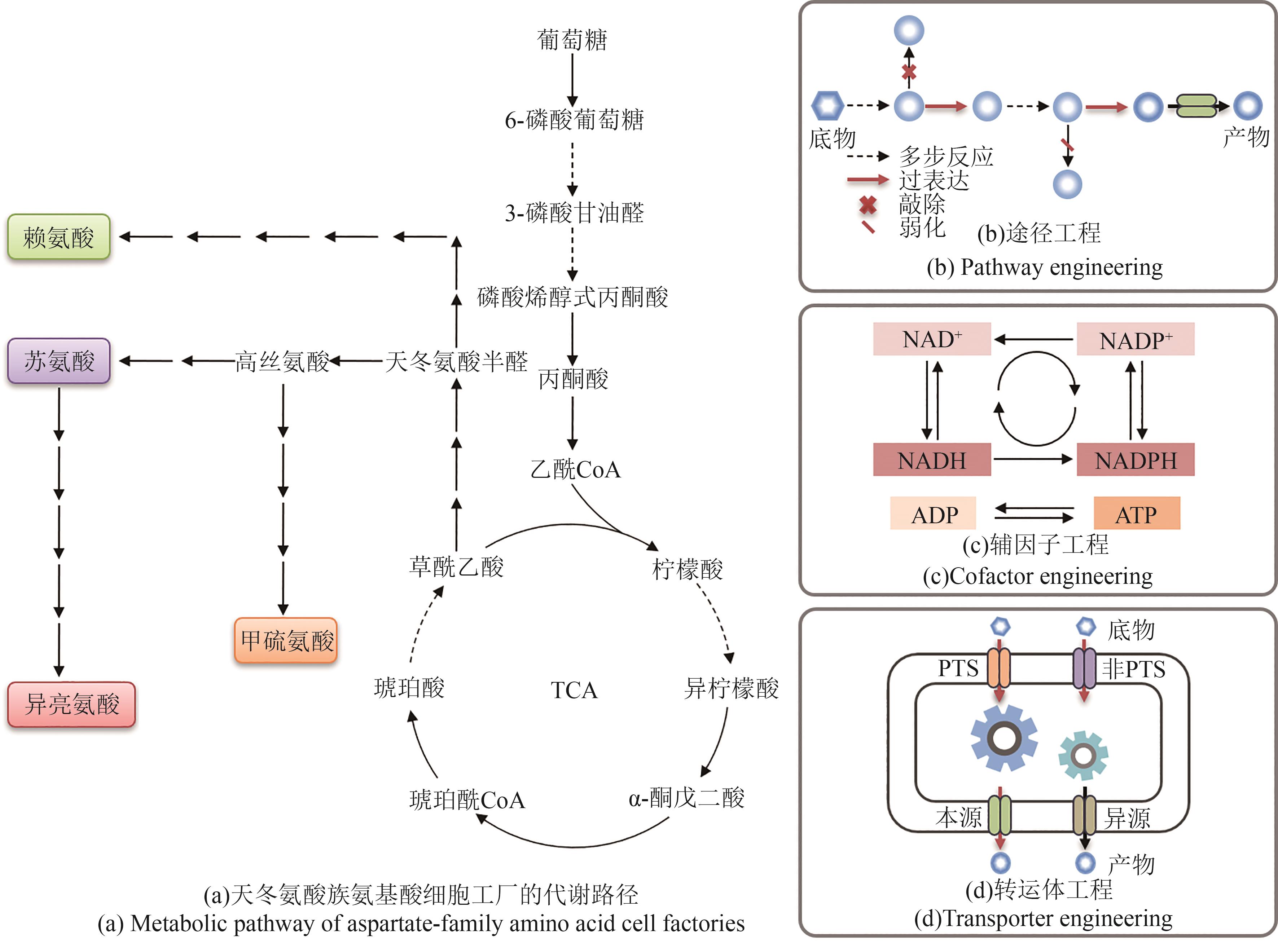
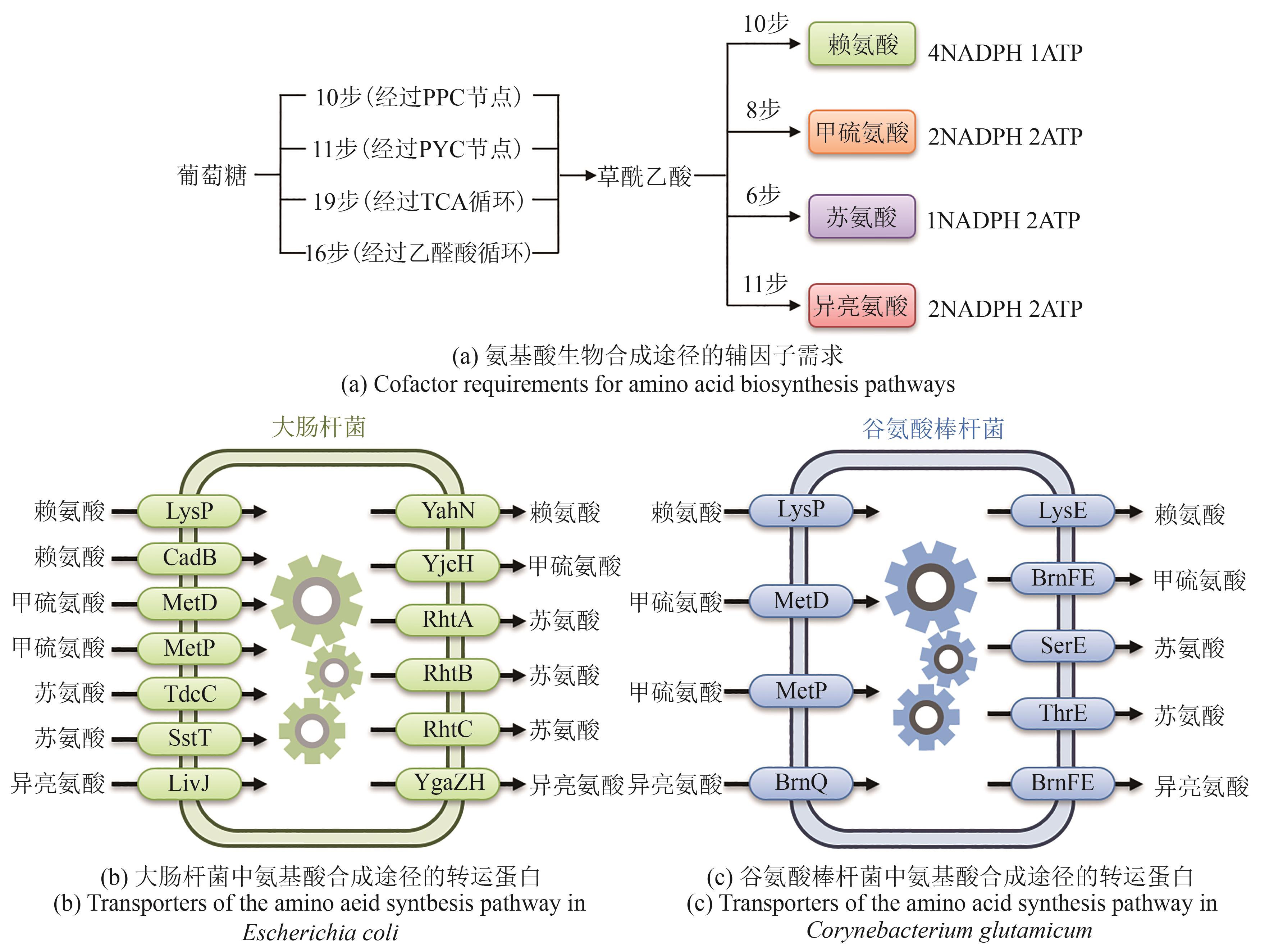
合成生物学 ›› 2025, Vol. 6 ›› Issue (5): 1184-1202.DOI: 10.12211/2096-8280.2025-032
天冬氨酸族饲用氨基酸微生物细胞工厂的创制
赵欣雨, 盛琦, 刘开放, 刘佳, 刘立明
- 江南大学生物工程学院,工业生物技术教育部重点实验室,江苏 无锡 214122
-
收稿日期:2025-04-02修回日期:2025-06-18出版日期:2025-10-31发布日期:2025-11-05 -
通讯作者:刘佳,刘立明 -
作者简介:赵欣雨 (2001—),女,硕士研究生。研究方向为微生物细胞工厂。E-mail:zxy_201710@163.com刘佳 (1988—),女,高级实验师。研究方向为酶催化工程技术。E-mail:liujia@jiangnan.edu.cn刘立明 (1976—),男,教授,博士生导师,教育部高层次人才。研究方向为合成生物学技术和微生物细胞工厂。E-mail:mingll@jiangnan.edu.cn -
基金资助:江苏省农业科技自主创新资金项目(CX(23)2005)
Construction of microbial cell factories for aspartate-family feed amino acids
ZHAO Xinyu, SHENG Qi, LIU Kaifang, LIU Jia, LIU Liming
- Key Laboratory of Industrial Biotechnology,Ministry of Education,School of Biotechnology,Jiangnan University,Wuxi 214122,Jiangsu,China
-
Received:2025-04-02Revised:2025-06-18Online:2025-10-31Published:2025-11-05 -
Contact:LIU Jia, LIU Liming
摘要:
氨基酸作为动物饲料的重要组成部分,是提高畜禽消化机能、禽肉品质、蛋白转化效率以及降低豆粕使用量的关键要素。合成生物技术的快速发展为氨基酸高产菌株构建和优化铺平了道路,极大地提升了氨基酸生产效率,显著降低了生产成本。本文在分析L-赖氨酸、L-甲硫氨酸、L-苏氨酸和L-异亮氨酸等四种天冬氨酸族氨基酸合成途径的基础上,详细介绍了菌种改造方法和策略,包括代谢路径重构、代谢途径优化、辅因子供应和增强产物外排等四个方面,未来要从工业环境抗逆性、底物利用范围的拓展以及动态调控系统的优化三个方面进行突破,才能为高性能氨基酸生产菌株的创制提供理论指导和技术支撑。
中图分类号:
引用本文
赵欣雨, 盛琦, 刘开放, 刘佳, 刘立明. 天冬氨酸族饲用氨基酸微生物细胞工厂的创制[J]. 合成生物学, 2025, 6(5): 1184-1202.
ZHAO Xinyu, SHENG Qi, LIU Kaifang, LIU Jia, LIU Liming. Construction of microbial cell factories for aspartate-family feed amino acids[J]. Synthetic Biology Journal, 2025, 6(5): 1184-1202.
| 产品 | 菌种 | |
|---|---|---|
| 液体L-赖氨酸碱 | 大肠杆菌KCCM 80190 | 大肠杆菌NITE BP-02917 |
| 谷氨酸棒状杆菌KCCM 80216 | 谷氨酸棒状杆菌KCTC 12307BP | |
| 谷氨酸棒状杆菌NRRL-B-67439 | 谷氨酸棒状杆菌NRRL-B-67535 | |
| 液体L-赖氨酸单盐酸盐 | 大肠杆菌NITE BP-02917 | |
| 技术纯L-赖氨酸单盐酸盐 | 大肠杆菌NITE BP-02917 | 谷氨酸棒状杆菌NRRL-B-67439 |
| 谷氨酸棒状杆菌DSM 32932 | 谷氨酸棒状杆菌CGMCC 7.266 | |
| 谷氨酸棒状杆菌KCCM 80183 | 谷氨酸棒状杆菌CGMCC 17927 | |
| 谷氨酸棒状杆菌NRRL B-67535 | 谷氨酸棒状杆菌CCTCC M 2015595 | |
| L-赖氨酸硫酸盐 | 大肠杆菌CGMCC 7.398 | 谷氨酸棒状杆菌CGMCC 7.266 |
| 谷氨酸棒状杆菌KFCC 11043 | 谷氨酸棒状杆菌CGMCC 17927 | |
| 谷氨酸棒状杆菌KCCM 80227 | 谷氨酸棒状杆菌CCTCC M 2015595 | |
| L-苏氨酸 | 大肠杆菌DSM 25085 | 大肠杆菌FERM BP-11383 |
| 大肠杆菌DSM 25086 | 大肠杆菌FERM BP-10942 | |
| 大肠杆菌CGMCC 3703 | 大肠杆菌NRRL B-30843 | |
| 大肠杆菌CGMCC 7.58 | 大肠杆菌KCCM 11133P | |
| 大肠杆菌CGMCC 7.232 | 谷氨酸棒状杆菌KCCM 80117 | |
| 大肠杆菌CGMCC 13325 | 谷氨酸棒状杆菌KCCM 80118 | |
| L-甲硫氨酸 | 大肠杆菌KCCM 80245 | 谷氨酸棒状杆菌KCCM 80246 |
| L-异亮氨酸 | 大肠杆菌FERM ABP-1064 | 谷氨酸棒杆菌KCCM 80189 |
| 谷氨酸棒杆菌KCCM 80185 | 谷氨酸棒杆菌CGMCC 20437 | |
表1 欧盟授权允许可在饲料中使用的天冬氨酸族氨基酸生产菌种
Table 1 EU-authorized microbial strains for production of aspartate-family amino acids approved as feed additives
| 产品 | 菌种 | |
|---|---|---|
| 液体L-赖氨酸碱 | 大肠杆菌KCCM 80190 | 大肠杆菌NITE BP-02917 |
| 谷氨酸棒状杆菌KCCM 80216 | 谷氨酸棒状杆菌KCTC 12307BP | |
| 谷氨酸棒状杆菌NRRL-B-67439 | 谷氨酸棒状杆菌NRRL-B-67535 | |
| 液体L-赖氨酸单盐酸盐 | 大肠杆菌NITE BP-02917 | |
| 技术纯L-赖氨酸单盐酸盐 | 大肠杆菌NITE BP-02917 | 谷氨酸棒状杆菌NRRL-B-67439 |
| 谷氨酸棒状杆菌DSM 32932 | 谷氨酸棒状杆菌CGMCC 7.266 | |
| 谷氨酸棒状杆菌KCCM 80183 | 谷氨酸棒状杆菌CGMCC 17927 | |
| 谷氨酸棒状杆菌NRRL B-67535 | 谷氨酸棒状杆菌CCTCC M 2015595 | |
| L-赖氨酸硫酸盐 | 大肠杆菌CGMCC 7.398 | 谷氨酸棒状杆菌CGMCC 7.266 |
| 谷氨酸棒状杆菌KFCC 11043 | 谷氨酸棒状杆菌CGMCC 17927 | |
| 谷氨酸棒状杆菌KCCM 80227 | 谷氨酸棒状杆菌CCTCC M 2015595 | |
| L-苏氨酸 | 大肠杆菌DSM 25085 | 大肠杆菌FERM BP-11383 |
| 大肠杆菌DSM 25086 | 大肠杆菌FERM BP-10942 | |
| 大肠杆菌CGMCC 3703 | 大肠杆菌NRRL B-30843 | |
| 大肠杆菌CGMCC 7.58 | 大肠杆菌KCCM 11133P | |
| 大肠杆菌CGMCC 7.232 | 谷氨酸棒状杆菌KCCM 80117 | |
| 大肠杆菌CGMCC 13325 | 谷氨酸棒状杆菌KCCM 80118 | |
| L-甲硫氨酸 | 大肠杆菌KCCM 80245 | 谷氨酸棒状杆菌KCCM 80246 |
| L-异亮氨酸 | 大肠杆菌FERM ABP-1064 | 谷氨酸棒杆菌KCCM 80189 |
| 谷氨酸棒杆菌KCCM 80185 | 谷氨酸棒杆菌CGMCC 20437 | |
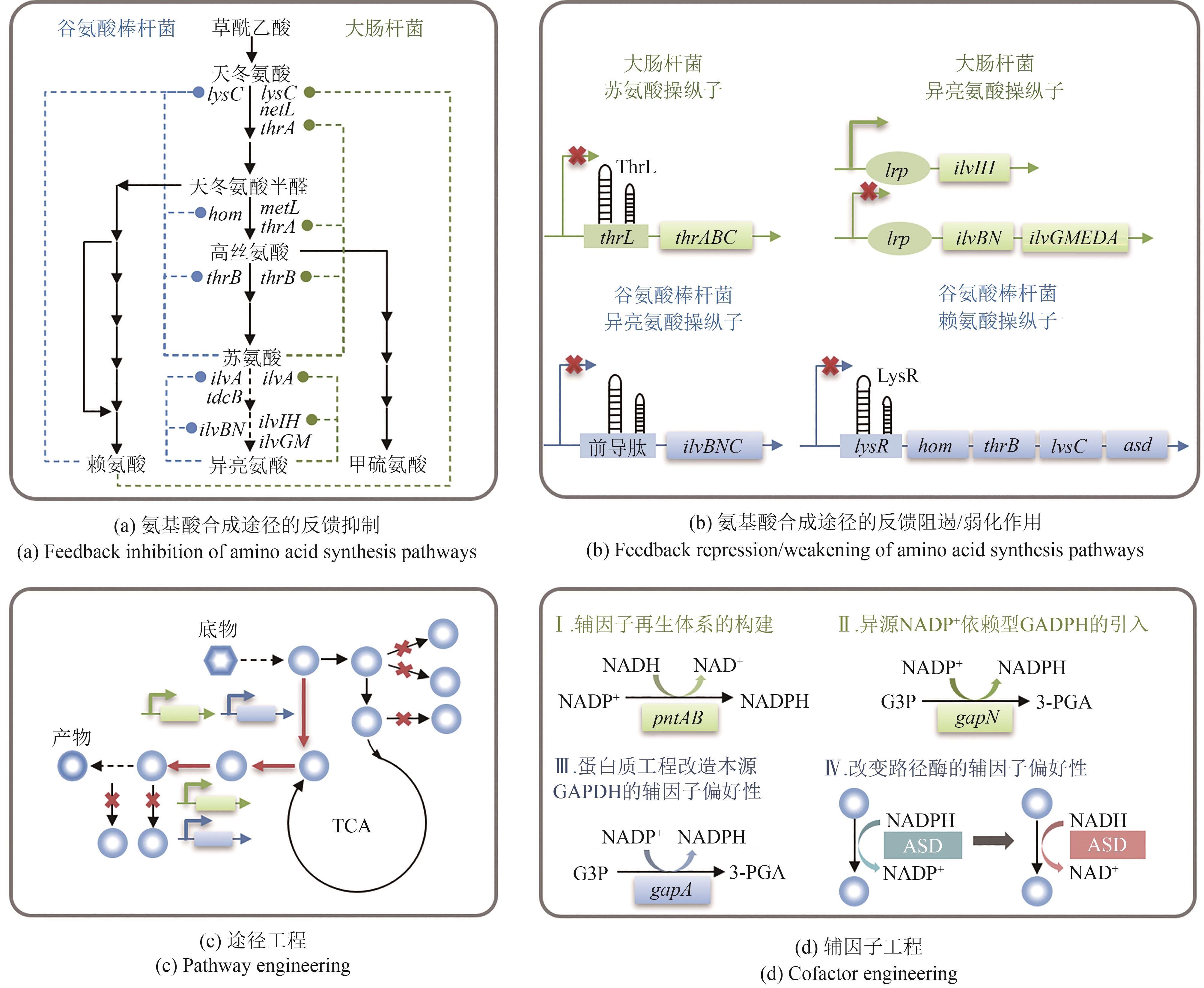
图3 天冬氨酸族氨基酸的反馈调控机制及代谢路径优化策略(thrA—编码天冬氨酸激酶Ⅰ;metL—编码天冬氨酸激酶Ⅱ;lysC—编码天冬氨酸激酶Ⅲ;hom—编码高丝氨酸脱氢酶;thrB—编码高丝氨酸激酶;ilvA—编码苏氨酸脱水酶Ⅰ;tdcB—编码苏氨酸脱水酶Ⅱ;ilvBN—编码乙酰乳酸合酶Ⅰ;ilvGM—编码乙酰乳酸合酶Ⅱ;ilvIH—编码乙酰乳酸合酶Ⅲ;asd—编码天冬氨酸半醛脱氢酶;pntAB—编码NAD(P)转氢酶亚基;gapN/gapA—编码甘油醛-3-磷酸脱氢酶;lrp—编码转录调控因子Lrp;G3P—3-磷酸甘油醛;3-PGA—3-磷酸甘油酸;ASD—天冬氨酸半醛脱氢酶)
Fig. 3 Feedback regulation mechanism and metabolic pathway optimization strategy of aspartate-family amino acids(thrA—Encoding aspartate kinase Ⅰ; metL—Encoding aspartate kinase Ⅱ; lysC—Encoding aspartate kinase Ⅲ; hom—Encoding homoserine dehydrogenase; thrB—Encoding homoserine kinase; thrB—Encoding homoserine kinase; ilvA—Encoding threonine dehydrase Ⅰ; tdcB—Encoding threonine dehydrase Ⅱ; ilvBN—Encoding acetolactate synthase Ⅰ; ilvGM—Encoding acetolactate synthase Ⅱ; ilvIH—Encoding acetolactate synthase Ⅲ; asd—Encoding aspartate semialdehyde dehydrogenase; pntAB—Encoding subunits of NAD(P) transhydrogenase; gapN/gapA—Encoding glyceraldehyde-3-phosphate dehydrogenase; lrp—Encoding transcriptional regulatory factor Lrp; G3P—Glyceraldehyde 3-phosphate; 3-PGA—3-Phosphoglyceric acid; ASD—Aspartate semialdehyde dehydrogenase)
| 酶/前导肽/阻遏蛋白 | 基因 | 菌株 | 策略 | 效果 | 参考文献 |
|---|---|---|---|---|---|
| AK | lysC | E. coli | T344M | L-赖氨酸产量6.3 g/L | [ |
| T352I | L-苏氨酸产量14.4 g/L,提升30.9% | [ | |||
| C. glutamicum | T311I | L-苏氨酸产量0.27 g/L | [ | ||
| S301F | L-苏氨酸产量14.17 g/L,提升4.2% | [ | |||
| thrA | E. coli | S345F | L-苏氨酸产量82.4 mmol/L | [ | |
| G433R | L-苏氨酸产量36.61 mg/L | [ | |||
| AHAS | ilvH | E. coli | G14D, S17F | L-异亮氨酸产量0.322 g/L | [ |
| ilvN | E. coli | H47L | L-异亮氨酸产量5.1 g/L,提升59.4% | [ | |
| ilvB | E. coli | K30Q, N156D, V233I | L-异亮氨酸产量5.1 g/L,提升59.4% | [ | |
| C. glutamicum | P176S, D426E, L575W | L-异亮氨酸产量4.17 g/L,提升61.0% | [ | ||
| I47Y | L-异亮氨酸产量22.7 g/L,提升10.2% | [ | |||
| TD | ilvA | E. coli | S97F | L-异亮氨酸产量0.322 g/L | [ |
| C. glutamicum | G96D | L-异亮氨酸产量12 g/L | [ | ||
| ThrL | thrL | E. coli | 敲除thrL基因 | L-苏氨酸产量1.63 g/L | [ |
| MetJ | metJ | E. coli | 敲除metJ基因 | L-甲硫氨酸产量提升11% | [ |
表2 解除反馈调节机制的策略
Table 2 Strategies for relieving feedback regulation mechanisms
| 酶/前导肽/阻遏蛋白 | 基因 | 菌株 | 策略 | 效果 | 参考文献 |
|---|---|---|---|---|---|
| AK | lysC | E. coli | T344M | L-赖氨酸产量6.3 g/L | [ |
| T352I | L-苏氨酸产量14.4 g/L,提升30.9% | [ | |||
| C. glutamicum | T311I | L-苏氨酸产量0.27 g/L | [ | ||
| S301F | L-苏氨酸产量14.17 g/L,提升4.2% | [ | |||
| thrA | E. coli | S345F | L-苏氨酸产量82.4 mmol/L | [ | |
| G433R | L-苏氨酸产量36.61 mg/L | [ | |||
| AHAS | ilvH | E. coli | G14D, S17F | L-异亮氨酸产量0.322 g/L | [ |
| ilvN | E. coli | H47L | L-异亮氨酸产量5.1 g/L,提升59.4% | [ | |
| ilvB | E. coli | K30Q, N156D, V233I | L-异亮氨酸产量5.1 g/L,提升59.4% | [ | |
| C. glutamicum | P176S, D426E, L575W | L-异亮氨酸产量4.17 g/L,提升61.0% | [ | ||
| I47Y | L-异亮氨酸产量22.7 g/L,提升10.2% | [ | |||
| TD | ilvA | E. coli | S97F | L-异亮氨酸产量0.322 g/L | [ |
| C. glutamicum | G96D | L-异亮氨酸产量12 g/L | [ | ||
| ThrL | thrL | E. coli | 敲除thrL基因 | L-苏氨酸产量1.63 g/L | [ |
| MetJ | metJ | E. coli | 敲除metJ基因 | L-甲硫氨酸产量提升11% | [ |
| 产品 | 竞争/降解路径 | 菌株 | 策略 | 参考文献 |
|---|---|---|---|---|
| L-苏氨酸 | L-赖氨酸、L-甲硫氨酸 | E. coli | 敲除lysA和metA基因 | [ |
| L-赖氨酸、L-甲硫氨酸、L-异亮氨酸、L-甘氨酸 | E. coli | 敲除lysA、metA和tdh基因,引入ilvAS97F基因 | [ | |
| L-赖氨酸、L-异亮氨酸 | C. glutamicum | 异源表达Streptococcus pneumoniae来源的dapA基因和E. coli来源的ilvA基因 | [ | |
| L-赖氨酸、L-异亮氨酸、L-丙氨酸 | C. glutamicum | 引入ilvAG96D和dapAK68H基因,敲除alaT和avtA基因 | [ |
表3 阻断竞争路径和弱化降解路径的策略
Table 3 Strategies for blocking competing pathways and attenuating degradation pathways
| 产品 | 竞争/降解路径 | 菌株 | 策略 | 参考文献 |
|---|---|---|---|---|
| L-苏氨酸 | L-赖氨酸、L-甲硫氨酸 | E. coli | 敲除lysA和metA基因 | [ |
| L-赖氨酸、L-甲硫氨酸、L-异亮氨酸、L-甘氨酸 | E. coli | 敲除lysA、metA和tdh基因,引入ilvAS97F基因 | [ | |
| L-赖氨酸、L-异亮氨酸 | C. glutamicum | 异源表达Streptococcus pneumoniae来源的dapA基因和E. coli来源的ilvA基因 | [ | |
| L-赖氨酸、L-异亮氨酸、L-丙氨酸 | C. glutamicum | 引入ilvAG96D和dapAK68H基因,敲除alaT和avtA基因 | [ |
| 产品 | 菌株 | 策略 | 参考文献 |
|---|---|---|---|
| L-苏氨酸 | E. coli | 过表达thrB、thrC和编码抗反馈抑制突变体的thrAS345F基因 | [ |
| P cysJ 启动子动态调控asd和thrABC基因的表达量 | [ | ||
| C. glutamicum | 过表达asd、thrB以及编码抗反馈抑制突变体的lysCT311I和homG378E基因 | [ | |
| L-甲硫氨酸 | E. coli | 过表达metC和编码抗反馈抑制突变体的metAR27C,I296S,P298L基因 | [ |
| C. glutamicum | 过表达lysC、asd、hom、metH、aecD和metYX基因 | [ | |
| L-赖氨酸 | E. coli | 过表达lysC和lysA基因 | [ |
| C. glutamicum | 过表达lysC、asd、dapA、dapB、ddh和lysA基因 | [ | |
| L-异亮氨酸 | E. coli | 过表达编码抗反馈抑制突变体的ilvAP363L、ilvNK30Q,N156D,V233I、metAG189C和metBQ5R, L29H, G69D, F87I, E136G, N148Y, K273G, A346T基因 | [ |
| C. glutamicum | 过表达编码抗反馈抑制突变体的ilvAF383V和ilvBP176S,D426EE,L575W基因 | [ |
表4 重构主合成代谢路径的策略
Table 4 Strategies for reconstructing primary anabolic pathways
| 产品 | 菌株 | 策略 | 参考文献 |
|---|---|---|---|
| L-苏氨酸 | E. coli | 过表达thrB、thrC和编码抗反馈抑制突变体的thrAS345F基因 | [ |
| P cysJ 启动子动态调控asd和thrABC基因的表达量 | [ | ||
| C. glutamicum | 过表达asd、thrB以及编码抗反馈抑制突变体的lysCT311I和homG378E基因 | [ | |
| L-甲硫氨酸 | E. coli | 过表达metC和编码抗反馈抑制突变体的metAR27C,I296S,P298L基因 | [ |
| C. glutamicum | 过表达lysC、asd、hom、metH、aecD和metYX基因 | [ | |
| L-赖氨酸 | E. coli | 过表达lysC和lysA基因 | [ |
| C. glutamicum | 过表达lysC、asd、dapA、dapB、ddh和lysA基因 | [ | |
| L-异亮氨酸 | E. coli | 过表达编码抗反馈抑制突变体的ilvAP363L、ilvNK30Q,N156D,V233I、metAG189C和metBQ5R, L29H, G69D, F87I, E136G, N148Y, K273G, A346T基因 | [ |
| C. glutamicum | 过表达编码抗反馈抑制突变体的ilvAF383V和ilvBP176S,D426EE,L575W基因 | [ |
| 前体 | 产品 | 菌株 | 策略 | 参考文献 |
|---|---|---|---|---|
| 草酰乙酸 | L-赖氨酸 | C. glutamicum | 过表达ppc和pyc基因,敲除pck基因 | [ |
| L-苏氨酸 | E. coli | 过表达ppc基因,异源表达Rhizobium etli来源的pyc基因,利用P flic 启动子动态调控gltA基因 | [ | |
| 过表达ppc和acs基因,敲除iclR基因 | [ | |||
| C. glutamicum | 过表达ppc、pyc和aspB基因,异源表达E. coli来源的aspA基因 | [ | ||
| L-甲硫氨酸 | E. coli | 敲除pykA和pykF基因 | [ | |
| 过表达sucA基因,敲除sucD基因 | [ | |||
| C. glutamicum | 过表达pycP458S基因,敲除pyk2基因 | [ | ||
| L-异亮氨酸 | E. coli | 敲除ptsG、pykF和iclR基因,过表达ppc基因,异源表达Methanococcus jannaschii来源的cimAI47V, E114V, H126Q, T204A, L238S, V373STOP基因 | [ | |
| 敲除aceA和sucCD基因,过表达metA和metB基因 | [ | |||
| C. glutamicum | 敲除alaT和alr基因 | [ |
表5 强化关键代谢节点前体供应的策略
Table 5 Strategies for enhancing precursor supply at key metabolic nodes
| 前体 | 产品 | 菌株 | 策略 | 参考文献 |
|---|---|---|---|---|
| 草酰乙酸 | L-赖氨酸 | C. glutamicum | 过表达ppc和pyc基因,敲除pck基因 | [ |
| L-苏氨酸 | E. coli | 过表达ppc基因,异源表达Rhizobium etli来源的pyc基因,利用P flic 启动子动态调控gltA基因 | [ | |
| 过表达ppc和acs基因,敲除iclR基因 | [ | |||
| C. glutamicum | 过表达ppc、pyc和aspB基因,异源表达E. coli来源的aspA基因 | [ | ||
| L-甲硫氨酸 | E. coli | 敲除pykA和pykF基因 | [ | |
| 过表达sucA基因,敲除sucD基因 | [ | |||
| C. glutamicum | 过表达pycP458S基因,敲除pyk2基因 | [ | ||
| L-异亮氨酸 | E. coli | 敲除ptsG、pykF和iclR基因,过表达ppc基因,异源表达Methanococcus jannaschii来源的cimAI47V, E114V, H126Q, T204A, L238S, V373STOP基因 | [ | |
| 敲除aceA和sucCD基因,过表达metA和metB基因 | [ | |||
| C. glutamicum | 敲除alaT和alr基因 | [ |
| 辅因子 | 菌株 | 产品 | 策略 | 参考文献 |
|---|---|---|---|---|
| NADPH | E. coli | L-苏氨酸 | 过表达pntAB基因,异源表达Tistrella mobilis来源的asd基因 | [ |
| L-异亮氨酸 | 过表达aspA、bcd和pntAB基因 | [ | ||
| C. glutamicum | L-赖氨酸 | 过表达pntAB和zwf基因,P lysE 启动子动态调控gapN基因 | [ | |
| L-甲硫氨酸 | 过表达zwf fbr和gnd fbr基因,异源表达Clostridium acetobutylicum来源的gapC基因 | [ | ||
| ATP | E. coli | L-苏氨酸 | 异源表达Tistrella mobilis来源的asd基因和Pseudomonas aeruginosa来源的adh基因,敲除amn基因,异源表达Mannheimia succiniciproducens来源的pckA基因和Vitreoscilla来源的血红蛋白突变体编码基因vgbH36R, Q66R | [ |
| C. glutamicum | L-赖氨酸 | 过表达pgk和pyk基因,敲除amn基因 | [ | |
| 异源表达M. maripaludis来源的glk/pfk基因,过表达ndh基因,敲除sigH基因 | [ |
表6 辅因子工程
Table 6 Cofactor engineering
| 辅因子 | 菌株 | 产品 | 策略 | 参考文献 |
|---|---|---|---|---|
| NADPH | E. coli | L-苏氨酸 | 过表达pntAB基因,异源表达Tistrella mobilis来源的asd基因 | [ |
| L-异亮氨酸 | 过表达aspA、bcd和pntAB基因 | [ | ||
| C. glutamicum | L-赖氨酸 | 过表达pntAB和zwf基因,P lysE 启动子动态调控gapN基因 | [ | |
| L-甲硫氨酸 | 过表达zwf fbr和gnd fbr基因,异源表达Clostridium acetobutylicum来源的gapC基因 | [ | ||
| ATP | E. coli | L-苏氨酸 | 异源表达Tistrella mobilis来源的asd基因和Pseudomonas aeruginosa来源的adh基因,敲除amn基因,异源表达Mannheimia succiniciproducens来源的pckA基因和Vitreoscilla来源的血红蛋白突变体编码基因vgbH36R, Q66R | [ |
| C. glutamicum | L-赖氨酸 | 过表达pgk和pyk基因,敲除amn基因 | [ | |
| 异源表达M. maripaludis来源的glk/pfk基因,过表达ndh基因,敲除sigH基因 | [ |
| 产品 | 菌株 | 策略 | 参考文献 |
|---|---|---|---|
| L-赖氨酸 | C. glutamicum | 敲除pgi基因,过表达ptsG基因 | [ |
| 过表达lysE基因,敲除lysP基因,异源表达E. coli来源的ybjE基因 | [ | ||
| L-苏氨酸 | E. coli | 敲除crr、sstT和tdcC基因,过表达glk、rhtC和rhtA基因,异源表达Zymomonas mobilis来源的glf基因 | [ |
| C. glutamicum | 过表达thrE基因,异源表达E. coli来源的rhtC基因 | [ | |
| L-甲硫氨酸 | E. coli | 过表达yjeH基因,敲除metD和rhtA基因 | [ |
| C. glutamicum | 过表达brnFE基因,敲除metD基因 | [ | |
| L-异亮氨酸 | E. coli | 过表达brnFE基因,敲除brnQ基因 | [ |
| 过表达glk、ygaZH基因,敲除ptsG和rhtC基因 | [ | ||
| 过表达ygaZH基因,敲除rhtC和livJ基因 | [ |
表7 转运体工程
Table 7 Transporter engineering
| 产品 | 菌株 | 策略 | 参考文献 |
|---|---|---|---|
| L-赖氨酸 | C. glutamicum | 敲除pgi基因,过表达ptsG基因 | [ |
| 过表达lysE基因,敲除lysP基因,异源表达E. coli来源的ybjE基因 | [ | ||
| L-苏氨酸 | E. coli | 敲除crr、sstT和tdcC基因,过表达glk、rhtC和rhtA基因,异源表达Zymomonas mobilis来源的glf基因 | [ |
| C. glutamicum | 过表达thrE基因,异源表达E. coli来源的rhtC基因 | [ | |
| L-甲硫氨酸 | E. coli | 过表达yjeH基因,敲除metD和rhtA基因 | [ |
| C. glutamicum | 过表达brnFE基因,敲除metD基因 | [ | |
| L-异亮氨酸 | E. coli | 过表达brnFE基因,敲除brnQ基因 | [ |
| 过表达glk、ygaZH基因,敲除ptsG和rhtC基因 | [ | ||
| 过表达ygaZH基因,敲除rhtC和livJ基因 | [ |
| [31] | ZHANG Q Q, WANG Y H, WANG X L, et al. Metabolic engineering of Escherichia coli for efficient L-isoleucine production based on the citramalate pathway[J]. Journal of Agricultural and Food Chemistry, 2025, 73(19): 11900-11911. |
| [32] | GUILLOUET S, RODAL A, AN G H, et al. Metabolic redirection of carbon flow toward isoleucine by expressing a catabolic threonine dehydratase in a threonine-overproducing Corynebacterium glutamicum [J]. Applied Microbiology and Biotechnology, 2001, 57(5): 667-673. |
| [33] | YU S Z, ZHENG B, CHEN Z Y, et al. Metabolic engineering of Corynebacterium glutamicum for producing branched chain amino acids[J]. Microbial Cell Factories, 2021, 20(1): 230. |
| [34] | GUO Y F, HAN M, XU J Z, et al. Analysis of acetohydroxyacid synthase variants from branched-chain amino acids-producing strains and their effects on the synthesis of branched-chain amino acids in Corynebacterium glutamicum [J]. Protein Expression and Purification, 2015, 109: 106-112. |
| [35] | YIN L H, HU X Q, XU D Q, et al. Co-expression of feedback-resistant threonine dehydratase and acetohydroxy acid synthase increase L-isoleucine production in Corynebacterium glutamicum [J]. Metabolic Engineering, 2012, 14(5): 542-550. |
| [36] | SHI J, FENG Z Z, SONG Q, et al. Structural and functional insights into transcription activation of the essential LysR-type transcriptional regulators[J]. Protein Science, 2024, 33(6): e5012. |
| [37] | DE GRAAF A A, EGGELING L, SAHM H. Metabolic engineering for L-lysine production by Corynebacterium glutamicum [J]. Advances in Biochemical Engineering/Biotechnology, 2001, 73: 9-29. |
| [38] | GRUZDEV N, HACHAM Y, HAVIV H, et al. Conversion of methionine biosynthesis in Escherichia coli from trans- to direct-sulfurylation enhances extracellular methionine levels[J]. Microbial Cell Factories, 2023, 22(1): 151. |
| [39] | ZHOU Z, ZHANG X Y, WU J, et al. Targeting cofactors regeneration in methylation and hydroxylation for high level production of Ferulic acid[J]. Metabolic Engineering, 2022, 73: 247-255. |
| [40] | RHEE K Y, PAREKH B S, HATFIELD G W. Leucine-responsive regulatory protein-DNA interactions in the leader region of the ilvGMEDA operon of Escherichia coli [J]. Journal of Biological Chemistry, 1996, 271(43): 26499-26507. |
| [41] | JAFRI S, CHEN S L, CALVO J M. ilvIH operon expression in Escherichia coli requires Lrp binding to two distinct regions of DNA[J]. Journal of Bacteriology, 2002, 184(19): 5293-5300. |
| [42] | MORBACH S, JUNGER C, SAHM H, et al. Attenuation control of ilvBNC in Corynebacterium glutamicum: evidence of leader peptide formation without the presence of a ribosome binding site[J]. Journal of Bioscience and Bioengineering, 2000, 90(5): 501-507. |
| [1] | FANG Q C, ZHANG X Y, DAI G C, et al. Low-opportunity-cost feed can reduce land-use-related environmental impacts by about one-third in China[J]. Nature Food, 2023, 4(8): 677-685. |
| [2] | MA Q, ZHANG Q W, XU Q Y, et al. Systems metabolic engineering strategies for the production of amino acids[J]. Synthetic and Systems Biotechnology, 2017, 2(2): 87-96. |
| [3] | 刘佳, 盛琦, 刘开放, 等. 微生物制造饲用氨基酸助力豆粕减量替代[J]. 中国科学院院刊, 2025, 40(1): 25-35. |
| LIU J, SHENG Q, LIU K F, et al. Microbial production of feed amino acids promotes reduction and replacement of soybean meal[J]. Bulletin of Chinese Academy of Sciences, 2025, 40(1): 25-35. | |
| [4] | KIM S Y. 9 What would be the next feed amino acid based on a microbial point of view?[J]. Journal of Animal Science, 2021, 99(S1): 12-13. |
| [5] | LIAO J L, ZHANG P G, YIN J D, et al. New insights into the effects of dietary amino acid composition on meat quality in pigs: a review[J]. Meat Science, 2025, 221: 109721. |
| [6] | OLUWABIYI C T, SONG Z G. Branched-chain amino acids supplementation in low-protein broiler diets: a review[J]. Animal Feed Science and Technology, 2024, 318: 116114. |
| [7] | LI M H, LI H, ZHANG X, et al. Metabolic engineering of Corynebacterium glutamicum: unlocking its potential as a key cell factory platform for organic acid production[J]. Biotechnology Advances, 2024, 77: 108475. |
| [8] | LIU J, XU J Z, RAO Z M, et al. Industrial production of L-lysine in Corynebacterium glutamicum: progress and prospects[J]. Microbiological Research, 2022, 262: 127101. |
| [9] | LI C Y, ZHANG R H, WANG J, et al. Protein engineering for improving and diversifying natural product biosynthesis[J]. Trends in Biotechnology, 2020, 38(7): 729-744. |
| [10] | DONG X Y, ZHAO Y, HU J Y, et al. Attenuating L-lysine production by deletion of ddh and lysE and their effect on L-threonine and L-isoleucine production in Corynebacterium glutamicum [J]. Enzyme and Microbial Technology, 2016, 93-94: 70-78. |
| [11] | BOMMAREDDY R R, CHEN Z, RAPPERT S, et al. A de novo NADPH generation pathway for improving lysine production of Corynebacterium glutamicum by rational design of the coenzyme specificity of glyceraldehyde 3-phosphate dehydrogenase[J]. Metabolic Engineering, 2014, 25: 30-37. |
| [43] | BOOB A G, CHEN J Y, ZHAO H M. Enabling pathway design by multiplex experimentation and machine learning[J]. Metabolic Engineering, 2024, 81: 70-87. |
| [44] | DING D Q, ZHU Y R, BAI D Y, et al. Monitoring and dynamically controlling glucose uptake rate and central metabolism[J]. Nature Chemical Engineering, 2025, 2(1): 50-62. |
| [45] | SHENG Q, YI L X, ZHONG B, et al. Shikimic acid biosynthesis in microorganisms: current status and future direction[J]. Biotechnology Advances, 2023, 62: 108073. |
| [46] | XU J Z, HAN M, REN X D, et al. Modification of aspartokinase Ⅲ and dihydrodipicolinate synthetase increases the production of L-lysine in Escherichia coli [J]. Biochemical Engineering Journal, 2016, 114: 79-86. |
| [47] | LIU J H, LIU J, LI J J, et al. Reconstruction the feedback regulation of amino acid metabolism to develop a non-auxotrophic L-threonine producing Corynebacterium glutamicum [J]. Bioresources and Bioprocessing, 2024, 11(1): 43. |
| [48] | LEE J H, LEE D E, LEE B U, et al. Global analyses of transcriptomes and proteomes of a parent strain and an L-threonine-overproducing mutant strain[J]. Journal of Bacteriology, 2003, 185(18): 5442-5451. |
| [49] | ZHAO Z Q, YOU J J, SHI X P, et al. Engineering Escherichia coli for L-threonine hyperproduction based on multidimensional optimization strategies[J]. Journal of Agricultural and Food Chemistry, 2024, 72(41): 22682-22691. |
| [50] | PARK J H, OH J E, LEE K H, et al. Rational design of Escherichia coli for L-isoleucine production[J]. ACS Synthetic Biology, 2012, 1(11): 532-540. |
| [51] | ZHANG Y C, LIU Y D, ZHANG S Y, et al. Metabolic engineering of Corynebacterium glutamicum WM001 to improve L-isoleucine production[J]. Biotechnology and Applied Biochemistry, 2021, 68(3): 568-584. |
| [52] | LIU Y D, LI Y Y, WANG X Y. Acetohydroxyacid synthases: evolution, structure, and function[J]. Applied Microbiology and Biotechnology, 2016, 100(20): 8633-8649. |
| [53] | DUAN M, CHEN S, LIU X L, et al. The application of Corynebacterium glutamicum in L-threonine biosynthesis[J]. Fermentation, 2023, 9(9): 822. |
| [54] | JIN X, WANG S M, GAO Y P, et al. Combinatorial metabolic engineering of Escherichia coli to efficiently produce L-threonine from untreated cane molasses[J]. Bioresource Technology, 2025, 419: 132058. |
| [12] | CHEN Y Y, HUANG L G, YU T, et al. Balancing the AspC and AspA pathways of Escherichia coli by systematic metabolic engineering strategy for high-efficient L-homoserine production[J]. ACS Synthetic Biology, 2024, 13(8): 2457-2469. |
| [13] | BASSALO M C, GARST A D, CHOUDHURY A, et al. Deep scanning lysine metabolism in Escherichia coli [J]. Molecular Systems Biology, 2018, 14(11): e8371. |
| [14] | OSIRE T, YANG T W, XU M J, et al. Integrated gene engineering synergistically improved substrate-product transport, cofactor generation and gene translation for cadaverine biosynthesis in E. coli [J]. International Journal of Biological Macromolecules, 2021, 169: 8-17. |
| [15] | XIAO J, WANG D T, WANG L, et al. Increasing L-lysine production in Corynebacterium glutamicum by engineering amino acid transporters[J]. Amino Acids, 2020, 52(10): 1363-1374. |
| [16] | LIU J, OU Y, XU J Z, et al. L-lysine production by systems metabolic engineering of an NADPH auto-regulated Corynebacterium glutamicum [J]. Bioresource Technology, 2023, 387: 129701. |
| [17] | MALLA S, VAN DER HELM E, DARBANI B, et al. A novel efficient L-lysine exporter identified by functional metagenomics[J]. Frontiers in Microbiology, 2022, 13: 855736. |
| [18] | LI Z C, LIU Q, SUN J H, et al. Multivariate modular metabolic engineering for enhanced L-methionine biosynthesis in Escherichia coli [J]. Biotechnology for Biofuels and Bioproducts, 2023, 16(1): 101. |
| [19] | LI Y, CONG H, LIU B N, et al. Metabolic engineering of Corynebacterium glutamicum for methionine production by removing feedback inhibition and increasing NADPH level[J]. Antonie Van Leeuwenhoek, 2016, 109(9): 1185-1197. |
| [20] | ZHOU B Z, ZHAO G H, YU J, et al. Multi-step metabolic engineering Corynebacterium glutamicum ATCC13032 to produce L-methionine[J]. Systems Microbiology and Biomanufacturing, 2025, 5(2): 593-610. |
| [21] | ZHAO Z Q, YOU J J, SHI X P, et al. Multi-module engineering to guide the development of an efficient L-threonine-producing cell factory[J]. Bioresource Technology, 2025, 416: 131802. |
| [22] | LEE K H, PARK J H, KIM T Y, et al. Systems metabolic engineering of Escherichia coli for L-threonine production[J]. Molecular Systems Biology, 2007, 3: 149. |
| [23] | VO T M, PARK J Y, KIM D, et al. Use of acetate as substrate for sustainable production of homoserine and threonine by Escherichia coli W3110: a modular metabolic engineering approach[J]. Metabolic Engineering, 2024, 84: 13-22. |
| [55] | YE C, LUO Q L, GUO L, et al. Improving lysine production through construction of an Escherichia coli enzyme-constrained model[J]. Biotechnology and Bioengineering, 2020, 117(11): 3533-3544. |
| [56] | LV Q L, HU M K, TIAN L Z, et al. Enhancing L-glutamine production in Corynebacterium glutamicum by rational metabolic engineering combined with a two-stage pH control strategy[J]. Bioresource Technology, 2021, 341: 125799. |
| [57] | WANG L J, GUO Y Y, LI M Y, et al. Antibiotic-free high-level L-methionine production in engineered Escherichia coli [J]. Journal of Agricultural and Food Chemistry, 2024, 72(46): 25791-25800. |
| [58] | JIANG S, WANG R R, WANG D H, et al. Metabolic reprogramming and biosensor-assisted mutagenesis screening for high-level production of L-arginine in Escherichia coli [J]. Metabolic Engineering, 2023, 76: 146-157. |
| [59] | YE J W, HU D K, CHE X M, et al. Engineering of Halomonas bluephagenesis for low cost production of poly (3-hydroxybutyrate-co-4-hydroxybutyrate) from glucose[J]. Metabolic Engineering, 2018, 47: 143-152. |
| [60] | YE J W, HU D K, YIN J, et al. Stimulus response-based fine-tuning of polyhydroxyalkanoate pathway in Halomonas [J]. Metabolic Engineering, 2020, 57: 85-95. |
| [61] | LI T, YE J W, SHEN R, et al. Semirational approach for ultrahigh poly(3-hydroxybutyrate) accumulation in Escherichia coli by combining one-step library construction and high-throughput screening[J]. ACS Synthetic Biology, 2016, 5(11): 1308-1317. |
| [62] | SHEN R, YIN J, YE J W, et al. Promoter engineering for enhanced P(3HB-co-4HB) production by Halomonas bluephagenesis [J]. ACS Synthetic Biology, 2018, 7(8): 1897-1906. |
| [63] | DU F, LI Z J, LI X, et al. Optimizing multicopy chromosomal integration for stable high-performing strains[J]. Nature Chemical Biology, 2024, 20(12): 1670-1679. |
| [64] | BAEK J M, MAZUMDAR S, LEE S W, et al. Butyrate production in engineered Escherichia coli with synthetic scaffolds[J]. Biotechnology and Bioengineering, 2013, 110(10): 2790-2794. |
| [65] | DUEBER J E, WU G C, MALMIRCHEGINI G R, et al. Synthetic protein scaffolds provide modular control over metabolic flux[J]. Nature Biotechnology, 2009, 27(8): 753-759. |
| [66] | ZHOU H, VONK B, ROUBOS J A, et al. Algorithmic co-optimization of genetic constructs and growth conditions: application to 6-ACA, a potential nylon-6 precursor[J]. Nucleic Acids Research, 2015, 43(21): 10560-10570. |
| [67] | VENAYAK N, ANESIADIS N, CLUETT W R, et al. Engineering metabolism through dynamic control[J]. Current Opinion in Biotechnology, 2015, 34: 142-152. |
| [68] | CRESS B F, TRANTAS E A, VERVERIDIS F, et al. Sensitive cells: enabling tools for static and dynamic control of microbial metabolic pathways[J]. Current Opinion in Biotechnology, 2015, 36: 205-214. |
| [69] | IMMETHUN C M, DELORENZO D M, FOCHT C M, et al. Physical, chemical, and metabolic state sensors expand the synthetic biology toolbox for Synechocystis sp. PCC 6803[J]. Biotechnology and Bioengineering, 2017, 114(7): 1561-1569. |
| [70] | SHEN X L, WANG J, LI C Y, et al. Dynamic gene expression engineering as a tool in pathway engineering[J]. Current Opinion in Biotechnology, 2019, 59: 122-129. |
| [71] | LALWANI M A, ZHAO E M, AVALOS J L. Current and future modalities of dynamic control in metabolic engineering[J]. Current Opinion in Biotechnology, 2018, 52: 56-65. |
| [72] | PINTO D, VECCHIONE S, WU H, et al. Engineering orthogonal synthetic timer circuits based on extracytoplasmic function σ factors[J]. Nucleic Acids Research, 2018, 46(14): 7450-7464. |
| [73] | MEYER A J, SEGALL-SHAPIRO T H, GLASSEY E, et al. Escherichia coli “Marionette” strains with 12 highly optimized small-molecule sensors[J]. Nature Chemical Biology, 2019, 15(2): 196-204. |
| [74] | 叶健文, 陈江楠, 张旭, 等. 动态调控: 一种高效的细胞工厂工程化代谢改造策略[J]. 生物技术通报, 2020, 36(6): 1-12. |
| YE J W, CHEN J N, ZHANG X, et al. Dynamic control: an efficient strategy for metabolically engineering microbial cell factories[J]. Biotechnology Bulletin, 2020, 36(6): 1-12. | |
| [75] | BORKOWSKI O, BRICIO C, MURGIANO M, et al. Cell-free prediction of protein expression costs for growing cells[J]. Nature Communications, 2018, 9: 1457. |
| [76] | GUPTA A, REIZMAN I M B, REISCH C R, et al. Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit[J]. Nature Biotechnology, 2017, 35(3): 273-279. |
| [77] | DING X H, YANG W J, DU X B, et al. High-level and-yield production of L-leucine in engineered Escherichia coli by multistep metabolic engineering[J]. Metabolic Engineering, 2023, 78: 128-136. |
| [78] | YANG L Y, MU X Q, NIE Y, et al. Improving the production of NAD+ via multi-strategy metabolic engineering in Escherichia coli [J]. Metabolic Engineering, 2021, 64: 122-133. |
| [79] | BOUZON M, DÖRING V, DUBOIS I, et al. Change in cofactor specificity of oxidoreductases by adaptive evolution of an Escherichia coli NADPH-auxotrophic strain[J]. mBio, 2021, 12(4): e0032921. |
| [80] | XU J Z, RUAN H Z, YU H B, et al. Metabolic engineering of carbohydrate metabolism systems in Corynebacterium glutamicum for improving the efficiency of L-lysine production from mixed sugar[J]. Microbial Cell Factories, 2020, 19(1): 39. |
| [81] | WANG K, SONG X T, CUI B Y, et al. Metabolic engineering of Escherichia coli for efficient production of ectoine[J]. Journal of Agricultural and Food Chemistry, 2025, 73(1): 646-654. |
| [82] | LIU B N, SUN X Y, LIU Y, et al. Increased NADPH supply enhances glycolysis metabolic flux and L-methionine production in Corynebacterium glutamicum [J]. Foods, 2022, 11(7): 1031. |
| [83] | YU F, ZHAO X R, ZHOU J W, et al. Biosynthesis of high-active hemoproteins by the efficient heme-supply Pichia pastoris chassis[J]. Advanced Science, 2023, 10(30): 2302826. |
| [84] | WANG Y S, BAI Y L, ZENG Q, et al. Recent advances in the metabolic engineering and physiological opportunities for microbial synthesis of L-aspartic acid family amino acids: a review[J]. International Journal of Biological Macromolecules, 2023, 253: 126916. |
| [85] | 赵阔, 程金宇, 郭亮, 等. 谷氨酸棒杆菌代谢工程高效生产L-缬氨酸[J]. 生物工程学报, 2023, 39(8): 3253-3272. |
| ZHAO K, CHENG J Y, GUO L, et al. Highly efficient production of L-valine by multiplex metabolic engineering of Corynebacterium glutamicum [J]. Chinese Journal of Biotechnology, 2023, 39(8): 3253-3272. | |
| [86] | XU S Y, ZHOU L, XU Y, et al. Recent advances in structure-based enzyme engineering for functional reconstruction[J]. Biotechnology and Bioengineering, 2023, 120(12): 3427-3445. |
| [87] | 贾男, 臧国伟, 李春, 等. 辅因子在微生物细胞工厂中的代谢调控与应用[J]. 中国生物工程杂志, 2022, 42(7): 79-89. |
| JIA N, ZANG G W, LI C, et al. Metabolic regulations and applications of cofactors in microbial cell factories[J]. China Biotechnology, 2022, 42(7): 79-89. | |
| [88] | 陈修来, 刘佳, 罗秋玲, 等. 微生物辅因子平衡的代谢调控[J]. 生物工程学报, 2017, 33(1): 16-26. |
| CHEN X L, LIU J, LUO Q L, et al. Manipulation of cofactor balance in microorganisms[J]. Chinese Journal of Biotechnology, 2017, 33(1): 16-26. | |
| [89] | 陈雅维. ATP调控策略及其在微生物代谢产物合成中的应用[J]. 生物工程学报, 2020, 36(8): 1515-1527. |
| CHEN Y W. ATP regulation strategy and its application in the synthesis of microbial metabolites[J]. Chinese Journal of Biotechnology, 2020, 36(8): 1515-1527. | |
| [90] | WEUSTHUIS R A, FOLCH P L, POZO-RODRÍGUEZ A, et al. Applying non-canonical redox cofactors in fermentation processes[J]. iScience, 2020, 23(9): 101471. |
| [91] | 张鸿伟, 王鹏超. 微生物辅因子工程研究进展[J]. 中国生物工程杂志, 2023, 43(4): 112-122. |
| ZHANG H W, WANG P C. Research progress of microbial cofactor engineering[J]. China Biotechnology, 2023, 43(4): 112-122. | |
| [92] | GAO C, SONG W, YE C, et al. Bifunctional optogenetic switch powered NADPH availability for improving L-valine production in Escherichia coli [J]. ACS Sustainable Chemistry & Engineering, 2024, 12(41): 15103-15113. |
| [93] | 刘开放. 新型微生物辅因子系统的开发与应用[D]. 无锡: 江南大学, 2024. |
| LIU K F. Development and application of novel cofactor systems in microorganisms[D]. Wuxi: Jiangnan University, 2024. | |
| [94] | BLACK W B, ZHANG L Y, MAK W S, et al. Engineering a nicotinamide mononucleotide redox cofactor system for biocatalysis[J]. Nature Chemical Biology, 2020, 16(1): 87-94. |
| [95] | 于袁欢, 周阳, 王欣怡, 等. 光遗传学照进生物医学研究进展[J]. 合成生物学, 2023, 4(1): 102-140. |
| YU Y H, ZHOU Y, WANG X Y, et al. Advances in optogenetics for biomedical research[J]. Synthetic Biology Journal, 2023, 4(1): 102-140. | |
| [96] | CHEN R B, YANG S, ZHANG L, et al. Advanced strategies for production of natural products in yeast[J]. iScience, 2020, 23(3): 100879. |
| [97] | CHEN R B, GAO J Q, YU W, et al. Engineering cofactor supply and recycling to drive phenolic acid biosynthesis in yeast[J]. Nature Chemical Biology, 2022, 18(5): 520-529. |
| [98] | ZHANG Y F, ZHANG G Y, ZHANG H F, et al. Efficient fermentative production of β-alanine from glucose through multidimensional engineering of Escherichia coli [J]. Journal of Agricultural and Food Chemistry, 2024, 72(25): 14274-14283. |
| [99] | LINDNER S N, PETROV D P, HAGMANN C T, et al. Phosphotransferase system-mediated glucose uptake is repressed in phosphoglucoisomerase-deficient Corynebacterium glutamicum strains[J]. Applied and Environmental Microbiology, 2013, 79(8): 2588-2595. |
| [100] | MOLINA-VÁZQUEZ E R, CASPETA L, GOSSET G, et al. Tailoring Escherichia coli BL21 (DE3) for preferential xylose utilization via metabolic and regulatory engineering[J]. Applied Microbiology and Biotechnology, 2025, 109(1): 54. |
| [101] | ZHU X N, FAN F Y, QIU H N, et al. New xylose transporters support the simultaneous consumption of glucose and xylose in Escherichia coli [J]. mLife, 2022, 1(2): 156-170. |
| [102] | LI J, QIU Z T, ZHAO G R. Modular engineering of E. coli coculture for efficient production of resveratrol from glucose and arabinose mixture[J]. Synthetic and Systems Biotechnology, 2022, 7(2): 718-729. |
| [103] | HALLE L, HÖPPNER D, DOSER M, et al. From molasses to purified α-ketoglutarate with engineered Corynebacterium glutamicum [J]. Bioresource Technology, 2025, 416: 131803. |
| [104] | MOON M W, PARK S Y, CHOI S K, et al. The phosphotransferase system of Corynebacterium glutamicum: features of sugar transport and carbon regulation[J]. Journal of Molecular Microbiology and Biotechnology, 2007, 12(1-2): 43-50. |
| [105] | 刘冬冬. 强化葡萄糖代谢途径提高L-赖氨酸发酵水平的研究[D]. 无锡: 江南大学, 2017. |
| LIU D D. Increasing the fermentation level of L-lysine via enhancing glucose metabolism pathways[D]. Wuxi: Jiangnan University, 2017. | |
| [106] | PARCHE S, BURKOVSKI A, SPRENGER G A, et al. Corynebacterium glutamicum: a dissection of the PTS[J]. Journal of Molecular Microbiology and Biotechnology, 2001, 3(3): 423-428. |
| [107] | WANG L, LI N, YU S Q, et al. Enhancing caffeic acid production in Escherichia coli by engineering the biosynthesis pathway and transporter[J]. Bioresource Technology, 2023, 368: 128320. |
| [108] | 郭亮, 高聪, 柳亚迪, 等. 大肠杆菌生产饲用氨基酸的研究进展[J]. 合成生物学, 2021, 2(6): 964-981. |
| GUO L, GAO C, LIU Y D, et al. Advances in bioproduction of feed amino acid by Escherichia coli [J]. Synthetic Biology Journal, 2021, 2(6): 964-981. | |
| [109] | 于勇, 朱欣娜, 毕昌昊, 等. 大肠杆菌细胞工厂的创建技术[J]. 生物工程学报, 2021, 37(5): 1564-1577. |
| YU Y, ZHU X N, BI C H, et al. Construction of Escherichia coli cell factories[J]. Chinese Journal of Biotechnology, 2021, 37(5): 1564-1577. | |
| [110] | 赵静. 几种主要饲用氨基酸的营养研究进展[J]. 中国饲料添加剂, 2016(1): 10-13. |
| ZHAO J. Research progress on nutrition of several main feed amino acids[J]. China Feed Additive, 2016(1): 10-13. | |
| [111] | ISOGAI S, TAKAGI H. Enhancement of lysine biosynthesis confers high-temperature stress tolerance to Escherichia coli cells[J]. Applied Microbiology and Biotechnology, 2021, 105(18): 6899-6908. |
| [112] | LIU J, ZHAO X J, CHENG H J, et al. Comprehensive screening of industrially relevant components at genome scale using a high-quality gene overexpression collection of Corynebacterium glutamicum [J]. Trends in Biotechnology, 2025, 43(1): 220-247. |
| [113] | ZHANG Y W, YANG J J, QIAN F H, et al. Engineering a xylose fermenting yeast for lignocellulosic ethanol production[J]. Nature Chemical Biology, 2025, 21(3): 443-450. |
| [114] | GAO J Q, LI Y X, YU W, et al. Rescuing yeast from cell death enables overproduction of fatty acids from sole methanol[J]. Nature Metabolism, 2022, 4(7): 932-943. |
| [115] | GE C, YU Z, SHENG H K, et al. Redesigning regulatory components of quorum-sensing system for diverse metabolic control[J]. Nature Communications, 2022, 13: 2182. |
| [24] | ZHAO G H, ZHANG D Z, ZHOU B Z, et al. Fine-regulating the carbon flux of L-isoleucine producing Corynebacterium glutamicum WM001 for efficient L-threonine production[J]. ACS Synthetic Biology, 2024, 13(10): 3446-3460. |
| [25] | PU W, FENG J H, CHEN J Z, et al. Engineering of L-threonine and L-proline biosensors by directed evolution of transcriptional regulator SerR and application for high-throughput screening[J]. Bioresources and Bioprocessing, 2025, 12(1): 4. |
| [26] | SONG J, ZHUANG M M, DU C Y, et al. Metabolic engineering of Escherichia coli for self-induced production of L-isoleucine[J]. ACS Synthetic Biology, 2025, 14(1): 179-192. |
| [27] | SHI C R, HUO X J, YOU R, et al. High yield production of L-isoleucine through readjusting the ratio of two direct precursors in Escherichia coli [J]. Bioresource Technology, 2025, 418: 131889. |
| [28] | LU N, WEI M H, YANG X J, et al. Growth-coupled production of L-isoleucine in Escherichia coli via metabolic engineering[J]. Metabolic Engineering, 2024, 86: 181-193. |
| [29] | LI Y J, WEI H B, WANG T, et al. Current status on metabolic engineering for the production of L-aspartate family amino acids and derivatives[J]. Bioresource Technology, 2017, 245(Pt B): 1588-1602. |
| [30] | LIU Z Y, LIU J, ZHANG F, et al. Modifying Corynebacterium glutamicum by metabolic engineering for efficient synthesis of L-lysine[J]. Systems Microbiology and Biomanufacturing, 2025, 5(1): 288-299. |
| [1] | 宋开南, 张礼文, 王超, 田平芳, 李广悦, 潘国辉, 徐玉泉. 小分子生物农药及其生物合成研究进展[J]. 合成生物学, 2025, 6(5): 1203-1223. |
| [2] | 于文文, 吕雪芹, 李兆丰, 刘龙. 植物合成生物学与母乳低聚糖生物制造[J]. 合成生物学, 2025, 6(5): 992-997. |
| [3] | 颜钊涛, 周鹏飞, 汪阳忠, 张鑫, 谢雯燕, 田晨菲, 王勇. 植物合成生物学:植物细胞大规模培养的新机遇[J]. 合成生物学, 2025, 6(5): 1107-1125. |
| [4] | 孙扬, 陈立超, 石艳云, 王珂, 吕丹丹, 徐秀美, 张立新. 作物光合作用合成生物学的策略与展望[J]. 合成生物学, 2025, 6(5): 1025-1040. |
| [5] | 何杨昱, 杨凯, 王玮琳, 黄茜, 丘梓樱, 宋涛, 何流赏, 姚金鑫, 甘露, 何玉池. 国际基因工程机器大赛中植物合成生物学主题的设计与实践[J]. 合成生物学, 2025, 6(5): 1243-1254. |
| [6] | 张学博, 朱成姝, 陈睿雲, 金庆姿, 刘晓, 熊燕, 陈大明. 农业合成生物学:政策规划与产业发展协同推进[J]. 合成生物学, 2025, 6(5): 1224-1242. |
| [7] | 刘婕, 郜钰, 马永硕, 尚轶. 合成生物学在农业中的进展及挑战[J]. 合成生物学, 2025, 6(5): 998-1024. |
| [8] | 郑雷, 郑棋腾, 张天骄, 段鲲, 张瑞福. 构建根际合成微生物菌群促进作物养分高效吸收利用[J]. 合成生物学, 2025, 6(5): 1058-1071. |
| [9] | 李超, 张焕, 杨军, 王二涛. 固氮合成生物学研究进展[J]. 合成生物学, 2025, 6(5): 1041-1057. |
| [10] | 魏家秀, 嵇佩云, 节庆雨, 黄秋燕, 叶浩, 戴俊彪. 植物人工染色体的构建与应用[J]. 合成生物学, 2025, 6(5): 1093-1106. |
| [11] | 方馨仪, 孙丽超, 霍毅欣, 王颖, 岳海涛. 微生物合成高级醇的发展趋势与挑战[J]. 合成生物学, 2025, 6(4): 873-898. |
| [12] | 朱欣悦, 陈恬恬, 邵恒煊, 唐曼玉, 华威, 程艳玲. 益生菌辅助防治恶性肿瘤的研究进展[J]. 合成生物学, 2025, 6(4): 899-919. |
| [13] | 吴晓燕, 宋琪, 许睿, 丁陈君, 陈方, 郭勍, 张波. 合成生物学研发竞争态势对比分析[J]. 合成生物学, 2025, 6(4): 940-955. |
| [14] | 王宏, 陆孔泳, 郑洋洋, 陈涛, 王智文. 基于转录因子生物传感器的构建与应用进展[J]. 合成生物学, 2025, 6(4): 829-845. |
| [15] | 张建康, 王文君, 郭洪菊, 白北辰, 张亚飞, 袁征, 李彦辉, 李航. 基于机器视觉的高通量微生物克隆挑选工作站研制及应用[J]. 合成生物学, 2025, 6(4): 956-971. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||