合成生物学 ›› 2025, Vol. 6 ›› Issue (5): 1041-1057.DOI: 10.12211/2096-8280.2025-081
固氮合成生物学研究进展
李超, 张焕, 杨军, 王二涛
- 中国科学院分子植物科学卓越创新中心,植物生理生态研究所,植物分子遗传国家重点实验室,上海 200032
-
收稿日期:2025-08-01修回日期:2025-09-10出版日期:2025-10-31发布日期:2025-11-05 -
通讯作者:杨军,王二涛 -
作者简介:李超 (1999—),男,博士研究生。研究方向为非豆科植物水稻的高效固氮共生改造。E-mail:lchao@cemps.ac.cn张焕 (1993—),女,博士。研究方向为利用合成生物技术,实现水稻、玉米和小麦等非豆科植物的高效共生固氮。E-mail:zhanghuan@cemps.ac.cn杨军 (1973—),男,正高级工程师,硕士生导师。研究方向为蒺藜首蓿-根瘤菌共生的分子机制,以及非豆科植物生物固氮改造的可行性探索。E-mail:jyang@cemps.ac.cn王二涛 (1979—),男,研究员,博士生导师。研究方向为植物-微生物共生及相关激素信号转导,从事模式植物水稻、苜蓿-菌根真菌共生的分子机理;豆科植物-根瘤菌共生固氮的分子机理;植物根际微生物群组装和利用;非豆科植物水稻、玉米和小麦结瘤固氮可行性的探讨方向的研究。E-mail:etwang@cemps.ac.cn -
基金资助:中国科学院战略性先导科技专项(XDB1240000)
Research advances in nitrogen fixation synthetic biology
LI Chao, ZHANG Huan, YANG Jun, WANG Ertao
- National Key Laboratory of Plant Molecular Genetics,CAS Center for Excellence in Molecular Plant Sciences,Institute of Plant Physiology and Ecology,Chinese Academy of Sciences,Shanghai 200032,China
-
Received:2025-08-01Revised:2025-09-10Online:2025-10-31Published:2025-11-05 -
Contact:YANG Jun, WANG Ertao -
Supported by:Ⅰ.Engineer nitrogen-fixing bacteria to enhance nitrogenase activity and optimize ammonium excretion efficiency;Ⅱ.Strengthen crops’ ability to recruit beneficial diazotrophs in the rhizosphere and regulate their nitrogen-fixing activity;Ⅲ.Decipher nodulation mechanisms to achieve symbiotic nitrogen fixation in non-leguminous plants;Ⅳ.Express nitrogenase in plant organelles to enable autonomous nitrogen fixation in crops
摘要:
自然界中,豆科植物可以通过与根瘤菌的共生,利用其固氮能力将空气中的氮气(N2)还原为植物可直接利用的氨(NH3),从而降低了豆科植物对化学氮肥的需求。然而,玉米和水稻等非豆科作物缺乏根瘤共生固氮的能力,其高产稳产严重依赖化学氮肥的施用。过量施用氮肥导致土壤板结酸化,温室气体排放及水体富营养化等严峻的环境问题,严重威胁农业可持续发展和粮食安全。本文综述了固氮合成生物学的研究历史与现状,为降低非豆科作物对化学氮肥的依赖,固氮合成生物学提出了多种策略:改造根际固氮菌以增强对宿主的氮素供给;增强作物根际招募有益固氮微生物的能力以提高氮素利用效率;工程化改造非豆科植物形成类根瘤器官实现共生固氮;或将固氮酶系统直接导入植物细胞以创制自主固氮作物。近年来,该领域在提升作物产量和部分替代化学氮肥方面已取得显著进展,推动了生物固氮技术在可持续农业与生态环境保护中的创新应用。本文最后对固氮合成生物学的未来发展方向进行了展望,旨在为相关研究提供理论参考与技术指导。
中图分类号:
引用本文
李超, 张焕, 杨军, 王二涛. 固氮合成生物学研究进展[J]. 合成生物学, 2025, 6(5): 1041-1057.
LI Chao, ZHANG Huan, YANG Jun, WANG Ertao. Research advances in nitrogen fixation synthetic biology[J]. Synthetic Biology Journal, 2025, 6(5): 1041-1057.
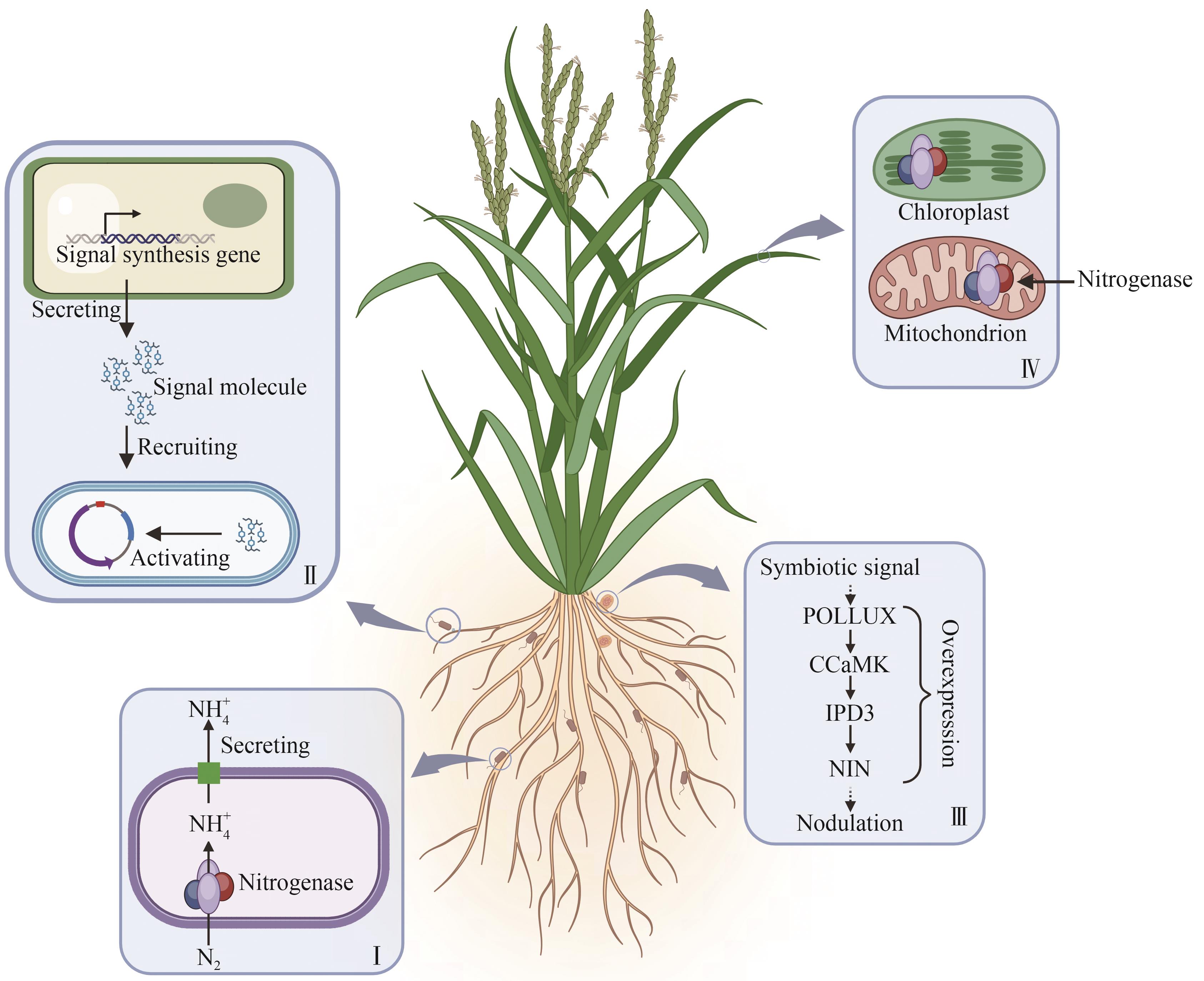
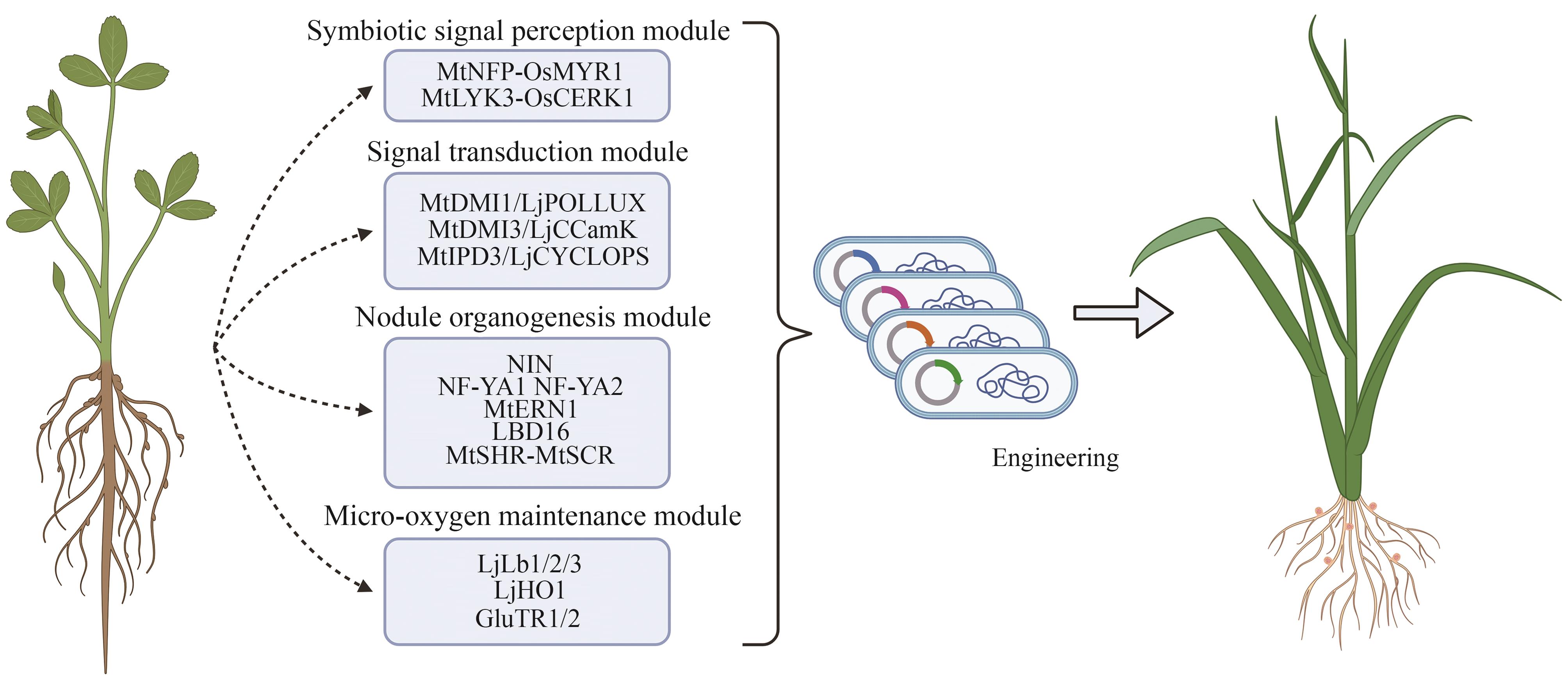
图1 设计或改进生物固氮的策略(Ⅰ.Engineer nitrogen-fixing bacteria to enhance nitrogenase activity and optimize ammonium excretion efficiency; Ⅱ.Strengthen crops’ ability to recruit beneficial diazotrophs in the rhizosphere and regulate their nitrogen-fixing activity; Ⅲ.Decipher nodulation mechanisms to achieve symbiotic nitrogen fixation in non-leguminous plants; Ⅳ.Express nitrogenase in plant organelles to enable autonomous nitrogen fixation in crops.)
Fig. 2 Main approaches to engineer or improve biological nitrogen fixation
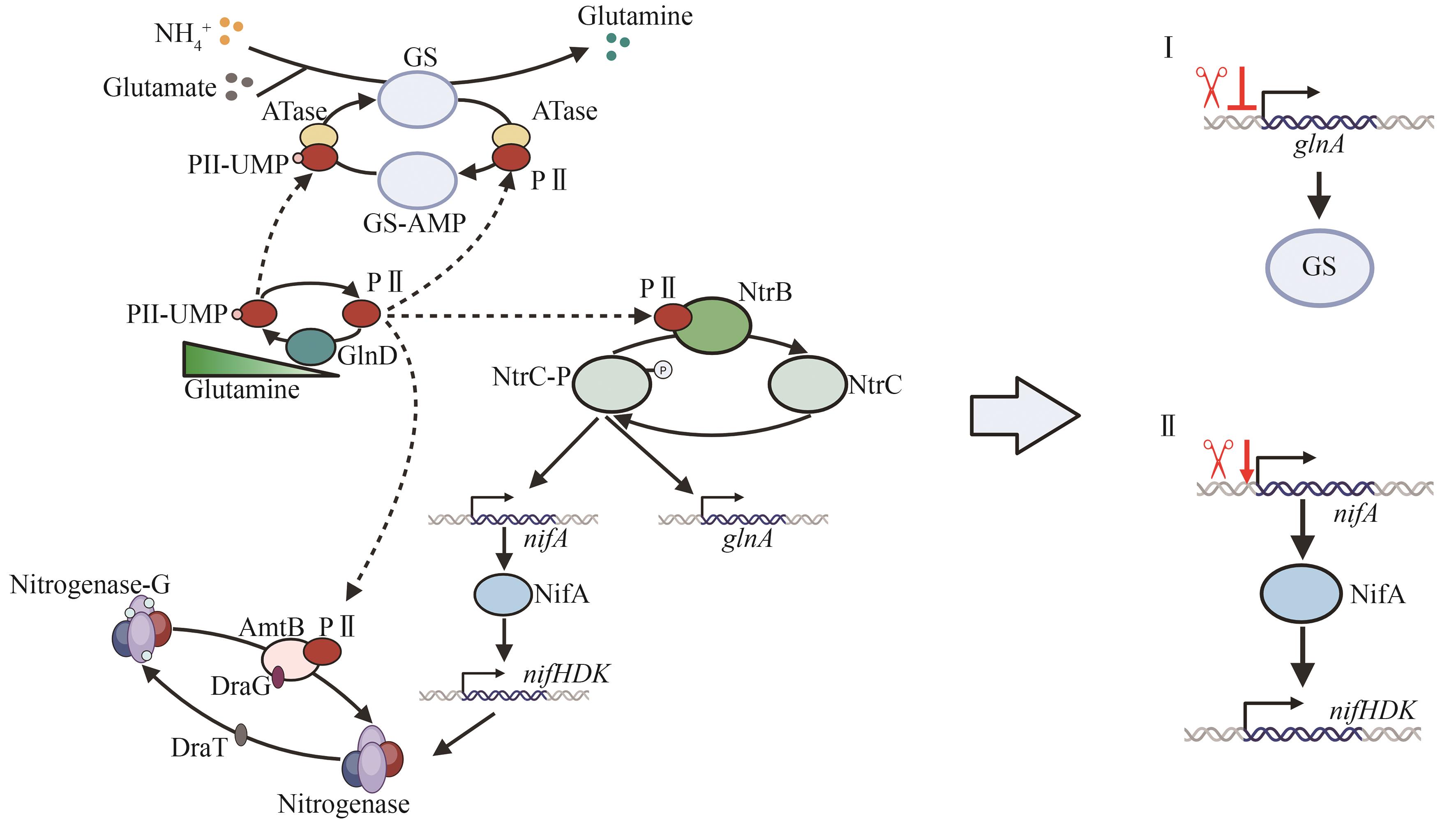
图2 固氮菌中以谷氨酰胺为核心的氮代谢调控通路与增强其固氮能力的策略(Ⅰ删除或调控glnA基因,使游离的NH4+被释放;Ⅱ调控nifA的表达,增强NifA对固氮基因簇的激活,增加固氮酶活性)
Fig. 2 The glutamine-core nitrogen metabolism regulatory pathway in diazotrophs and strategies for enhancing nitrogen fixation capacity
| [1] | ERISMAN J W, SUTTON M A, GALLOWAY J, et al. How a century of ammonia synthesis changed the world[J]. Nature Geoscience, 2008, 1(10): 636-639. |
| [2] | SMIL V. Enriching the earth: Fritz Haber, Carl Bosch, and the transformation of world food production[M]. Cambridge, Mass.: MIT Press, 2000. |
| [3] | CREWS T E, PEOPLES M B. Legume versus fertilizer sources of nitrogen: ecological tradeoffs and human needs[J]. Agriculture, Ecosystems & Environment, 2004, 102(3): 279-297. |
| [4] | GALLOWAY J N, TOWNSEND A R, ERISMAN J W, et al. Transformation of the nitrogen cycle: recent trends, questions, and potential solutions[J]. Science, 2008, 320(5878): 889-892. |
| [5] | ZHANG X, DAVIDSON E A, MAUZERALL D L, et al. Managing nitrogen for sustainable development[J]. Nature, 2015, 528(7580): 51-59. |
| [6] | BULEN W A, LECOMTE J R. The nitrogenase system from Azotobacter: two-enzyme requirement for N2 reduction, ATP-dependent H2 evolution, and ATP hydrolysis[J]. Proceedings of the National Academy of Sciences of the United States of America, 1966, 56(3): 979-986. |
| [7] | XU P, WANG E T. Diversity and regulation of symbiotic nitrogen fixation in plants[J]. Current Biology, 2023, 33(11): R543-R559. |
| [8] | DOS SANTOS P C, FANG Z, MASON S W, et al. Distribution of nitrogen fixation and nitrogenase-like sequences amongst microbial genomes[J]. BMC Genomics, 2012, 13(1): 162. |
| [9] | PATRIARCA E J, TATÈ R, IACCARINO M. Key role of bacterial NH4 + metabolism in Rhizobium-plant symbiosis[J]. Microbiology and Molecular Biology Reviews, 2002, 66(2): 203-222. |
| [10] | BANO S A, IQBAL S M. Biological nitrogen fixation to improve plant growth and productivity[J]. International journal of agriculture innovations and research, 2016, 4(4): 596. |
| [11] | MUS F, CROOK M B, GARCIA K, et al. Symbiotic nitrogen fixation and the challenges to its extension to nonlegumes[J]. Applied and Environmental Microbiology, 2016, 82(13): 3698-3710. |
| [12] | PANKIEVICZ V C S, IRVING T B, MAIA L G S, et al. Are we there yet? The long walk towards the development of efficient symbiotic associations between nitrogen-fixing bacteria and non-leguminous crops[J]. BMC Biology, 2019, 17(1): 99. |
| [13] | SOUMARE A, DIEDHIOU A G, THUITA M, et al. Exploiting biological nitrogen fixation: a route towards a sustainable agriculture[J]. Plants, 2020, 9(8): 1011. |
| [14] | BUENO BATISTA M, DIXON R. Manipulating nitrogen regulation in diazotrophic bacteria for agronomic benefit[J]. Biochemical Society Transactions, 2019, 47(2): 603-614. |
| [15] | MONTAÑEZ A, BLANCO A R, BARLOCCO C, et al. Characterization of cultivable putative endophytic plant growth promoting bacteria associated with maize cultivars (Zea mays L.) and their inoculation effects in vitro [J]. Applied Soil Ecology, 2012, 58: 21-28. |
| [16] | ROSENBLUETH M, ORMEÑO-ORRILLO E, LÓPEZ-LÓPEZ A, et al. Nitrogen fixation in cereals[J]. Frontiers in Microbiology, 2018, 9: 1794. |
| [17] | VAN DEYNZE A, ZAMORA P, DELAUX P M, et al. Nitrogen fixation in a Landrace of maize is supported by a mucilage-associated diazotrophic microbiota[J]. PLoS Biology, 2018, 16(8): e2006352. |
| [18] | DE LAJUDIE P M, ANDREWS M, ARDLEY J, et al. Minimal standards for the description of new Genera and species of rhizobia and agrobacteria[J]. International Journal of Systematic and Evolutionary Microbiology, 2019, 69(7): 1852-1863. |
| [19] | MATHESIUS U. Are legumes different? Origins and consequences of evolving nitrogen fixing symbioses[J]. Journal of Plant Physiology, 2022, 276: 153765. |
| [20] | ZHAO Y Y, ZHANG R, JIANG K W, et al. Nuclear phylotranscriptomics and phylogenomics support numerous polyploidization events and hypotheses for the evolution of rhizobial nitrogen-fixing symbiosis in Fabaceae[J]. Molecular Plant, 2021, 14(5): 748-773. |
| [21] | ANDREWS M, ANDREWS M E. Specificity in legume-rhizobia symbioses[J]. International Journal of Molecular Sciences, 2017, 18(4): 705. |
| [22] | CREWS T E, PEOPLES M B. Can the synchrony of nitrogen supply and crop demand be improved in legume and fertilizer-based agroecosystems? A review[J]. Nutrient Cycling in Agroecosystems, 2005, 72(2): 101-120. |
| [23] | GAGE D J. Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes[J]. Microbiology and Molecular Biology Reviews, 2004, 68(2): 280-300. |
| [24] | BARTLEY B A, KIM K, MEDLEY J K, et al. Synthetic biology: engineering living systems from biophysical principles[J]. Biophysical Journal, 2017, 112(6): 1050-1058. |
| [25] | GUPTA D, SHARMA G, SARASWAT P, et al. Synthetic biology in plants, a boon for coming decades[J]. Molecular Biotechnology, 2021, 63(12): 1138-1154. |
| [26] | DAI S Y, FENG W Y, SONG F H, et al. Review of biological algal fertilizer technology: alleviating salinization, sequestering carbon, and improving crop productivity[J]. Bioresource Technology, 2025, 429: 132507. |
| [27] | SHARMA A, BORA P. Engineering synthetic microbial communities to restructure the phytobiome for plant health and productivity[J]. World Journal of Microbiology and Biotechnology, 2025, 41(7): 228. |
| [28] | ROCCO C, SUZUKI M, VILAR R, et al. Enhancing zinc bioavailability in rice using the novel synthetic siderophore ligand proline-2'-deoxymugineic acid (PDMA): critical insights from metal binding studies and geochemical speciation modeling[J]. Journal of Agricultural and Food Chemistry, 2025, 73(14): 8243-8253. |
| [29] | CHOUDHARY D K, VARMA A. Nitrogenase (a Key Enzyme): structure and function[M]//Rhizobium biology and biotechnology. Cham: Springer International Publishing, 2017: 293-307. |
| [30] | SCHMIDT F V, SCHULZ L, ZARZYCKI J, et al. Structural insights into the iron nitrogenase complex[J]. Nature Structural & Molecular Biology, 2024, 31(1): 150-158. |
| [31] | SHAH V K, BRILL W J. Isolation of an iron-molybdenum cofactor from nitrogenase[J]. Proceedings of the National Academy of Sciences of the United States of America, 1977, 74(8): 3249-3253. |
| [32] | KIRN J S, REES D C. Crystallographic structure and functional implications of the nitrogenase molybdenum-iron protein from Azotobacter vinelandii [J]. Nature, 1992, 360(6404): 553-560. |
| [33] | REES D C, AKIF TEZCAN F, HAYNES C A, et al. Structural basis of biological nitrogen fixation[J]. Philosophical Transactions Series A, Mathematical, Physical, and Engineering Sciences, 2005, 363(1829): 971-984. |
| [34] | DIXON R A, POSTGATE J R. Transfer of nitrogen-fixation genes by conjugation in Klebsiella pneumoniae [J]. Nature, 1971, 234(5323): 47-48. |
| [35] | ZHAN Y H, YAN Y L, DENG Z P, et al. The novel regulatory ncRNA, NfiS, optimizes nitrogen fixation via base pairing with the nitrogenase gene nifK mRNA in Pseudomonas stutzeri A1501[J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(30): E4348-E4356. |
| [36] | WRIGHT G S A, SAEKI A, HIKIMA T, et al. Architecture of the complete oxygen-sensing FixL-FixJ two-component signal transduction system[J]. Science Signaling, 2018, 11(525): eaaq0825. |
| [37] | ZHANG W Y, CHEN Y H, HUANG K Y, et al. Molecular mechanism and agricultural application of the NifA-NifL system for nitrogen fixation[J]. International Journal of Molecular Sciences, 2023, 24(2): 907. |
| [38] | LEE S, RETH A, MELETZUS D, et al. Characterization of a major cluster of nif, fix, and associated genes in a sugarcane endophyte, Acetobacter diazotrophicus [J]. Journal of Bacteriology, 2000, 182(24): 7088-7091. |
| [39] | NAREN N, ZHANG X X. Role of a local transcription factor in governing cellular carbon/nitrogen homeostasis in Pseudomonas fluorescens [J]. Nucleic Acids Research, 2021, 49(6): 3204-3216. |
| [40] | LEE C C, GÓRECKI K, STANG M, et al. Cofactor maturase NifEN: a prototype ancient nitrogenase?[J]. Science Advances, 2024, 10(24): eado6169. |
| [41] | XU Y-Y, JIANG X-L, CHAI J-L, et al. Synthetic models of the nitrogenase FeMo cofactor[J]. Proceedings of the National Academy of Sciences of the United States of America, 2025, 122(24): e2419655122. |
| [42] | TEMME K, ZHAO D H, VOIGT C A. Refactoring the nitrogen fixation gene cluster from Klebsiella oxytoca [J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(18): 7085-7090. |
| [43] | YANG J G, XIE X Q, XIANG N, et al. Polyprotein strategy for stoichiometric assembly of nitrogen fixation components for synthetic biology[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(36): E8509-E8517. |
| [44] | VAN HEESWIJK W C, WESTERHOFF H V, BOOGERD F C. Nitrogen assimilation in Escherichia coli: putting molecular data into a systems perspective[J]. Microbiology and Molecular Biology Reviews, 2013, 77(4): 628-695. |
| [45] | POOLE P, ALLAWAY D. Carbon and nitrogen metabolism in Rhizobium [J]. Advances in Microbial Physiology, 2000, 43: 117-163. |
| [46] | ORTIZ-MARQUEZ J C F, NASCIMENTO M DO, CURATTI L. Metabolic engineering of ammonium release for nitrogen-fixing multispecies microbial cell-factories[J]. Metabolic Engineering, 2014, 23: 154-164. |
| [47] | DE ZAMAROCZY M, PAQUELIN A, ELMERICH C. Functional organization of the glnB-glnA cluster of Azospirillum brasilense [J]. Journal of Bacteriology, 1993, 175(9): 2507-2515. |
| [48] | DE ZAMAROCZY M. Structural homologues P(Ⅱ) and P(Z) of Azospirillum brasilense provide intracellular signalling for selective regulation of various nitrogen-dependent functions[J]. Molecular Microbiology, 1998, 29(2): 449-463. |
| [49] | JAGGI R, VAN HEESWIJK W C, WESTERHOFF H V, et al. The two opposing activities of adenylyl transferase reside in distinct homologous domains, with intramolecular signal transduction[J]. The EMBO Journal, 1997, 16(18): 5562-5571. |
| [50] | JIANG P, PIOSZAK A A, NINFA A J. Structure-function analysis of glutamine synthetase adenylyltransferase (ATase, EC 2.7.7.49) of Escherichia coli [J]. Biochemistry, 2007, 46(13): 4117-4132. |
| [51] | WANG Y L, LIU F, WANG W. Kinetics of transcription initiation directed by multiple cis-regulatory elements on the glnAp2 promoter[J]. Nucleic Acids Research, 2016, 44(22): 10530-10538. |
| [52] | HUERGO L F, SOUZA E M, STEFFENS M B R, et al. Regulation of glnB gene promoter expression in Azospirillum brasilense by the NtrC protein[J]. FEMS Microbiology Letters, 2003, 223(1): 33-40. |
| [53] | HUERGO L F, PEDROSA F O, MULLER-SANTOS M, et al. PⅡ signal transduction proteins: pivotal players in post-translational control of nitrogenase activity[J]. Microbiology, 2012, 158(Pt 1): 176-190. |
| [54] | MOURE V R, SIÖBERG C L B, VALDAMERI G, et al. The ammonium transporter AmtB and the PⅡ signal transduction protein GlnZ are required to inhibit DraG in Azospirillum brasilense [J]. The FEBS Journal, 2019, 286(6): 1214-1229. |
| [55] | MOURE V R, DANYAL K, YANG Z Y, et al. The nitrogenase regulatory enzyme dinitrogenase reductase ADP-ribosyltransferase (DraT) is activated by direct interaction with the signal transduction protein GlnB[J]. Journal of Bacteriology, 2013, 195(2): 279-286. |
| [56] | ZHANG Y P, POHLMANN E L, ROBERTS G P. GlnD is essential for NifA activation, NtrB/NtrC-regulated gene expression, and posttranslational regulation of nitrogenase activity in the photosynthetic, nitrogen-fixing bacterium Rhodospirillum rubrum [J]. Journal of Bacteriology, 2005, 187(4): 1254-1265. |
| [57] | AMBROSIO R, ORTIZ-MARQUEZ J C F, CURATTI L. Metabolic engineering of a diazotrophic bacterium improves ammonium release and biofertilization of plants and microalgae[J]. Metabolic Engineering, 2017, 40: 59-68. |
| [58] | MICHEL-REYDELLET N, KAMINSKI P A. Azorhizobium caulinodans PⅡ and GlnK proteins control nitrogen fixation and ammonia assimilation[J]. Journal of Bacteriology, 1999, 181(8): 2655-2658. |
| [59] | MUS F, TSENG A, DIXON R, et al. Diazotrophic growth allows Azotobacter vinelandii to overcome the deleterious effects of a glnE deletion[J]. Applied and Environmental Microbiology, 2017, 83(13): e00808-17. |
| [60] | SCHNABEL T, SATTELY E. Engineering posttranslational regulation of glutamine synthetase for controllable ammonia production in the plant symbiont Azospirillum brasilense [J]. Applied and Environmental Microbiology, 2021, 87(14): e00582-21. |
| [61] | HASKETT T L, KARUNAKARAN R, BUENO BATISTA M, et al. Control of nitrogen fixation and ammonia excretion in Azorhizobium caulinodans [J]. PLoS Genetics, 2022, 18(6): e1010276. |
| [62] | ZHANG T, YAN Y L, HE S, et al. Involvement of the ammonium transporter AmtB in nitrogenase regulation and ammonium excretion in Pseudomonas stutzeri A1501[J]. Research in Microbiology, 2012, 163(5): 332-339. |
| [63] | BALI A, BLANCO G, HILL S, et al. Excretion of ammonium by a nifL mutant of Azotobacter vinelandii fixing nitrogen[J]. Applied and Environmental Microbiology, 1992, 58(5): 1711-1718. |
| [64] | BREWIN B, WOODLEY P, DRUMMOND M. The basis of ammonium release in nifL mutants of Azotobacter vinelandii [J]. Journal of Bacteriology, 1999, 181(23): 7356-7362. |
| [65] | LI Q, ZHANG H W, SONG Y, et al. Alanine synthesized by alanine dehydrogenase enables ammonium-tolerant nitrogen fixation in Paenibacillus sabinae T27[J]. Proceedings of the National Academy of Sciences of the United States of America, 2022, 119(49): e2215855119. |
| [66] | TANG Y Q, QIN D B, TIAN Z X, et al. Diurnal switches in diazotrophic lifestyle increase nitrogen contribution to cereals[J]. Nature Communications, 2023, 14: 7516. |
| [67] | BRAUTASET T, LALE R, VALLA S. Positively regulated bacterial expression systems[J]. Microbial Biotechnology, 2009, 2(1): 15-30. |
| [68] | KENT R, DIXON N. Contemporary tools for regulating gene expression in bacteria[J]. Trends in Biotechnology, 2020, 38(3): 316-333. |
| [69] | FRAY R G, THROUP J P, DAYKIN M, et al. Plants genetically modified to produce N-acylhomoserine lactones communicate with bacteria[J]. Nature Biotechnology, 1999, 17(10): 1017-1020. |
| [70] | MURPHY P J, WEXLER W, GRZEMSKI W, et al. Rhizopines: their role in symbiosis and competition[J]. Soil Biology and Biochemistry, 1995, 27(4-5): 525-529. |
| [71] | MURPHY P J, TRENZ S P, GRZEMSKI W, et al. The Rhizobium meliloti rhizopine mos locus is a mosaic structure facilitating its symbiotic regulation[J]. Journal of Bacteriology, 1993, 175(16): 5193-5204. |
| [72] | GORDON D M A N, RYDER M H, HEINRICH K, et al. An experimental test of the rhizopine concept in Rhizobium meliloti [J]. Applied and Environmental Microbiology, 1996, 62(11): 3991-3996. |
| [73] | WEXLER M, GORDON D, MURPHY P J. The distribution of inositol rhizopine genes in Rhizobium populations[J]. Soil Biology and Biochemistry, 1995, 27(4-5): 531-537. |
| [74] | MURPHY P J, HEYCKE N, BANFALVI Z, et al. Genes for the catabolism and synthesis of an opine-like compound in Rhizobium meliloti are closely linked and on the Sym plasmid[J]. Proceedings of the National Academy of Sciences of the United States of America, 1987, 84(2): 493-497. |
| [75] | GEDDES B A, PARAMASIVAN P, JOFFRIN A, et al. Engineering transkingdom signalling in plants to control gene expression in rhizosphere bacteria[J]. Nature Communications, 2019, 10: 3430. |
| [76] | HASKETT T L, PARAMASIVAN P, MENDES M D, et al. Engineered plant control of associative nitrogen fixation[J]. Proceedings of the National Academy of Sciences of the United States of America, 2022, 119(16): e2117465119. |
| [77] | ROY S, LIU W, NANDETY R S, et al. Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation[J]. The Plant Cell, 2020, 32(1): 15-41. |
| [78] | YANG J, LAN L Y, JIN Y, et al. Mechanisms underlying legume—Rhizobium symbioses[J]. Journal of Integrative Plant Biology, 2022, 64(2): 244-267. |
| [79] | SUBRAMANIAN S, STACEY G, YU O. Distinct, crucial roles of flavonoids during legume nodulation[J]. Trends in Plant Science, 2007, 12(7): 282-285. |
| [80] | VENKATESHWARAN M, VOLKENING J D, SUSSMAN M R, et al. Symbiosis and the social network of higher plants[J]. Current Opinion in Plant Biology, 2013, 16(1): 118-127. |
| [81] | GENRE A, RUSSO G. Does a common pathway transduce symbiotic signals in plant-microbe interactions?[J]. Frontiers in Plant Science, 2016, 7: 96. |
| [82] | MADSEN E B, MADSEN L H, RADUTOIU S, et al. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals[J]. Nature, 2003, 425(6958): 637-640. |
| [83] | SMIT P, LIMPENS E, GEURTS R, et al. Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling[J]. Plant Physiology, 2007, 145(1): 183-191. |
| [84] | OLDROYD G E D. Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants[J]. Nature Reviews Microbiology, 2013, 11(4): 252-263. |
| [85] | MARTIN F M, UROZ S, BARKER D G. Ancestral alliances: plant mutualistic symbioses with fungi and bacteria[J]. Science, 2017, 356(6340): eaad4501. |
| [86] | LÉVY J, BRES C, GEURTS R, et al. A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses[J]. Science, 2004, 303(5662): 1361-1364. |
| [87] | GLEASON C, CHAUDHURI S, YANG T B, et al. Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition[J]. Nature, 2006, 441(7097): 1149-1152. |
| [88] | TIRICHINE L, IMAIZUMI-ANRAKU H, YOSHIDA S, et al. Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development[J]. Nature, 2006, 441(7097): 1153-1156. |
| [89] | JIN Y, LIU H, LUO D X, et al. DELLA proteins are common components of symbiotic rhizobial and mycorrhizal signalling pathways[J]. Nature Communications, 2016, 7: 12433. |
| [90] | SINGH S, KATZER K, LAMBERT J, et al. CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development[J]. Cell Host & Microbe, 2014, 15(2): 139-152. |
| [91] | YANO K, YOSHIDA S, MÜLLER J, et al. CYCLOPS, a mediator of symbiotic intracellular accommodation[J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(51): 20540-20545. |
| [92] | DELWICHE C F, COOPER E D. The evolutionary origin of a terrestrial flora[J]. Current Biology, 2015, 25(19): R899-R910. |
| [93] | PARNISKE M. Arbuscular mycorrhiza: the mother of plant root endosymbioses[J]. Nature Reviews Microbiology, 2008, 6(10): 763-775. |
| [94] | GENRE A, LANFRANCO L, PEROTTO S, et al. Unique and common traits in mycorrhizal symbioses[J]. Nature Reviews Microbiology, 2020, 18(11): 649-660. |
| [95] | SHI J C, WANG X L, WANG E T. Mycorrhizal symbiosis in plant growth and stress adaptation: from genes to ecosystems[J]. Annual Review of Plant Biology, 2023, 74: 569-607. |
| [96] | WANG W, XIE Z P, STAEHELIN C. Functional analysis of chimeric lysin motif domain receptors mediating Nod factor-induced defense signaling in Arabidopsis thaliana and chitin-induced nodulation signaling in Lotus japonicus [J]. The Plant Journal, 2014, 78(1): 56-69. |
| [97] | HE J M, ZHANG C, DAI H L, et al. A LysM receptor heteromer mediates perception of arbuscular mycorrhizal symbiotic signal in rice[J]. Molecular Plant, 2019, 12(12): 1561-1576. |
| [98] | WANG D P, JIN R, SHI X B, et al. A kinase mediator of rhizobial symbiosis and immunity in Medicago [J]. Nature, 2025, 643(8072): 768-775. |
| [99] | GAMAS P, BRAULT M, JARDINAUD M F, et al. Cytokinins in symbiotic nodulation: when, where, what for?[J]. Trends in Plant Science, 2017, 22(9): 792-802. |
| [100] | ARIEL F, BRAULT-HERNANDEZ M, LAFFONT C, et al. Two direct targets of cytokinin signaling regulate symbiotic nodulation in Medicago truncatula [J]. The Plant Cell, 2012, 24(9): 3838-3852. |
| [101] | HECKMANN A B, SANDAL N, BEK A S, et al. Cytokinin induction of root nodule primordia in Lotus japonicus is regulated by a mechanism operating in the root cortex[J]. Molecular Plant-Microbe Interactions, 2011, 24(11): 1385-1395. |
| [102] | HIRSCH A M, FANG Y, ASAD S, et al. The role of phytohormones in plant-microbe symbioses[J]. Plant and Soil, 1997, 194(1): 171-184. |
| [103] | ARORA N, SKOOG F, ALLEN O N. Kinetin-induced pseudonodules on tobacco roots[J]. American Journal of Botany, 1959, 46(8): 610-613. |
| [104] | RODRIGUEZ-BARRUECO C, DE CASTRO F B. Cytokinin-induced pseudonodules on Alnus glutinosa [J]. Physiologia Plantarum, 1973, 29(2): 277-280. |
| [105] | SOYANO T, KOUCHI H, HIROTA A, et al. Nodule inception directly targets NF-Y subunit genes to regulate essential processes of root nodule development in Lotus japonicus [J]. PLoS Genetics, 2013, 9(3): e1003352. |
| [106] | LALOUM T, BAUDIN M, FRANCES L, et al. Two CCAAT-box-binding transcription factors redundantly regulate early steps of the legume-rhizobia endosymbiosis[J]. The Plant Journal, 2014, 79(5): 757-768. |
| [107] | GOH T, JOI S, MIMURA T, et al. The establishment of asymmetry in Arabidopsis lateral root founder cells is regulated by LBD16/ASL18 and related LBD/ASL proteins[J]. Development, 2012, 139(5): 883-893. |
| [108] | SCHIESSL K, LILLEY J L S, LEE T, et al. NODULE INCEPTION recruits the lateral root developmental program for symbiotic nodule organogenesis in Medicago truncatula [J]. Current Biology, 2019, 29(21): 3657-3668.e5. |
| [109] | SOYANO T, SHIMODA Y, KAWAGUCHI M, et al. A shared gene drives lateral root development and root nodule symbiosis pathways in Lotus [J]. Science, 2019, 366(6468): 1021-1023. |
| [110] | DI LAURENZIO L, WYSOCKA-DILLER J, MALAMY J E, et al. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root[J]. Cell, 1996, 86(3): 423-433. |
| [111] | HELARIUTTA Y, FUKAKI H, WYSOCKA-DILLER J, et al. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling[J]. Cell, 2000, 101(5): 555-567. |
| [112] | DONG W T, ZHU Y Y, CHANG H Z, et al. An SHR-SCR module specifies legume cortical cell fate to enable nodulation[J]. Nature, 2021, 589(7843): 586-590. |
| [113] | WANG L L, RUBIO M C, XIN X, et al. CRISPR/Cas9 knockout of leghemoglobin genes in Lotus japonicus uncovers their synergistic roles in symbiotic nitrogen fixation[J]. New Phytologist, 2019, 224(2): 818-832. |
| [114] | ZHOU Y, WANG L L, RUBIO M C, et al. Heme catabolism mediated by heme oxygenase in uninfected interstitial cells enables efficient symbiotic nitrogen fixation in Lotus japonicus nodules[J]. New Phytologist, 2023, 239(5): 1989-2006. |
| [115] | WANG L L, TIAN T, DENG Y, et al. Plant glutamyl-tRNA reductases coordinate plant and rhizobial heme biosynthesis in nitrogen-fixing nodules[J]. The Plant Cell, 2025, 37(5): koaf095. |
| [116] | HOFFMAN B M, LUKOYANOV D, YANG Z Y, et al. Mechanism of nitrogen fixation by nitrogenase: the next stage[J]. Chemical Reviews, 2014, 114(8): 4041-4062. |
| [117] | RIBBE M W, HU Y L, HODGSON K O, et al. Biosynthesis of nitrogenase metalloclusters[J]. Chemical Reviews, 2014, 114(8): 4063-4080. |
| [118] | HU Y L, FAY A W, DOS SANTOS P C, et al. Characterization of Azotobacter vinelandii nifZ deletion strains. Indication of stepwise MoFe protein assembly[J]. The Journal of Biological Chemistry, 2004, 279(52): 54963-54971. |
| [119] | HU Y L, FAY A W, LEE C C, et al. P-cluster maturation on nitrogenase MoFe protein[J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(25): 10424-10429. |
| [120] | WIIG J A, HU Y L, RIBBE M W. Refining the pathway of carbide insertion into the nitrogenase M-cluster[J]. Nature Communications, 2015, 6: 8034. |
| [121] | FAY A W, BLANK M A, REBELEIN J G, et al. Assembly scaffold NifEN: a structural and functional homolog of the nitrogenase catalytic component[J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(34): 9504-9508. |
| [122] | FRANKE P, FREIBERGER S, ZHANG L, et al. Conformational protection of molybdenum nitrogenase by Shethna protein Ⅱ[J]. Nature, 2025, 637(8047): 998-1004. |
| [123] | GALLON J R. The oxygen sensitivity of nitrogenase: a problem for biochemists and micro-organisms[J]. Trends in Biochemical Sciences, 1981, 6: 19-23. |
| [124] | NAREHOOD S M, COOK B D, SRISANTITHAM S, et al. Structural basis for the conformational protection of nitrogenase from O2 [J]. Nature, 2025, 637(8047): 991-997. |
| [125] | POOLE R K, HILL S. Respiratory protection of nitrogenase activity in Azotobacter vinelandii: roles of the terminal oxidases[J]. Bioscience Reports, 1997, 17(3): 303-317. |
| [126] | ROBSON R L, POSTGATE J R. Oxygen and hydrogen in biological nitrogen fixation[J]. Annual Review of Microbiology, 1980, 34: 183-207. |
| [127] | WITTENBERG J B. Facilitated oxygen diffusion. The role of leghemoglobin in nitrogen fixation by bacteroids isolated from soybean root nodules[J]. The Journal of Biological Chemistry, 1974, 249(13): 4057-4066. |
| [128] | TAKIMOTO R, TATEMICHI Y, AOKI W, et al. A critical role of an oxygen-responsive gene for aerobic nitrogenase activity in Azotobacter vinelandii and its application to Escherichia coli [J]. Scientific Reports, 2022, 12: 4182. |
| [129] | GOOD A. Toward nitrogen-fixing plants[J]. Science, 2018, 359(6378): 869-870. |
| [130] | XIANG N, GUO C Y, LIU J W, et al. Using synthetic biology to overcome barriers to stable expression of nitrogenase in eukaryotic organelles[J]. Proceedings of the National Academy of Sciences of the United States of America, 2020, 117(28): 16537-16545. |
| [131] | SOTO G, FOX A R, AYUB N D. Exploring the intrinsic limits of nitrogenase transfer from bacteria to eukaryotes[J]. Journal of Molecular Evolution, 2013, 77(1): 3-7. |
| [132] | LÓPEZ-TORREJÓN G, JIMÉNEZ-VICENTE E, BUESA J M, et al. Expression of a functional oxygen-labile nitrogenase component in the mitochondrial matrix of aerobically grown yeast[J]. Nature Communications, 2016, 7: 11426. |
| [133] | BURÉN S, YOUNG E M, SWEENY E A, et al. Formation of nitrogenase NifDK tetramers in the mitochondria of Saccharomyces cerevisiae [J]. ACS Synthetic Biology, 2017, 6(6): 1043-1055. |
| [134] | ALLEN R S, TILBROOK K, WARDEN A C, et al. Expression of 16 nitrogenase proteins within the plant mitochondrial matrix[J]. Frontiers in Plant Science, 2017, 8: 287. |
| [135] | HE W S, BURÉN S, BAYSAL C, et al. Nitrogenase cofactor maturase NifB isolated from transgenic rice is active in FeMo-co synthesis[J]. ACS Synthetic Biology, 2022, 11(9): 3028-3036. |
| [136] | JIANG X, COROIAN D, BARAHONA E, et al. Functional nitrogenase cofactor maturase NifB in mitochondria and chloroplasts of Nicotiana benthamiana [J]. mBio, 2022, 13(3): e00268-22 |
| [137] | YANG J G, XIE X Q, YANG M X, et al. Modular electron-transport chains from eukaryotic organelles function to support nitrogenase activity[J]. Proceedings of the National Academy of Sciences of the United States of America, 2017, 114(12): E2460-E2465. |
| [138] | YANG J G, XIANG N, LIU Y H, et al. Organelle-dependent polyprotein designs enable stoichiometric expression of nitrogen fixation components targeted to mitochondria[J]. Proceedings of the National Academy of Sciences of the United States of America, 2023, 120(34): e2305142120. |
| [139] | YAO Q H, PENG R H, WANG B, et al. Endowing plants with the capacity for autogenic nitrogen fixation[EB/OL]. Research Square, 2021. (2021-05-07)[2025-09-01]. . |
| [140] | SHANG Y M, SHI H W, LIU M Z, et al. Using synthetic biology to express nitrogenase biosynthesis pathway in rice and to overcome barriers of nitrogenase instability in plant cytosol[J]. Trends in Biotechnology, 2025, 43(4): 946-968. |
| [141] | SCHULTE C C M, BORAH K, WHEATLEY R M, et al. Metabolic control of nitrogen fixation in Rhizobium-legume symbioses[J]. Science Advances, 2021, 7(31): eabh2433. |
| [142] | ZHAO Q, WANG J P, HE Q Q, et al. Carbon type and quantity regulate soil free-living nitrogen fixation through restructuring diazotrophic community[J]. Applied Soil Ecology, 2024, 202: 105586. |
| [143] | LV F Y, ZHAN Y H, LU W, et al. Regulation of hierarchical carbon substrate utilization, nitrogen fixation, and root colonization by the Hfq/Crc/CrcZY genes in Pseudomonas stutzeri [J]. iScience, 2022, 25(12): 105663. |
| [144] | WITTE I P, LAMPE G D, EITZINGER S, et al. Programmable gene insertion in human cells with a laboratory-evolved CRISPR-associated transposase[J]. Science, 2025, 388(6748): eadt5199. |
| [145] | DOMAN J L, PANDEY S, NEUGEBAUER M E, et al. Phage-assisted evolution and protein engineering yield compact, efficient prime editors[J]. Cell, 2023, 186(18): 3983-4002.e26. |
| [146] | MILLER S M, WANG T N, LIU D R. Phage-assisted continuous and non-continuous evolution[J]. Nature Protocols, 2020, 15(12): 4101-4127. |
| [147] | BEATTY P H, GOOD A G. Plant science. Future prospects for cereals that fix nitrogen[J]. Science, 2011, 333(6041): 416-417. |
| [148] | BLOCH S E, RYU M H, OZAYDIN B, et al. Harnessing atmospheric nitrogen for cereal crop production[J]. Current Opinion in Biotechnology, 2020, 62: 181-188. |
| [149] | 燕永亮, 田长富, 杨建国, 等. 人工高效生物固氮体系创建及其农业应用[J]. 生命科学, 2021, 33(12): 1532-1543. |
| YAN Y L, TIAN C F, YANG J G, et al. Establishment of artificial efficiency biological nitrogen fixation system and its agricultural application[J]. Chinese Bulletin of Life Sciences, 2021, 33(12): 1532-1543. | |
| [150] | COOK N M, GOBBATO G, JACOTT C N, et al. Autoactive CNGC15 enhances root endosymbiosis in legume and wheat[J]. Nature, 2025, 638(8051): 752-759. |
| [151] | GAUTRAT P, LAFFONT C, FRUGIER F, et al. Nitrogen systemic signaling: from symbiotic nodulation to root acquisition[J]. Trends in Plant Science, 2021, 26(4): 392-406. |
| [152] | TSCHITSCHKO B, ESTI M, PHILIPPI M, et al. Rhizobia-diatom symbiosis fixes missing nitrogen in the ocean[J]. Nature, 2024, 630(8018): 899-904. |
| [153] | COALE T H, LOCONTE V, TURK-KUBO K A, et al. Nitrogen-fixing organelle in a marine Alga [J]. Science, 2024, 384(6692): 217-222. |
| [154] | LI J J, CHEN W X, LU Z Z, et al. Nanoengineered Azotobacter Pseudomonas stutzeri A1501 for soil ecology restoration and biological nitrogen fixation[J]. ACS Nano, 2025, 19(19): 18143-18155. |
| [1] | 宋开南, 张礼文, 王超, 田平芳, 李广悦, 潘国辉, 徐玉泉. 小分子生物农药及其生物合成研究进展[J]. 合成生物学, 2025, 6(5): 1203-1223. |
| [2] | 于文文, 吕雪芹, 李兆丰, 刘龙. 植物合成生物学与母乳低聚糖生物制造[J]. 合成生物学, 2025, 6(5): 992-997. |
| [3] | 颜钊涛, 周鹏飞, 汪阳忠, 张鑫, 谢雯燕, 田晨菲, 王勇. 植物合成生物学:植物细胞大规模培养的新机遇[J]. 合成生物学, 2025, 6(5): 1107-1125. |
| [4] | 孙扬, 陈立超, 石艳云, 王珂, 吕丹丹, 徐秀美, 张立新. 作物光合作用合成生物学的策略与展望[J]. 合成生物学, 2025, 6(5): 1025-1040. |
| [5] | 赵欣雨, 盛琦, 刘开放, 刘佳, 刘立明. 天冬氨酸族饲用氨基酸微生物细胞工厂的创制[J]. 合成生物学, 2025, 6(5): 1184-1202. |
| [6] | 何杨昱, 杨凯, 王玮琳, 黄茜, 丘梓樱, 宋涛, 何流赏, 姚金鑫, 甘露, 何玉池. 国际基因工程机器大赛中植物合成生物学主题的设计与实践[J]. 合成生物学, 2025, 6(5): 1243-1254. |
| [7] | 张学博, 朱成姝, 陈睿雲, 金庆姿, 刘晓, 熊燕, 陈大明. 农业合成生物学:政策规划与产业发展协同推进[J]. 合成生物学, 2025, 6(5): 1224-1242. |
| [8] | 刘婕, 郜钰, 马永硕, 尚轶. 合成生物学在农业中的进展及挑战[J]. 合成生物学, 2025, 6(5): 998-1024. |
| [9] | 郑雷, 郑棋腾, 张天骄, 段鲲, 张瑞福. 构建根际合成微生物菌群促进作物养分高效吸收利用[J]. 合成生物学, 2025, 6(5): 1058-1071. |
| [10] | 魏家秀, 嵇佩云, 节庆雨, 黄秋燕, 叶浩, 戴俊彪. 植物人工染色体的构建与应用[J]. 合成生物学, 2025, 6(5): 1093-1106. |
| [11] | 方馨仪, 孙丽超, 霍毅欣, 王颖, 岳海涛. 微生物合成高级醇的发展趋势与挑战[J]. 合成生物学, 2025, 6(4): 873-898. |
| [12] | 朱欣悦, 陈恬恬, 邵恒煊, 唐曼玉, 华威, 程艳玲. 益生菌辅助防治恶性肿瘤的研究进展[J]. 合成生物学, 2025, 6(4): 899-919. |
| [13] | 吴晓燕, 宋琪, 许睿, 丁陈君, 陈方, 郭勍, 张波. 合成生物学研发竞争态势对比分析[J]. 合成生物学, 2025, 6(4): 940-955. |
| [14] | 张建康, 王文君, 郭洪菊, 白北辰, 张亚飞, 袁征, 李彦辉, 李航. 基于机器视觉的高通量微生物克隆挑选工作站研制及应用[J]. 合成生物学, 2025, 6(4): 956-971. |
| [15] | 李全飞, 陈乾, 刘浩, 贺坤东, 潘亮, 雷鹏, 谷益安, 孙良, 李莎, 邱益彬, 王瑞, 徐虹. 高黏性蛋白材料的合成生物学及应用[J]. 合成生物学, 2025, 6(4): 806-828. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||