 | SCHOFIELD D A, WESTWATER C. Phage-mediated bioluminescent detection of Bacillus anthracis [J]. Journal of Applied Microbiology, 2009, 107(5): 1468-1478. |
| 127 | O'DONNELL M R, PYM A, JAIN P, et al. A novel reporter phage to detect tuberculosis and rifampin resistance in a high-HIV-burden population [J]. Journal of Clinical Microbiology, 2015, 53(7): 2188-2194. |
| 128 | PIURI M, JACOBS W R JR, HATFULL G F. Fluoromycobacteriophages for rapid, specific, and sensitive antibiotic susceptibility testing of Mycobacterium tuberculosis [J]. PLoS One, 2009, 4(3): e4870. |
| 129 | KUMAR V, LOGANATHAN P, SIVARAMAKRISHNAN G, et al. Characterization of temperate phage Che12 and construction of a new tool for diagnosis of tuberculosis [J]. Tuberculosis, 2008, 88(6): 616-623. |
| 130 | DUSTHACKEER A, KUMAR V, SUBBIAN S, et al. Construction and evaluation of luciferase reporter phages for the detection of active and non-replicating tubercle bacilli [J]. Journal of Microbiological Methods, 2008, 73(1): 18-25. |
| 131 | PENG Yong, JIN Yanqiu, LIN Hong, et al. Application of the VPp1 bacteriophage combined with a coupled enzyme system in the rapid detection of Vibrio parahaemolyticus [J]. Journal of Microbiological Methods, 2014, 98: 99-104. |
| 132 | KIM Seongmi, KIM Minsik, Sangryeol RYU. Development of an engineered bioluminescent reporter phage for the sensitive detection of viable Salmonella typhimurium [J]. Analytical Chemistry, 2014, 86(12): 5858-5864. |
| 133 | VANDAMM JP, RAJANNA C, SHARP N J, et al. Rapid detection and simultaneous antibiotic susceptibility analysis of Yersinia pestis directly from clinical specimens by use of reporter phage [J]. Journal of Clinical Microbiology, 2014, 52(8): 2998-3003. |
| 134 | SCHOFIELD DA, MOLINEUX I J, WESTWATER C. Diagnostic bioluminescent phage for detection of Yersinia pestis [J]. Journal of Clinical Microbiology, 2009, 47(12): 3887-3894. |
| 135 | SCHOFIELD D A, WRAY D J, MOLINEUX I J. Isolation and development of bioluminescent reporter phages for bacterial dysentery [J]. European Journal of Clinical Microbiology and Infectious Diseases, 2015, 34(2): 395-403. |
| 136 | SHARP N J, VANDAMM J P, MOLINEUX I J, et al. Rapid detection of Bacillus anthracis in complex food matrices using phage-mediated bioluminescence [J]. Journal of Food Protection, 2015, 78(5): 963-968. |
| 137 | SHARP N J, MOLINEUX I J, PAGE M A, et al. Rapid detection of viable Bacillus anthracis spores in environmental samples by using engineered reporter phages [J]. Applied and Environmental Microbiology, 2016, 82(8): 2380-2387. |
| 138 | KIM Jinwoo, KIM Minsik, KIM Seongmi, et al. Sensitive detection of viable Escherichia coli O157: H7 from foods using a luciferase-reporter phage phiV10lux [J]. International Journal of Food Microbiology, 2017, 254: 11-17. |
| 139 | KANNAN P, YONG Ho Yao, REIMAN L, et al. Bacteriophage-based rapid and sensitive detection of Escherichia coli O157: H7 isolates from ground beef [J]. Foodborne Pathogens and Disease, 2010, 7(12): 1551-1558. |
| 140 | SCHOFIELD D A, SHARP N J, WESTWATER C. Phage-based platforms for the clinical detection of human bacterial pathogens [J]. Bacteriophage, 2012, 2(2): 105-283. |
| 141 | SCHMIDT A, RABSCH W, BROEKER N K, et al. Bacteriophage tailspike protein based assay to monitor phase variable glucosylations in Salmonella O-antigens [J]. BMC Microbiology, 2016, 16(1): 207. |
| 142 | GÓMEZ-TORRES N, DUNNE M, GARDE S, et al. Development of a specific fluorescent phage endolysin for in situ detection of Clostridium species associated with cheese spoilage [J]. Microbial Biotechnology, 2018, 11(2): 332-345. |
| 143 | BENEŠÍK M, NOVÁČEK J, JANDA L, et al. Role of SH3b binding domain in a natural deletion mutant of Kayvirus endolysin LysF1 with a broad range of lytic activity [J]. Virus Genes, 2018, 54(1): 130-139. |
| 144 | FEDERICI S, NOBS SP, ELINAV E. Phages and their potential to modulate the microbiome and immunity [J]. Cellular & Molecular Immunology, 2020. DOI: 10.1038/s41423-020-00532-4 . |
| 145 | WANG Xiaofang, WEI Zhong, YANG Keming, et al. Phage combination therapies for bacterial wilt disease in tomato [J]. Nature Biotechnology, 2019, 37(12): 1513-1520. |
| [1] | 高歌, 边旗, 王宝俊. 合成基因线路的工程化设计研究进展与展望[J]. 合成生物学, 2025, 6(1): 45-64. |
| [2] | 李冀渊, 吴国盛. 合成生物学视域下有机体的两种隐喻[J]. 合成生物学, 2025, 6(1): 190-202. |
| [3] | 焦洪涛, 齐蒙, 邵滨, 蒋劲松. DNA数据存储技术的法律治理议题[J]. 合成生物学, 2025, 6(1): 177-189. |
| [4] | 唐兴华, 陆钱能, 胡翌霖. 人类世中对合成生物学的哲学反思[J]. 合成生物学, 2025, 6(1): 203-212. |
| [5] | 徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176. |
| [6] | 石婷, 宋展, 宋世怡, 张以恒. 体外生物转化(ivBT):生物制造的新前沿[J]. 合成生物学, 2024, 5(6): 1437-1460. |
| [7] | 柴猛, 王风清, 魏东芝. 综合利用木质纤维素生物转化合成有机酸[J]. 合成生物学, 2024, 5(6): 1242-1263. |
| [8] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [9] | 陈雨, 张康, 邱以婧, 程彩云, 殷晶晶, 宋天顺, 谢婧婧. 微生物电合成技术转化二氧化碳研究进展[J]. 合成生物学, 2024, 5(5): 1142-1168. |
| [10] | 郑皓天, 李朝风, 刘良叙, 王嘉伟, 李恒润, 倪俊. 负碳人工光合群落的设计、优化与应用[J]. 合成生物学, 2024, 5(5): 1189-1210. |
| [11] | 夏孔晨, 徐维华, 吴起. 光酶催化混乱性反应的研究进展[J]. 合成生物学, 2024, 5(5): 997-1020. |
| [12] | 陈子苓, 向阳飞. 类器官技术与合成生物学协同研究进展[J]. 合成生物学, 2024, 5(4): 795-812. |
| [13] | 蔡冰玉, 谭象天, 李伟. 合成生物学在干细胞工程化改造中的研究进展[J]. 合成生物学, 2024, 5(4): 782-794. |
| [14] | 谢皇, 郑义蕾, 苏依婷, 阮静怡, 李永泉. 放线菌聚酮类化合物生物合成体系重构研究进展[J]. 合成生物学, 2024, 5(3): 612-630. |
| [15] | 查文龙, 卜兰, 訾佳辰. 中药药效成分群的合成生物学研究进展[J]. 合成生物学, 2024, 5(3): 631-657. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||