合成生物学 ›› 2022, Vol. 3 ›› Issue (6): 1218-1234.DOI: 10.12211/2096-8280.2022-044
微藻叶绿体细胞器工厂研究进展
朱振1,2, 田晶2, 江静1,3, 王旺银1, 曹旭鹏1
- 1.中国科学院大连化学物理研究所,催化国家重点实验室,洁净能源国家实验室(筹),辽宁 大连 116023
2.大连工业大学,生物工程学院,辽宁 大连 116034
3.中国科学技术大学化学与材料科学学院,安徽 合肥 230026
-
收稿日期:2022-08-08修回日期:2022-09-28出版日期:2022-12-31发布日期:2023-01-17 -
作者简介:朱振 (1991—),女,博士研究生。研究方向为合成生物学和微藻可控培养研究。E-mail:zhuzhen@dicp.ac.cn曹旭鹏 (1978—),男,研究员,博士生导师。致力于太阳能生物转化研究。。E-mail:c_x_p@dicp.ac.cn -
基金资助:国家自然科学基金(21878285);国家自然科学基金委“人工光合成”基础科学中心项目(22088102)
Progress in microalgae chloroplast organelle factory development
ZHU Zhen1,2, TIAN Jing2, JIANG Jing1,3, WANG Wangyin1, CAO Xupeng1
- 1.China State Key Laboratory of Catalysis,Dalian Institute of Chemical Physics,Chinese Academy of Sciences,Dalian National Laboratory for Clean Energy,Dalian 116023,Liaoning,China
2.School of Bioengineering,Dalian Polytechnic University,Dalian 116034,Liaoning,China
3.School of Chemical and Materials Science,University of Science and Technology of China,Hefei 230026,Anhui,China
-
Received:2022-08-08Revised:2022-09-28Online:2022-12-31Published:2023-01-17
摘要:
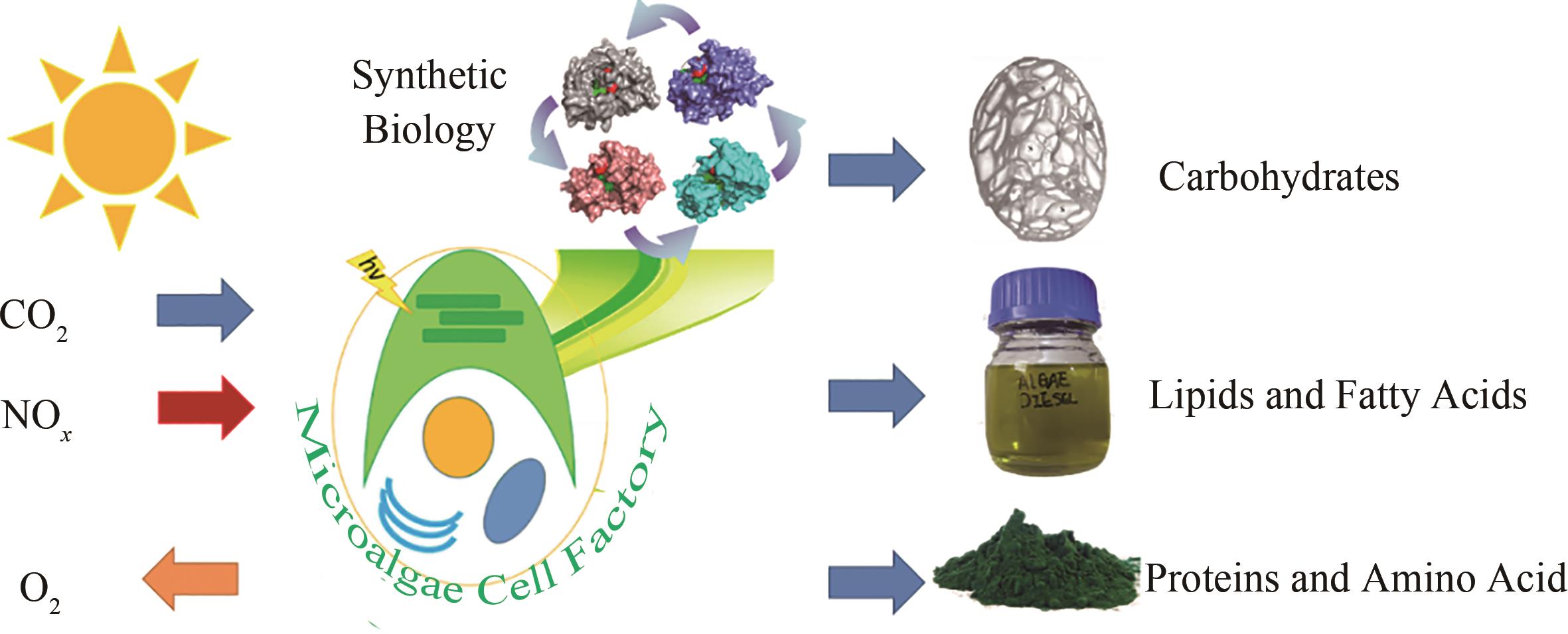
微藻是重要的太阳能驱动CO2生物转化的生物,建立微藻叶绿体细胞器工厂是通过合成生物学手段实现“碳中和”的潜在途径之一。这是因为微藻叶绿体是碳同化以及后续碳水化合物、脂肪酸、天然色素、氨基酸等重要合成器官,与高等植物细胞内具备多个相对较小的叶绿体不同,大部分微藻仅拥有一个占细胞体积50%以上的大叶绿体,更有利于获得纯净的株系,有望在食品、水产、医药、化学品、生物燃料等领域占据重要地位。微藻改造及微藻叶绿体细胞器工厂研究尚处于起步阶段,本文系统通过对现有转化、表达技术进展进行汇总和简要分析,对比了叶绿体直接转化和基于叶绿体转运肽的核转化叶绿体表达不同策略的优缺点,为后续发展提供借鉴。其中,靶向叶绿体基因组的直接转化策略应用较广泛,主要集中在莱茵衣藻中,已成功表达了100多种不同的蛋白质,但是叶绿体基因组可插入位点有限和调控手段相对缺乏;通过利用叶绿体转运肽,叶绿体中90%以上的蛋白都是核编码并被可控递送到叶绿体内,因此近年来基于叶绿体转运肽的核转化叶绿体定位表达技术的关注度得到了提升,并且已经在固碳、油脂生产调节方面展示出了一定优势。
中图分类号:
引用本文
朱振, 田晶, 江静, 王旺银, 曹旭鹏. 微藻叶绿体细胞器工厂研究进展[J]. 合成生物学, 2022, 3(6): 1218-1234.
ZHU Zhen, TIAN Jing, JIANG Jing, WANG Wangyin, CAO Xupeng. Progress in microalgae chloroplast organelle factory development[J]. Synthetic Biology Journal, 2022, 3(6): 1218-1234.
| 藻株 | 表达系统 | 表达水平 | 描述 | 文献 |
|---|---|---|---|---|
| C.reinhardtii | pACTBVP1vector (atpX-CTBVP1),atpA promoter, rbcL terminator | 3 % TSP | CTB-VP1融合基因保留了其蛋白的神经节苷脂GM1亲和力和抗原性,并证明绿藻叶绿体作为生产黏膜疫苗的潜力 | [ |
| C.reinhardtii | psbA promoter and 5′ UTR, psbA 3′ UTR | ND | 利用微藻生产具有可溶性和酶活性的抗毒素,并延长了植入人类B细胞肿瘤小鼠的生存时间 | [ |
| C.reinhardtii | pSRSapI and pASapI vector,psaA-1 promoter, rbcL 3′ UTR | 约1.3 mg/g DB | 利用莱茵衣藻叶绿体生产抗人类病原体肺炎链球菌特有的内溶素Cpl-1和Pal并具有抗菌活性 | [ |
| C.reinhardtii | psbA promoter and 5′ UTR | >5 % TSP | 利用莱茵衣藻叶绿体稳定表达具有生物活性且可溶性的哺乳类蛋白(MSAA) | [ |
| C.reinhardtii | atpA or rbcL promoters and 5′ UTR | 0.5 % TSP | 首次实现利用莱茵衣藻叶绿体积累可溶性的,正确折叠的lsc抗体 | [ |
| C.reinhardtii | pASapI vector, psbH, aadA | 6.77 mg/L 22 mg/L DA | 首次实现中试规模下的利用莱茵衣藻缺壁株的叶绿体表达细胞色素P450和二萜合酶TPS4 | [ |
| C.reinhardtii | psbA, atpApsbD promoter | 21% TCP | VP28基因的密码子优化会影响蛋白质的积累 | [ |
| H.pluvialis | 16S-trnI/trnA-23S region,psbA and rbcL promoter, terminator | ND | 首次实现利用雨生红球藻叶绿体成功表达抗菌肽 | [ |
| C.vulgaris | 16S-trnI/trnA-23S region, rbcL promoter, psbA terminator, aadA | ND | 首次在小球藻叶绿体中成功实现2个密码子优化后的抗菌肽共表达 | [ |
表1 利用微藻叶绿体表达目标蛋白
Tab. 1 Target proteins produced in algal chloroplasts
| 藻株 | 表达系统 | 表达水平 | 描述 | 文献 |
|---|---|---|---|---|
| C.reinhardtii | pACTBVP1vector (atpX-CTBVP1),atpA promoter, rbcL terminator | 3 % TSP | CTB-VP1融合基因保留了其蛋白的神经节苷脂GM1亲和力和抗原性,并证明绿藻叶绿体作为生产黏膜疫苗的潜力 | [ |
| C.reinhardtii | psbA promoter and 5′ UTR, psbA 3′ UTR | ND | 利用微藻生产具有可溶性和酶活性的抗毒素,并延长了植入人类B细胞肿瘤小鼠的生存时间 | [ |
| C.reinhardtii | pSRSapI and pASapI vector,psaA-1 promoter, rbcL 3′ UTR | 约1.3 mg/g DB | 利用莱茵衣藻叶绿体生产抗人类病原体肺炎链球菌特有的内溶素Cpl-1和Pal并具有抗菌活性 | [ |
| C.reinhardtii | psbA promoter and 5′ UTR | >5 % TSP | 利用莱茵衣藻叶绿体稳定表达具有生物活性且可溶性的哺乳类蛋白(MSAA) | [ |
| C.reinhardtii | atpA or rbcL promoters and 5′ UTR | 0.5 % TSP | 首次实现利用莱茵衣藻叶绿体积累可溶性的,正确折叠的lsc抗体 | [ |
| C.reinhardtii | pASapI vector, psbH, aadA | 6.77 mg/L 22 mg/L DA | 首次实现中试规模下的利用莱茵衣藻缺壁株的叶绿体表达细胞色素P450和二萜合酶TPS4 | [ |
| C.reinhardtii | psbA, atpApsbD promoter | 21% TCP | VP28基因的密码子优化会影响蛋白质的积累 | [ |
| H.pluvialis | 16S-trnI/trnA-23S region,psbA and rbcL promoter, terminator | ND | 首次实现利用雨生红球藻叶绿体成功表达抗菌肽 | [ |
| C.vulgaris | 16S-trnI/trnA-23S region, rbcL promoter, psbA terminator, aadA | ND | 首次在小球藻叶绿体中成功实现2个密码子优化后的抗菌肽共表达 | [ |

图1 莱茵衣藻中利用核与叶绿体的两种典型的表达策略5′ UTR—5′非翻译区; Rbc S2 promoter—Rbc S2启动子;intron-1 Rbc S2—Rbc S2第一内含子;cTP—叶绿体转运肽;cTP Ⅰ/Ⅱ type sequence—叶绿体Ⅰ/Ⅱ型转运肽序列;Rbc S2-3′ UTR—Rbc S2基因的3′非翻译区
Fig. 1 Two typical expression strategies using nuclear and chloroplast in C. reinhardtii5′ UTR—5′ untranslated region; cTP—choroplast transit peptide

图2 cTP及叶绿体/类囊体蛋白定位过程示意图TP—转运肽 ;Toc complex —Toc 复合物;Tic complex —Tic 复合物;Tat—双精氨酸转运
Fig. 2 Schematic diagram of the localization cTP and chloroplast/thylakoid proteinTP—transit peptide; Toc complex—translocon at the outer envelope membrane of chloroplasts;Tic complex—translocon at the inner envelope membrane of chloroplasts; Tat—twin arginine translocation
| 58 | MANUELL A L, BELIGNI M V, ELDER J H, et al. Robust expression of a bioactive mammalian protein in Chlamydomonas chloroplast[J]. Plant Biotechnology Journal, 2007, 5(3): 402-412. |
| 59 | MAYFIELD S P, FRANKLIN S E, LERNER R A. Expression and assembly of a fully active antibody in algae[J]. Proceedings of the National Academy of Sciences of the United States of America, 2003, 100(2): 438-442. |
| 60 | ZEDLER J A Z, GANGL D, GUERRA T, et al. Pilot-scale cultivation of wall-deficient transgenic Chlamydomonas reinhardtii strains expressing recombinant proteins in the chloroplast[J]. Applied Microbiology and Biotechnology, 2016, 100(16): 7061-7070. |
| 61 | WANG K, CUI Y L, WANG Y C, et al. Chloroplast genetic engineering of a unicellular green alga Haematococcus pluvialis with expression of an antimicrobial peptide[J]. Marine Biotechnology, 2020, 22(4): 572-580. |
| 62 | WANG K, GAO Z Q, WANG Y C, et al. The chloroplast genetic engineering of a unicellular green alga Chlorella vulgaris with two foreign peptides co-expression[J]. Algal Research, 2021, 54: 102214. |
| 63 | MA K, BAO Q, WU Y, et al. Evaluation of microalgae as immunostimulants and recombinant vaccines for diseases prevention and control in aquaculture[J]. Frontiers in Bioengineering and Biotechnology, 2020, 8: 590431. |
| 64 | CORAGLIOTTI A T, BELIGNI M V, FRANKLIN S E, et al. Molecular factors affecting the accumulation of recombinant proteins in the Chlamydomonas reinhardtii chloroplast[J]. Molecular Biotechnology, 2011, 48(1): 60-75. |
| 65 | RASALA B A, MUTO M, SULLIVAN J, et al. Improved heterologous protein expression in the chloroplast of Chlamydomonas reinhardtii through promoter and 5′ untranslated region optimization[J]. Plant Biotechnology Journal, 2011, 9(6): 674-683. |
| 66 | OEY M, ROSS I L, HANKAMER B. Gateway-assisted vector construction to facilitate expression of foreign proteins in the chloroplast of single celled algae[J]. PLoS One, 2014, 9(2): e86841. |
| 67 | ZEDLER J A Z, GANGL D, HAMBERGER B, et al. Stable expression of a bifunctional diterpene synthase in the chloroplast of Chlamydomonas reinhardtii [J]. Journal of Applied Phycology, 2015, 27(6): 2271-2277. |
| 68 | NAWKARKAR P, CHUGH S, SHARMA S, et al. Characterization of the chloroplast genome facilitated the transformation of parachlorella kessleri-I, a potential marine alga for biofuel production[J]. Current Genomics, 2020, 21(8): 610-623. |
| 69 | XIA D H, QIU W, WANG X X, et al. Recent advancements and future perspectives of microalgae-derived pharmaceuticals[J]. Marine Drugs, 2021, 19(12): 703. |
| 70 | ALMARAZ-DELGADO A L, FLORES-URIBE J, PÉREZ-ESPAÑA V H, et al. Production of therapeutic proteins in the chloroplast of Chlamydomonas reinhardtii [J]. AMB Express, 2014, 4: 57. |
| 71 | SHAMRIZ S, OFOGHI H. Outlook in the application of Chlamydomonas reinhardtii chloroplast as a platform for recombinant protein production[J]. Biotechnology and Genetic Engineering Reviews, 2016, 32(1/2): 92-106. |
| 72 | 李珺. 几种新型细胞器植物生物反应器的研究[J]. 生物技术通报, 2008(3): 30-32. |
| LI J. Research of several new plant cell bioreactor[J]. Biotechnology Bulletin, 2008(3): 30-32. | |
| 73 | RASALA B A, MAYFIELD S P. Photosynthetic biomanufacturing in green algae; production of recombinant proteins for industrial, nutritional, and medical uses[J]. Photosynthesis Research, 2015, 123(3): 227-239. |
| 74 | GOULD S B, WALLER R F, MCFADDEN G I. Plastid evolution[J]. Annual Review of Plant Biology, 2008, 59: 491-517. |
| 75 | GINGER M L, MCFADDEN G I, MICHELS P A M. The evolution of organellar metabolism in unicellular eukaryotes[J]. Philosophical Transactions of the Royal Society B: Biological Sciences, 2010, 365(1541): 693-698. |
| 76 | FRANZÉN L G, ROCHAIX J D, VON HEIJNE G. Chloroplast transit peptides from the green alga Chlamydomonas reinhardtii share features with both mitochondrial and higher plant chloroplast presequences[J]. FEBS Letters, 1990, 260(2): 165-168. |
| 77 | VILLAREJO A, BURÉN S, LARSSON S, et al. Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast[J]. Nature Cell Biology, 2005, 7(12): 1224-1231. |
| 78 | UNIACKE J, ZERGES W. Chloroplast protein targeting involves localized translation in Chlamydomonas [J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(5): 1439-1444. |
| 79 | SHI L X, THEG S M. The motors of protein import into chloroplasts[J]. Plant Signaling & Behavior, 2011, 6(9): 1397-1401. |
| 80 | VOJTA L, SOLL J, BÖLTER B. Protein transport in chloroplasts-targeting to the intermembrane space[J]. The FEBS Journal, 2007, 274(19): 5043-5054. |
| 81 | YANG B, LIU J, MA X, et al. Genetic engineering of the Calvin cycle toward enhanced photosynthetic CO2 fixation in microalgae[J]. Biotechnology for Biofuels, 2017, 10: 229. |
| 1 | BEHRENFELD M J, RANDERSON J T, MCCLAIN C R, et al. Biospheric primary production during an ENSO transition[J]. Science, 2001, 291(5513): 2594-2597. |
| 2 | SIMKIN A J, LÓPEZ-CALCAGNO P E, RAINES C A. Feeding the world: improving photosynthetic efficiency for sustainable crop production[J]. Journal of Experimental Botany, 2019, 70(4): 1119-1140. |
| 3 | LONG S P, ZHU X G, NAIDU S L, et al. Can improvement in photosynthesis increase crop yields? [J]. Plant, Cell & Environment, 2006, 29(3): 315-330. |
| 4 | PARRY M A J, REYNOLDS M, SALVUCCI M E, et al. Raising yield potential of wheat (Ⅱ): Increasing photosynthetic capacity and efficiency[J]. Journal of Experimental Botany, 2010, 62(2): 453-467. |
| 5 | BLANKENSHIP R E, TIEDE D M, BARBER J, et al. Comparing photosynthetic and photovoltaic efficiencies and recognizing the potential for improvement[J]. Science, 2011, 332(6031): 805-809. |
| 6 | KSELÍKOVÁ V, SINGH A, BIALEVICH V, et al. Improving microalgae for biotechnology-From genetics to synthetic biology-Moving forward but not there yet[J]. Biotechnology Advances, 2022, 58: 107885. |
| 7 | LU Y D, ZHANG X, GU X P, et al. Engineering microalgae: transition from empirical design to programmable cells[J]. Critical Reviews in Biotechnology, 2021, 41(8): 1233-1256. |
| 8 | EINHAUS A, BAIER T, ROSENSTENGEL M, et al. Rational promoter engineering enables robust terpene production in microalgae[J]. ACS Synthetic Biology, 2021, 10(4): 847-856. |
| 9 | MUÑOZ C F, SÜDFELD C, NADUTHODI M I S, et al. Genetic engineering of microalgae for enhanced lipid production[J]. Biotechnology Advances, 2021, 52: 107836. |
| 10 | WICHMANN J, BAIER T, WENTNAGEL E, et al. Tailored carbon partitioning for phototrophic production of (E)-α-bisabolene from the green microalga Chlamydomonas reinhardtii [J]. Metabolic Engineering, 2018, 45: 211-222. |
| 11 | TRAN N T, KALDENHOFF R. Metabolic engineering of ketocarotenoids biosynthetic pathway in Chlamydomonas reinhardtii strain CC-4102[J]. Scientific Reports, 2020, 10: 10688. |
| 12 | KUSMAYADI A, LEONG Y K, YEN H W, et al. Microalgae as sustainable food and feed sources for animals and humans - biotechnological and environmental aspects[J]. Chemosphere, 2021, 271: 129800. |
| 13 | GEROTTO C, NORICI A, GIORDANO M. Toward enhanced fixation of CO2 in aquatic biomass: focus on microalgae[J]. Frontiers in Energy Research, 2020, 8: 213. |
| 82 | PEROZENI F, CAZZANIGA S, BAIER T, et al. Turning a green alga red: Engineering astaxanthin biosynthesis by intragenic pseudogene revival in Chlamydomonas reinhardtii [J]. Plant Biotechnology Journal, 2020, 18(10): 2053-2067. |
| 83 | SPROLES A E, BERNDT A, FIELDS F J, et al. Improved high-throughput screening technique to rapidly isolate Chlamydomonas transformants expressing recombinant proteins[J]. Applied Microbiology and Biotechnology, 2022, 106(4): 1677-1689. |
| 84 | ZHANG Y T, JIANG J Y, SHI T Q, et al. Application of the CRISPR/Cas system for genome editing in microalgae[J]. Applied Microbiology and Biotechnology, 2019, 103(8): 3239-3248. |
| 85 | LUBBEN T H, BANSBERG J, KEEGSTRA K. Stop-transfer regions do not halt translocation of proteins into chloroplasts[J]. Science, 1987, 238(4830): 1112-1114. |
| 86 | KIM D H, LEE J E, XU Z Y, et al. Cytosolic targeting factor AKR2A captures chloroplast outer membrane-localized client proteins at the ribosome during translation[J]. Nature Communications, 2015, 6: 6843. |
| 87 | STAUB J M, GARCIA B, GRAVES J, et al. High-yield production of a human therapeutic protein in tobacco chloroplasts[J]. Nature Biotechnology, 2000, 18(3): 333-338. |
| 88 | KIKUCHI S, BÉDARD J, HIRANO M, et al. Uncovering the protein translocon at the chloroplast inner envelope membrane[J]. Science, 2013, 339(6119): 571-574. |
| 89 | DEPÈGE N, BELLAFIORE S, ROCHAIX J D. Role of chloroplast protein kinase Stt7 in LHCII phosphorylation and state transition in Chlamydomonas [J]. Science, 2003, 299(5612): 1572-1575. |
| 90 | STENGEL K F, HOLDERMANN I, CAIN P, et al. Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43[J]. Science, 2008, 321(5886): 253-256. |
| 91 | HIRSCH S, MUCKEL E, HEEMEYER F, et al. A receptor component of the chloroplast protein translocation machinery[J]. Science, 1994, 266(5193): 1989-1992. |
| 92 | LEE D W, HWANG I. Evolution and design principles of the diverse chloroplast transit peptides[J]. Molecules and Cells, 2018, 41(3): 161-167. |
| 93 | RADHAMONY R N, THEG S M. Evidence for an ER to Golgi to chloroplast protein transport pathway[J]. Trends in Cell Biology, 2006, 16(8): 385-387. |
| 14 | MUTALE-JOAN C, SBABOU L, HICHAM E A. Microalgae and cyanobacteria: How exploiting these microbial resources can address the underlying challenges related to food sources and sustainable agriculture: a review[J]. Journal of Plant Growth Regulation, 2022: 1-20. |
| 15 | AL-HAJ L, LUI Y T, ABED R M, et al. Cyanobacteria as chassis for industrial biotechnology: progress and prospects[J]. Life, 2016, 6(4): E42. |
| 16 | XUE Y, HE Q F. Cyanobacteria as cell factories to produce plant secondary metabolites[J]. Frontiers in Bioengineering and Biotechnology, 2015, 3: 57. |
| 17 | CASE A E, ATSUMI S. Cyanobacterial chemical production[J]. Journal of Biotechnology, 2016, 231: 106-114. |
| 18 | ZHU T, XIE X M, LI Z M, et al. Enhancing photosynthetic production of ethylene in genetically engineered Synechocystis sp. PCC 6803[J]. Green Chemistry, 2015, 17(1): 421-434. |
| 19 | QIAO Y, WANG W H, LU X F. Engineering Cyanobacteria as cell factories for direct trehalose production from CO2 [J]. Metabolic Engineering, 2020, 62: 161-171. |
| 20 | SONG K, TAN X M, LIANG Y J, et al. The potential of Synechococcus elongatus UTEX 2973 for sugar feedstock production[J]. Applied Microbiology and Biotechnology, 2016, 100(18): 7865-7875. |
| 21 | TAN C L, TAO F, XU P. Direct carbon capture for the production of high-performance biodegradable plastics by Cyanobacterial cell factories[J]. Green Chemistry, 2022, 24(11): 4470-4483. |
| 22 | GAO X, GAO F, LIU D, et al. Engineering the methylerythritol phosphate pathway in Cyanobacteria for photosynthetic isoprene production from CO2 [J]. Energy & Environmental Science, 2016, 9(4): 1400-1411. |
| 23 | DREESEN I A J, HAMRI G C E, FUSSENEGGER M. Heat-stable oral alga-based vaccine protects mice from Staphylococcus aureus infection[J]. Journal of Biotechnology, 2010, 145(3): 273-280. |
| 24 | XING J L, PAN J T, YI H, et al. The plastid-encoded protein Orf2971 is required for protein translocation and chloroplast quality control[J]. The Plant Cell, 2022, 34(9): 3383-3399. |
| 25 | 任聪, 王丹, 杨琳, 等. 叶绿体中表达外源蛋白的研究进展[J]. 科技通报, 2012, 28(3): 38-42. |
| 94 | BEHRENS M R, MUTLU N, CHAKRABORTY S, et al. Dicamba resistance: enlarging and preserving biotechnology-based weed management strategies[J]. Science, 2007, 316(5828): 1185-1188. |
| 95 | KOBAYASHI Y, MISUMI O, ODAHARA M, et al. Holliday junction resolvases mediate chloroplast nucleoid segregation[J]. Science, 2017, 356(6338): 631-634. |
| 96 | ZEDLER J A Z, MULLINEAUX C W, ROBINSON C. Efficient targeting of recombinant proteins to the thylakoid lumen in Chlamydomonas reinhardtii using a bacterial Tat signal peptide[J]. Algal Research, 2016, 19: 57-62. |
| 97 | FOTH B J, RALPH S A, TONKIN C J, et al. Dissecting apicoplast targeting in the malaria parasite Plasmodium falciparum [J]. Science, 2003, 299(5607): 705-708. |
| 98 | KINDLE K L. High-frequency nuclear transformation of Chlamydomonas reinhardtii [J]. Proceedings of the National Academy of Sciences of the United States of America, 1990, 87(3): 1228-1232. |
| 99 | RECUENCO-MUÑOZ L, OFFRE P, VALLEDOR L, et al. Targeted quantitative analysis of a diurnal RuBisCO subunit expression and translation profile in Chlamydomonas reinhardtii introducing a novel Mass Western approach[J]. Journal of Proteomics, 2015, 113: 143-153. |
| 100 | LLANSOLA-PORTOLES M J, LITVIN R, ILIOAIA C, et al. Pigment structure in the violaxanthin-chlorophyll-a-binding protein VCP[J]. Photosynthesis Research, 2017, 134(1): 51-58. |
| 101 | CERUTTI H, JOHNSON A M, GILLHAM N W, et al. A eubacterial gene conferring spectinomycin resistance on Chlamydomonas reinhardtii: integration into the nuclear genome and gene expression[J]. Genetics, 1997, 145(1): 97-110. |
| 102 | SCRANTON M A, OSTRAND J T, GEORGIANNA D R, et al. Synthetic promoters capable of driving robust nuclear gene expression in the green alga Chlamydomonas reinhardtii [J]. Algal Research, 2016, 15: 135-142. |
| 103 | SCHRODA M, BECK C F, VALLON O. Sequence elements within an HSP70 promoter counteract transcriptional transgene silencing in Chlamydomonas [J]. The Plant Journal: for Cell and Molecular Biology, 2002, 31(4): 445-455. |
| 104 | STRENKERT D, SCHMOLLINGER S, SCHRODA M. Heat shock factor 1 counteracts epigenetic silencing of nuclear transgenes in Chlamydomonas reinhardtii [J]. Nucleic Acids Research, 2013, 41(10): 5273-5289. |
| 105 | DORON L, SEGAL N, SHAPIRA M. Transgene expression in microalgae-from tools to applications[J]. Frontiers in Plant Science, 2016, 7: 505. |
| 25 | REN C, WANG D, YANG L, et al. Research advance on foreign protein production in chloroplast[J]. Bulletin of Science and Technology, 2012, 28(3): 38-42. |
| 26 | SHI Q W, CHEN C, ZHANG W, et al. Transgenic eukaryotic microalgae as green factories: providing new ideas for the production of biologically active substances[J]. Journal of Applied Phycology, 2021, 33(2): 705-728. |
| 27 | RUSSELL C, RODRIGUEZ C, YASEEN M. High-value biochemical products & applications of freshwater eukaryotic microalgae[J]. Science of the Total Environment, 2022, 809: 151111. |
| 28 | SPECHT E, MIYAKE-STONER S, MAYFIELD S. Micro-algae come of age as a platform for recombinant protein production[J]. Biotechnology Letters, 2010, 32(10): 1373-1383. |
| 29 | NOORDALLY Z B, ISHII K, ATKINS K A, et al. Circadian control of chloroplast transcription by a nuclear-encoded timing signal[J]. Science, 2013, 339(6125): 1316-1319. |
| 30 | GUTENSOHN M, FAN E G, FRIELINGSDORF S, et al. Toc, Tic, Tatet al.: structure and function of protein transport machineries in chloroplasts[J]. Journal of Plant Physiology, 2006, 163(3): 333-347. |
| 31 | SIDDIQUI A, WEI Z Y, BOEHM M, et al. Engineering microalgae through chloroplast transformation to produce high-value industrial products[J]. Biotechnology and Applied Biochemistry, 2020, 67(1): 30-40. |
| 32 | MAUL J E, LILLY J W, CUI L Y, et al. The Chlamydomonas reinhardtii plastid chromosome: islands of genes in a sea of repeats[J]. The Plant Cell, 2002, 14(11): 2659-2679. |
| 33 | BOYNTON J E, GILLHAM N W, HARRIS E H, et al. Chloroplast transformation in Chlamydomonas with high velocity microprojectiles[J]. Science, 1988, 240(4858): 1534-1538. |
| 34 | GOLDSCHMIDT-CLERMONT M. Transgenic expression of aminoglycoside adenine transferase in the chloroplast: a selectable marker for site-directed transformation of Chlamydomonas [J]. Nucleic Acids Research, 1991, 19(15): 4083-4089. |
| 35 | ROCHAIX J D. Chloroplast reverse genetics: new insights into the function of plastid genes[J]. Trends in Plant Science, 1997, 2(11): 419-425. |
| 36 | LARREA-ALVAREZ M, PURTON S. Multigenic engineering of the chloroplast genome in the green alga Chlamydomonas reinhardtii [J]. Microbiology, 2020, 166(6): 510-515. |
| 37 | KANG S, JEON S, KIM S, et al. Development of a pVEC peptide-based ribonucleoprotein (RNP) delivery system for genome editing using CRISPR/Cas9 in Chlamydomonas reinhardtii [J]. Scientific Reports, 2020, 10: 22158. |
| KANG S, JEON S, KIM S, et al. Development of a pVEC peptide-based ribonucleoprotein (RNP) delivery system for genome editing using CRISPR/Cas9 in Chlamydomonas reinhardtii[J]. Scientific Reports, 2020, 10: 22158. | |
| 38 | SHIN S E, LIM J M, KOH H G, et al. CRISPR/Cas9-induced knockout and knock-in mutations in Chlamydomonas reinhardtii [J]. Scientific Reports, 2016, 6: 27810. |
| 39 | NYMARK M, SHARMA A K, SPARSTAD T, et al. A CRISPR/Cas9 system adapted for gene editing in marine algae[J]. Scientific Reports, 2016, 6: 24951. |
| 40 | CHEN X, KINDLE K, STERN D. Initiation codon mutations in the Chlamydomonas chloroplast petD gene result in temperature-sensitive photosynthetic growth[J]. The EMBO Journal, 1993, 12(9): 3627-3635. |
| 41 | MAJERAN W, WOLLMAN F A, VALLON O. Evidence for a role of ClpP in the degradation of the chloroplast cytochrome b6f complex[J]. The Plant Cell, 2000, 12(1): 137-149. |
| 42 | SURZYCKI R, COURNAC L, PELTIER G, et al. Potential for hydrogen production with inducible chloroplast gene expression in Chlamydomonas [J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(44): 17548-17553. |
| 43 | RAMUNDO S, RAHIRE M, SCHAAD O, et al. Repression of essential chloroplast genes reveals new signaling pathways and regulatory feedback loops in Chlamydomonas [J]. The Plant Cell, 2013, 25(1): 167-186. |
| 44 | RAMUNDO S, CASERO D, MÜHLHAUS T, et al. Conditional depletion of the Chlamydomonas chloroplast ClpP protease activates nuclear genes involved in autophagy and plastid protein quality control[J]. The Plant Cell, 2014, 26(5): 2201-2222. |
| 45 | SURZYCKI R, GREENHAM K, KITAYAMA K, et al. Factors effecting expression of vaccines in microalgae[J]. Biologicals, 2009, 37(3): 133-138. |
| 46 | BARKAN A, GOLDSCHMIDT-CLERMONT M. Participation of nuclear genes in chloroplast gene expression[J]. Biochimie, 2000, 82(6/7): 559-572. |
| 47 | NICKELSEN J, VAN DILLEWIJN J, RAHIRE M, et al. Determinants for stability of the chloroplast psbD RNA are located within its short leader region in Chlamydomonas reinhardtii [J]. The EMBO Journal, 1994, 13(13): 3182-3191. |
| 106 | AIGNER H, WILSON R H, BRACHER A, et al. Plant RuBisCo assembly in E. coli with five chloroplast chaperones including BSD2[J]. Science, 2017, 358(6368): 1272-1278. |
| 107 | YAO C H, AI J N, CAO X P, et al. Enhancing starch production of a marine green microalga Tetraselmis subcordiformis through nutrient limitation[J]. Bioresource Technology, 2012, 118: 438-444. |
| 108 | HIRAI M Y, YANO M, GOODENOWE D B, et al. Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana [J]. Proceedings of the National Academy of Sciences of the United States of America, 2004, 101(27): 10205-10210. |
| 109 | 朱振, 曹旭鹏, 苑广泽, 等. 莱茵衣藻叶绿体型磷酸甘油醛脱氢酶过表达对其储能物质生产的影响[J]. 中国海洋大学学报(自然科学版), 2019, 49(9): 50-58. |
| ZHU Z, CAO X P, YUAN G Z, et al. Studies on the effect of overexpressed chloroplast glyceraldehyde-3-phosphate dehydrogenase on carbohydrate and fatty acid contents of Chlamydomonas reinhardtii [J]. Periodical of Ocean University of China, 2019, 49(9): 50-58. | |
| 110 | ZHU Z, YUAN G, FAN X, et al. The synchronous TAG production with the growth by the expression of chloroplast transit peptide-fused ScPDAT in Chlamydomonas reinhardtii [J]. Biotechnology for Biofuels, 2018, 11: 156. |
| 111 | YAMASAKI T, MIYASAKA H, OHAMA T. Unstable RNAi effects through epigenetic silencing of an inverted repeat transgene in Chlamydomonas reinhardtii [J]. Genetics, 2008, 180(4): 1927-1944. |
| 112 | TARDIF M, ATTEIA A, SPECHT M, et al. PredAlgo: a new subcellular localization prediction tool dedicated to green algae[J]. Molecular Biology and Evolution, 2012, 29(12): 3625-3639. |
| 48 | MERCHANT S, BOGORAD L. The Cu(II)-repressible plastidic cytochrome c. Cloning and sequence of a complementary DNA for the pre-apoprotein[J]. Journal of Biological Chemistry, 1987, 262(19): 9062-9067. |
| 49 | QUINN J M, ERIKSSON M, MOSELEY J L, et al. Oxygen deficiency responsive gene expression in Chlamydomonas reinhardtii through a copper-sensing signal transduction pathway[J]. Plant Physiology, 2002, 128(2): 463-471. |
| 50 | HELLIWELL K E, SCAIFE M A, SASSO S, et al. Unraveling vitamin B12-responsive gene regulation in algae[J]. Plant Physiology, 2014, 165(1): 388-397. |
| 51 | CROFT M T, MOULIN M, WEBB M E, et al. Thiamine biosynthesis in algae is regulated by riboswitches[J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(52): 20770-20775. |
| 52 | MICHELET L, LEFEBVRE-LEGENDRE L, BURR S E, et al. Enhanced chloroplast transgene expression in a nuclear mutant of Chlamydomonas [J]. Plant Biotechnology Journal, 2011, 9(5): 565-574. |
| 53 | RASALA B A, MUTO M, LEE P A, et al. Production of therapeutic proteins in algae, analysis of expression of seven human proteins in the chloroplast of Chlamydomonas reinhardtii [J]. Plant Biotechnology Journal, 2010, 8(6): 719-733. |
| 54 | DYO Y M, PURTON S. The algal chloroplast as a synthetic biology platform for production of therapeutic proteins[J]. Microbiology, 2018, 164(2): 113-121. |
| 55 | SUN M, QIAN K X, SU N, et al. Foot-and-mouth disease virus VP1 protein fused with cholera toxin B subunit expressed in Chlamydomonas reinhardtii chloroplast[J]. Biotechnology Letters, 2003, 25(13): 1087-1092. |
| 56 | TRAN M, VAN C, BARRERA D J, et al. Production of unique immunotoxin cancer therapeutics in algal chloroplasts[J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(1): E15-E22. |
| 57 | STOFFELS L, TAUNT H N, CHARALAMBOUS B, et al. Synthesis of bacteriophage lytic proteins against Streptococcus pneumoniae in the chloroplast of Chlamydomonas reinhardtii [J]. Plant Biotechnology Journal, 2017, 15(9): 1130-1140. |
| [1] | 高歌, 边旗, 王宝俊. 合成基因线路的工程化设计研究进展与展望[J]. 合成生物学, 2025, 6(1): 45-64. |
| [2] | 李冀渊, 吴国盛. 合成生物学视域下有机体的两种隐喻[J]. 合成生物学, 2025, 6(1): 190-202. |
| [3] | 焦洪涛, 齐蒙, 邵滨, 蒋劲松. DNA数据存储技术的法律治理议题[J]. 合成生物学, 2025, 6(1): 177-189. |
| [4] | 唐兴华, 陆钱能, 胡翌霖. 人类世中对合成生物学的哲学反思[J]. 合成生物学, 2025, 6(1): 203-212. |
| [5] | 徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176. |
| [6] | 石婷, 宋展, 宋世怡, 张以恒. 体外生物转化(ivBT):生物制造的新前沿[J]. 合成生物学, 2024, 5(6): 1437-1460. |
| [7] | 柴猛, 王风清, 魏东芝. 综合利用木质纤维素生物转化合成有机酸[J]. 合成生物学, 2024, 5(6): 1242-1263. |
| [8] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [9] | 陈雨, 张康, 邱以婧, 程彩云, 殷晶晶, 宋天顺, 谢婧婧. 微生物电合成技术转化二氧化碳研究进展[J]. 合成生物学, 2024, 5(5): 1142-1168. |
| [10] | 郑皓天, 李朝风, 刘良叙, 王嘉伟, 李恒润, 倪俊. 负碳人工光合群落的设计、优化与应用[J]. 合成生物学, 2024, 5(5): 1189-1210. |
| [11] | 夏孔晨, 徐维华, 吴起. 光酶催化混乱性反应的研究进展[J]. 合成生物学, 2024, 5(5): 997-1020. |
| [12] | 陈子苓, 向阳飞. 类器官技术与合成生物学协同研究进展[J]. 合成生物学, 2024, 5(4): 795-812. |
| [13] | 蔡冰玉, 谭象天, 李伟. 合成生物学在干细胞工程化改造中的研究进展[J]. 合成生物学, 2024, 5(4): 782-794. |
| [14] | 谢皇, 郑义蕾, 苏依婷, 阮静怡, 李永泉. 放线菌聚酮类化合物生物合成体系重构研究进展[J]. 合成生物学, 2024, 5(3): 612-630. |
| [15] | 查文龙, 卜兰, 訾佳辰. 中药药效成分群的合成生物学研究进展[J]. 合成生物学, 2024, 5(3): 631-657. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||