Engineering fungal antifreeze proteins through ‘EKylation’ and mechanism analysis
CUI Zhongxin1,2, WANG Yi1, ZHANG Lei1,2, QI Haishan1,2
- 1.School of Synthetic Biology and Biomanufacturing,Tianjin University,Tianjin,China.
2.State Key Laboratory of Synthetic Biology,Frontiers Science Center for Synthetic Biology,Tianjin University,Tianjin,China.
-
Received:2025-03-19Revised:2025-07-18Published:2025-08-01 -
Contact:ZHANG Lei, QI Haishan
“EKylation”策略改造真菌抗冻蛋白及机制解析
崔忠信1,2, 王怡1, 张雷1,2, 齐海山1,2
- 1.天津大学,合成生物与生物制造学院,天津 300072
2.天津大学,合成生物技术全国重点实验室,教育部合成生物学前沿科学中心,天津 300072
-
通讯作者:张雷,齐海山 -
作者简介:崔忠信 (1999—),男,博士在读,研究方向为功能蛋白的智能设计张雷 (1980—),男,博士,教授。研究方向为合成生物学、生物医学工程、生物材料与细胞冷冻保存技术。齐海山 (1986—),男,博士,副教授。研究方向为功能蛋白合成生物技术、细胞工厂智能创建。
CLC Number:
Cite this article
CUI Zhongxin, WANG Yi, ZHANG Lei, QI Haishan. Engineering fungal antifreeze proteins through ‘EKylation’ and mechanism analysis[J]. Synthetic Biology Journal, DOI: 10.12211/2096-8280.2025-019.
崔忠信, 王怡, 张雷, 齐海山. “EKylation”策略改造真菌抗冻蛋白及机制解析[J]. 合成生物学, DOI: 10.12211/2096-8280.2025-019.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2025-019
| 蛋白名称 | 5′端引物序列 | 3′端引物序列 |
|---|---|---|
| 5B5H | TGGGCAGCAGCCATCATCAT | TGGTGGTGGTGGTGCTC |
| 5B5H-EK | GTGGGCAGCAGCCATCATCAT | TGGTGGTGGTGGTGCTCT |
Table 1 Primer sequence
| 蛋白名称 | 5′端引物序列 | 3′端引物序列 |
|---|---|---|
| 5B5H | TGGGCAGCAGCCATCATCAT | TGGTGGTGGTGGTGCTC |
| 5B5H-EK | GTGGGCAGCAGCCATCATCAT | TGGTGGTGGTGGTGCTCT |
| 种类 | 序列 |
|---|---|
| 5B5H | AGPTAVPLGTAGNYAILASAGVSTVPQSVITGAVGLSPAAATFLTGFSLTMSSTGTFSTSTQVTGQLTAADYGTPTPSILTTAIGDMGTAYVNAATRSGPNFLEIYTGALGGKILPPGLYKWTSPVGASADFTIIGTSTDTWIFQIAGTLGLAAGKKIILAGGAQAKNIVWVVAGAVSIEAGAKFEGVILAKTAVTLKTGSSLNGRILSQTAVALQKATVVQK |
| 5B5H-EK | EKEKEKEKEKEKEKEKEKEKEKEKEKEKEKEKEKEKEKEKAGPTAVPLGTAGNYAILASAGVSTVPQSVITGAVGLSPAAATFLTGFSLTMSSTGTFSTSTQVTGQLTAADYGTPTPSILTTAIGDMGTAYVNAATRSGPNFLEIYTGALGGKILPPGLYKWTSPVGASADFTIIGTSTDTWIFQIAGTLGLAAGKKIILAGGAQAKNIVWVVAGAVSIEAGAKFEGVILAKTAVTLKTGSSLNGRILSQTAVALQKATVVQK |
Table 2 the amino acid sequence of 5B5H and 5B5H-EK
| 种类 | 序列 |
|---|---|
| 5B5H | AGPTAVPLGTAGNYAILASAGVSTVPQSVITGAVGLSPAAATFLTGFSLTMSSTGTFSTSTQVTGQLTAADYGTPTPSILTTAIGDMGTAYVNAATRSGPNFLEIYTGALGGKILPPGLYKWTSPVGASADFTIIGTSTDTWIFQIAGTLGLAAGKKIILAGGAQAKNIVWVVAGAVSIEAGAKFEGVILAKTAVTLKTGSSLNGRILSQTAVALQKATVVQK |
| 5B5H-EK | EKEKEKEKEKEKEKEKEKEKEKEKEKEKEKEKEKEKEKEKAGPTAVPLGTAGNYAILASAGVSTVPQSVITGAVGLSPAAATFLTGFSLTMSSTGTFSTSTQVTGQLTAADYGTPTPSILTTAIGDMGTAYVNAATRSGPNFLEIYTGALGGKILPPGLYKWTSPVGASADFTIIGTSTDTWIFQIAGTLGLAAGKKIILAGGAQAKNIVWVVAGAVSIEAGAKFEGVILAKTAVTLKTGSSLNGRILSQTAVALQKATVVQK |
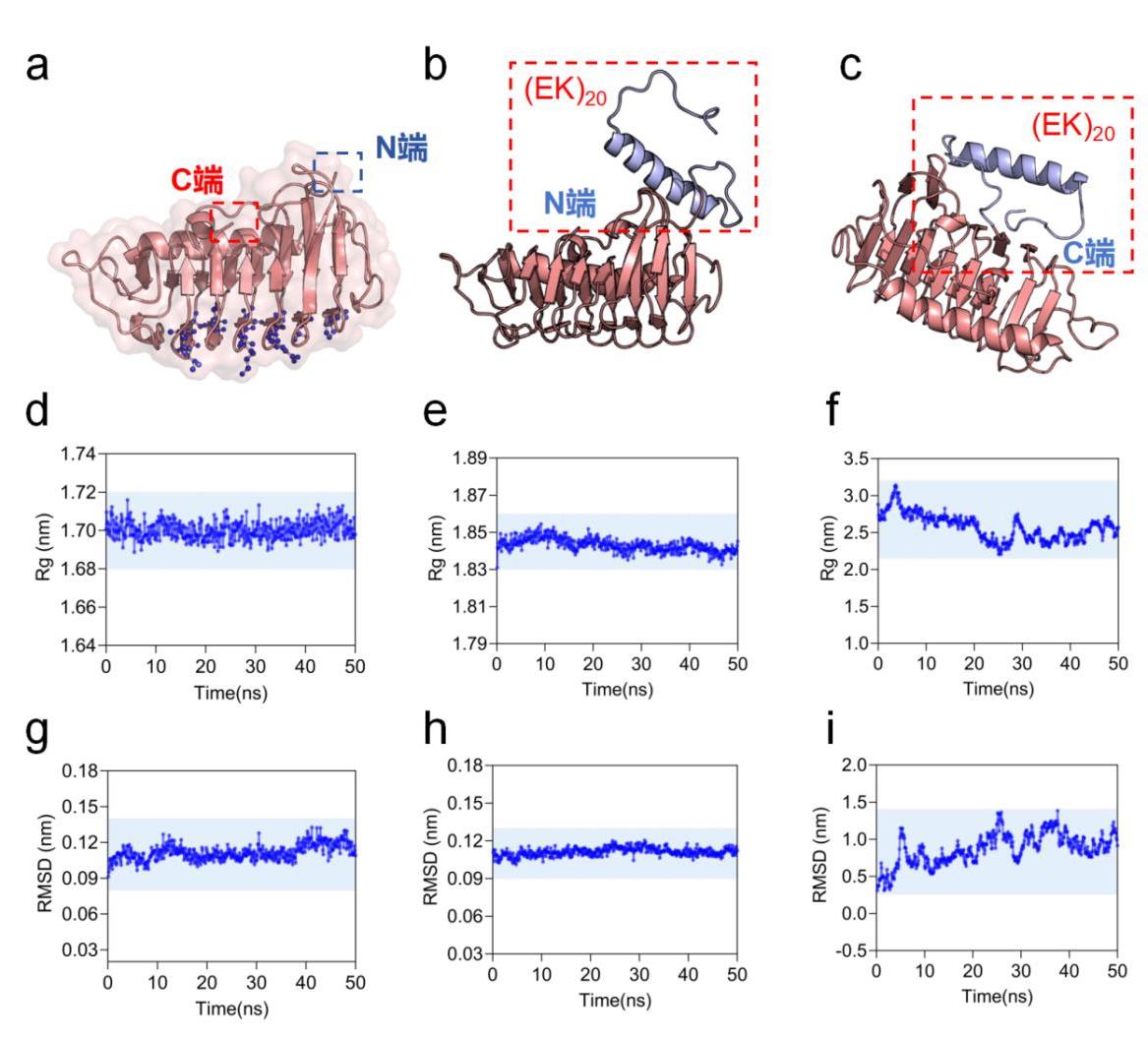
Fig. 1 Conformation resolution of EK, 5B5H and 5B5H-EK(a) 5B5H modification site exploration (b) 5B5H (N-terminal modification) spatial structure (c) 5B5H (C-terminal modification) spatial structure (d) 5B5H aqueous environment Rg simulation results (e) 5B5H (N-terminal modification) aqueous environment Rg simulation results (f) 5B5H (C-terminal modification) aqueous environment Rg simulation results (g) 5B5H aqueous environment RMSD simulation results (h) 5B5H (N-terminal modification) aqueous environment RMSD simulation results (i) 5B5H (C-terminal modification) aqueous environment RMSD simulation results
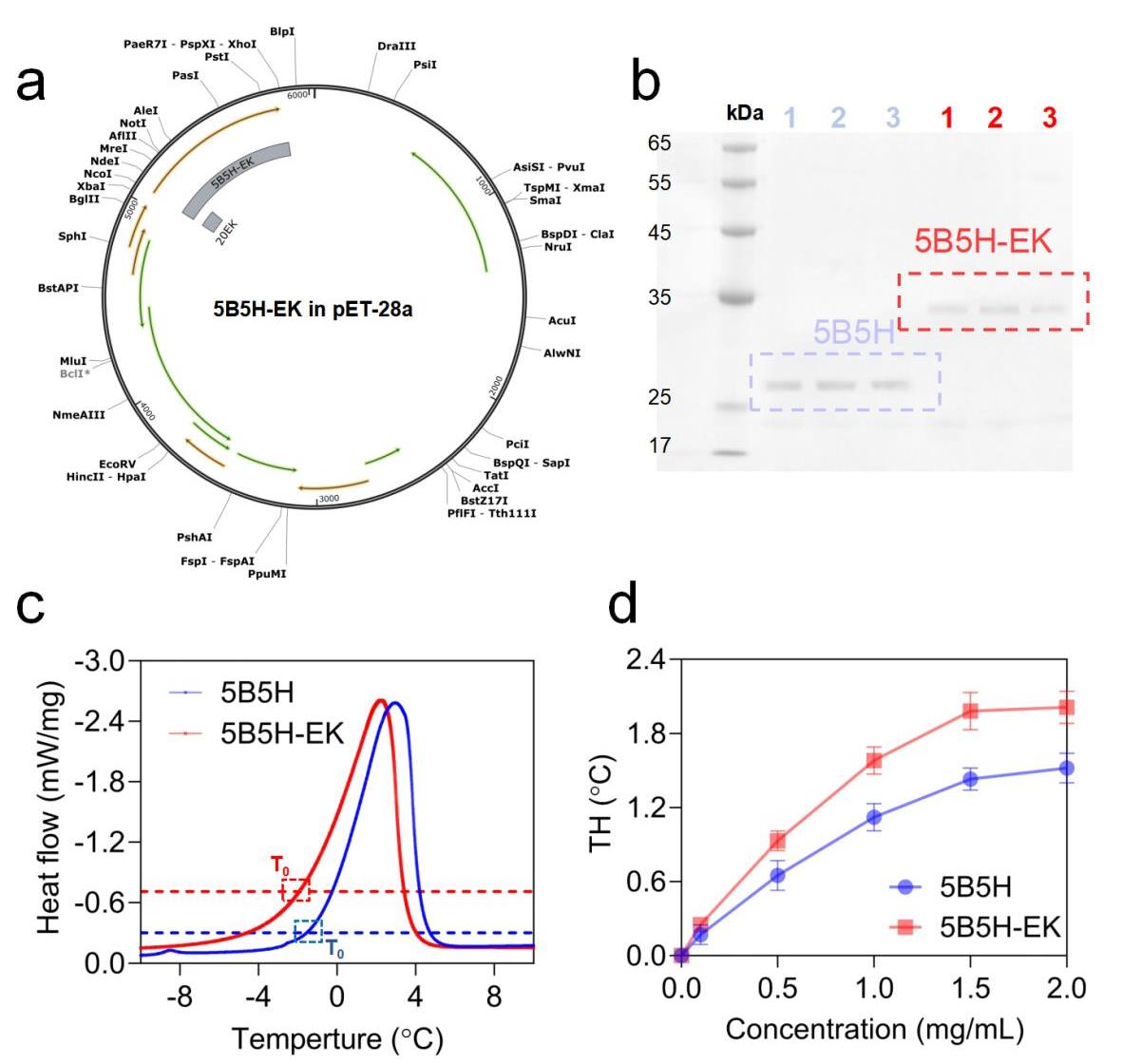
Fig. 2 Fermentation, purification and heat-lag activity measurements of 5B5H and 5B5H-EK(a) Plasmid of 5B5H-EK (b) Purification results of 5B5H and 5B5H-EK proteins (c) DSC results of 5B5H and 5B5H-EK at 2 mg/mL (d) Thermal hysteresis activities of 5B5H and 5B5H-EK at different concentrations
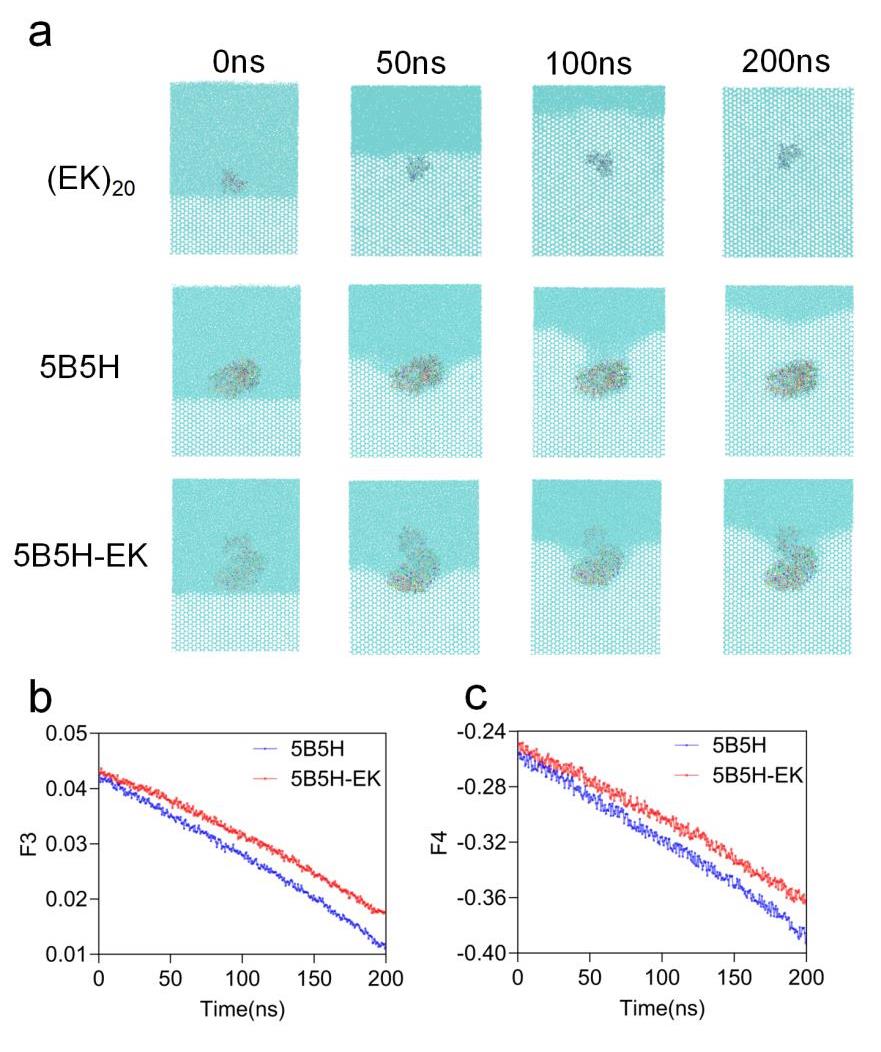
Fig. 3 5B5H and 5B5H-EK ice-water environment simulation results(a) Ice-water environment inhibition of ice crystal growth results of EK,5B5H and 5B5H-EK (b) F3 of 5B5H and 5B5H-EK in entire ice environment (c) F4 of 5B5H and 5B5H-EK in entire ice environment
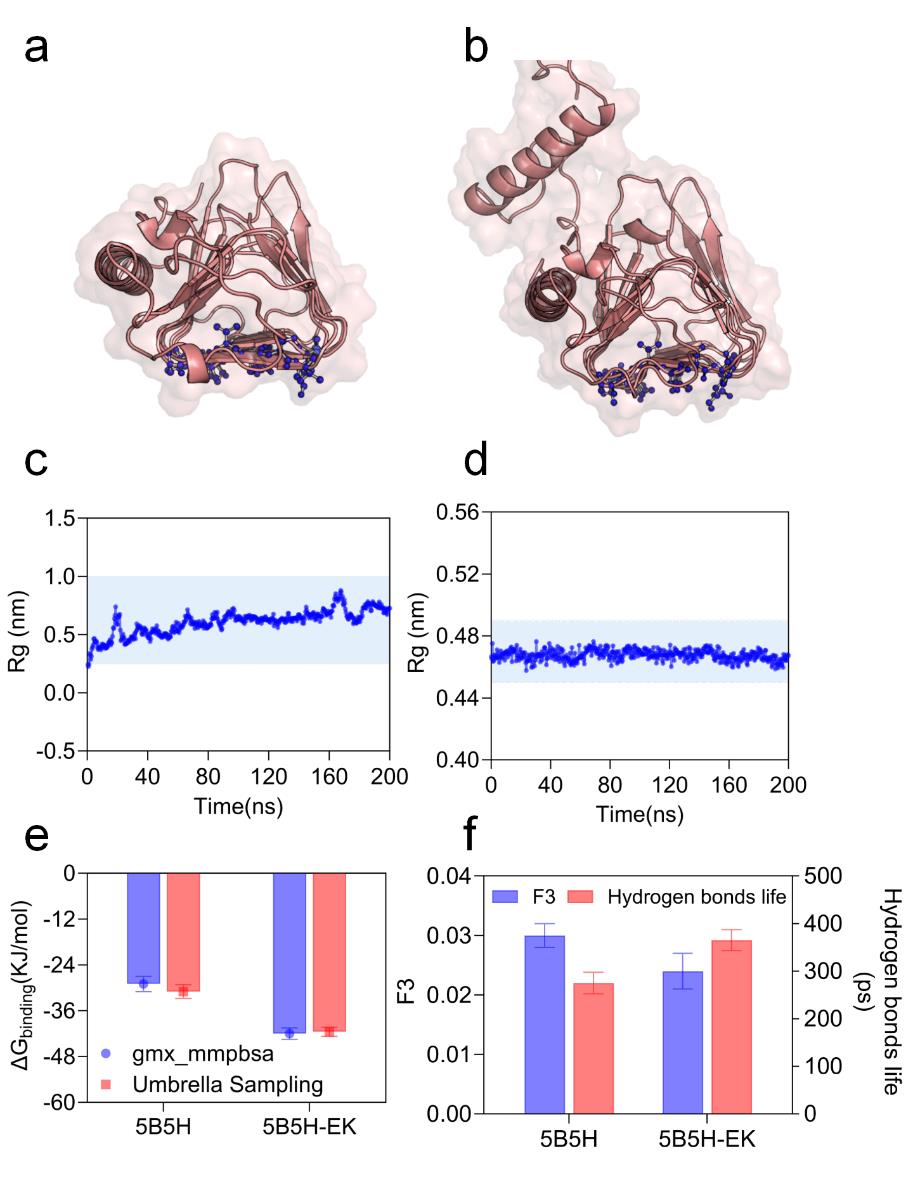
Fig. 4 5B5H and 5B5H-EK ice-crystal bonding surface resolution(a) Conformation of 5B5H's IBF (b) Conformation of 5B5H-EK's IBF (c) 5B5H ice-crystal bonding surface stability (d) 5B5H-EK ice-crystal bonding surface stability (e) 5B5H and 5B5H-EK with ice-crystal bonding energies (f) Hydrodynamics around the ice-crystal bonding surface of 5B5H and 5B5H-EK
| [1] | Yu H, Zheng H, Liu Y, Yang Q, Li W, Zhang Y, Fu F. Antifreeze protein from Ammopiptanthus nanus functions in temperature-stress through domain A. Sci Rep. 2021;11(1):8458. |
| [2] | Sreter JA, Foxall TL, Varga K. Intracellular and Extracellular Antifreeze Protein Significantly Improves Mammalian Cell Cryopreservation. Biomolecules. 2022;12(5):669. |
| [3] | Li X, Wang L, Yin C, Lin J, Wu Y, Chen D, Qiu C, Jia B, Huang J, Jiang X, Yang L, Liu L. Antifreeze protein from Anatolia polita (ApAFP914) improved outcome of vitrified in vitro sheep embryos. Cryobiology. 2020; 93:109-114. |
| [4] | Hudait A, Moberg DR, Qiu Y, Odendahl N, Paesani F, Molinero V. Preordering of water is not needed for ice recognition by hyperactive antifreeze proteins. Proc Natl Acad Sci U S A. 2018;115(33):8266-8271. |
| [5] | Zielkiewicz J. Solvation of molecules from the family of "domain of unknown function" 3494 and their ability to bind to ice. J Chem Phys. 2024;161(16):165101. |
| [6] | Chakraborty S, Jana B. Optimum Number of Anchored Clathrate Water and Its Instantaneous Fluctuations Dictate Ice Plane Recognition Specificities of Insect Antifreeze Protein. J Phys Chem B. 2018;122(12):3056-3067. |
| [7] | Drori R, Stevens CA. Divergent Mechanisms of Ice Growth Inhibition by Antifreeze Proteins. Methods Mol Biol. 2024;2730:169-181. |
| [8] | Kar RK, Bhunia A. Biophysical and biochemical aspects of antifreeze proteins: Using computational tools to extract atomistic information. Prog Biophys Mol Biol. 2015;119(2):194-204. |
| [9] | Chakraborty S, Jana B. Molecular Insight into the Adsorption of Spruce Budworm Antifreeze Protein to an Ice Surface: A Clathrate-Mediated Recognition Mechanism. Langmuir. 2017;33(28):7202-7214. |
| [10] | Grabowska J, Kuffel A, Zielkiewicz J. Molecular dynamics study on the role of solvation water in the adsorption of hyperactive AFP to the ice surface. Phys Chem Chem Phys. 2018;20(39):25365-25376. |
| [11] | Ekpo MD, Xie J, Hu Y, Liu X, Liu F, Xiang J, Zhao R, Wang B, Tan S. Antifreeze Proteins: Novel Applications and Navigation towards Their Clinical Application in Cryobanking. Int J Mol Sci. 2022;23(5):2639. |
| [12] | Elliott GD, Wang S, Fuller BJ. Cryoprotectants: A review of the actions and applications of cryoprotective solutes that modulate cell recovery from ultra-low temperatures. Cryobiology. 2017;76:74-91. |
| [13] | Xiang H, Yang X, Ke L, Hu Y. The properties, biotechnologies, and applications of antifreeze proteins. Int J Biol Macromol. 2020;153:661-675. |
| [14] | Kasahara K, Waku T, Wilson PW, Tonooka T, Hagiwara Y. The Inhibition of Icing and Frosting on Glass Surfaces by the Coating of Polyethylene Glycol and Polypeptide Mimicking Antifreeze Protein. Biomolecules, 2020;10(2):259. |
| [15] | Gao, Y, Qi, H, Fan, D, Yang, J, Zhang, L. Beetle and mussel-inspired chimeric protein for fabricating anti-icing coating. Colloids and surfaces. B, Biointerfaces, 2022;210:112252. |
| [16] | Liu Z, Yang W, Wei H, Deng S, Yu X, Huang T. The mechanisms and applications of cryoprotectants in aquatic products: An overview. Food Chem. 2023;408:135202. |
| [17] | Kim HJ, Lee JH, Hur YB, Lee CW, Park SH, Koo BW. Marine Antifreeze Proteins: Structure, Function, and Application to Cryopreservation as a Potential Cryoprotectant. Mar Drugs. 2017;15(2):27. |
| [18] | Diao H, Lin S, Li D, Li S, Feng Q, Sun N. Control on moisture distribution and protein changes of Antarctic krill meat by antifreeze protein during multiple freeze-thaw cycles. J Food Sci. 2022;87(10):4440-4452. |
| [19] | Ekpo, MD, Xie, J, Hu, Y, Liu, X, Liu, F, Xiang, J, Zhao, R, Wang, B, Tan, S. Antifreeze Proteins: Novel Applications and Navigation towards Their Clinical Application in Cryobanking. Int. J. Mol. Sci. 2022;23(5): 2639. |
| [20] | Elliott GD, Wang S, Fuller BJ. Cryoprotectants: A review of the actions and applications of cryoprotective solutes that modulate cell recovery from ultra-low temperatures. Cryobiology. 2017;76:74-91. |
| [21] | Kim HJ, Lee JH, Hur YB, Lee CW, Park SH, Koo BW. Marine Antifreeze Proteins: Structure, Function, and Application to Cryopreservation as a Potential Cryoprotectant. Mar Drugs. 2017;15(2):27. |
| [22] | Radziwon K, Weeks AM. Protein engineering for selective proteomics. Curr Opin Chem Biol. 2021;60:10-19. |
| [23] | Samineni L, Acharya B, Behera H, Oh H, Kumar M, Chowdhury R. Protein engineering of pores for separation, sensing, and sequencing. Cell Syst. 2023;14(8):676-691. |
| [24] | Lutz S, Iamurri SM. Protein Engineering: Past, Present, and Future. Methods Mol Biol. 2018;1685:1-12. |
| [25] | Rennella E, Sahtoe D D, Baker D, Kay L E. Exploiting conformational dynamics to modulate the function of designed proteins [J]. Proc Natl Acad Sci, 2023; 120(18):e2303149120. |
| [26] | Yang Q F, Tang C. On the necessity of an integrative approach to understand protein structural dynamics [J]. J. Zhejiang Univ. Sci. B, 2019;20(6):496-502. |
| [27] | Marshall C B, Daley M E, Sykes B D, Davies P L. Enhancing the activity of a beta-helical antifreeze protein by the engineered addition of coils [J]. Biochemistry, 2004; 43(37):11637-46. |
| [28] | Keefe AJ, Jiang S. Poly(zwitterionic)protein conjugates offer increased stability without sacrificing binding affinity or bioactivity. Nat Chem. 2011;4(1):59-63. |
| [29] | Cui Z, Wang Y, Zhang L, Qi H. Zwitterionic Peptides: From Mechanism, Design Strategies to Applications. ACS Appl Mater Interfaces. 2024;16(42):56497-56518. |
| [30] | Liu EJ, Sinclair A, Keefe AJ, et al. EKylation: Addition of an Alternating-Charge Peptide Stabilizes Proteins. Biomacromolecules. 2015;16(10):3357-3361. |
| [31] | Liu EJ, Jiang S. Expressing a Monomeric Organophosphate Hydrolase as an EK Fusion Protein. Bioconjug Chem. 2018;29(11):3686-3690. |
| [32] | Yuan Z, Li B, Niu L, et al. Zwitterionic Peptide Cloak Mimics Protein Surfaces for Protein Protection. Angew Chem Int Ed Engl. 2020;59(50):22378-22381. |
| [33] | Shao Q. Effect of conjugated (EK)10 peptide on structural and dynamic properties of ubiquitin protein: a molecular dynamics simulation study. J Mater Chem B. 2020;8(31):6934-6943. |
| [34] | Teng J, Liu Y, Shen Z, Lv W, Chen Y. Molecular simulation of zwitterionic polypeptides on protecting glucagon-like peptide-1 (GLP-1). Int J Biol Macromol. 2021;174:519-526. |
| [35] | Biswas AD, Barone V, Daidone I. High Water Density at Non-Ice-Binding Surfaces Contributes to the Hyperactivity of Antifreeze Proteins. J Phys Chem Lett. 2021;12(36):8777-8783.[36]FaragH, PetersB. Engulfment Avalanches and Thermal Hysteresis for Antifreeze Proteins on Supercooled Ice. J Phys Chem B. 2023;127(24):5422-5431. |
| [37] | Kim EJ, Lee JH, Lee SG, Han SJ. Improving thermal hysteresis activity of antifreeze protein from recombinant Pichia pastoris by removal of N-glycosylation. Prep Biochem Biotechnol. 2017;47(3):299-304. |
| [38] | Kim DE, Chivian D, Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32(Web Server issue):W526- W531. |
| [39] | Chivian D, Kim DE, Malmström L, et al. Automated prediction of CASP-5 structures using the Robetta server. Proteins. 2003;53 :524-533. |
| [40] | Park H, Kim DE, Ovchinnikov S, Baker D, DiMaio F. Automatic structure prediction of oligomeric assemblies using Robetta in CASP12. Proteins. 2018;86(Suppl 1):283-291. |
| [41] | Páll S, Zhmurov A, Bauer P, et al. Heterogeneous parallelization and acceleration of molecular dynamics simulations in GROMACS. J Chem Phys. 2020;153(13):134110. |
| [42] | Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J Chem Theory Comput. 2008;4(3):435-447. |
| [43] | Linse JB, Hub JS. Three- and four-site models for heavy water: SPC/E-HW, TIP3P-HW, and TIP4P/2005-HW. J Chem Phys. 2021;154(19):194501. |
| [44] | Bore SL, Piaggi PM, Car R, Paesani F. Phase diagram of the TIP4P/Ice water model by enhanced sampling simulations. J Chem Phys. 2022;157(5):054504. |
| [45] | Ngo ST, Pham MQ. Umbrella Sampling-Based Method to Compute Ligand-Binding Affinity. Methods Mol Biol. 2022;2385:313-323. |
| [46] | Sharpee TO, Destexhe A, Kawato M, et al. 25th Annual Computational Neuroscience Meeting: CNS-2016. BMC Neurosci. 2016;17(Suppl 1):54. |
| [47] | Kozuch DJ, Stillinger FH, Debenedetti PG. Combined molecular dynamics and neural network method for predicting protein antifreeze activity. Proc Natl Acad Sci U S A. 2018;115(52):13252-13257. |
| [48] | Park H, Kim DE, Ovchinnikov S, Baker D, DiMaio F. Automatic structure prediction of oligomeric assemblies using Robetta in CASP12. Proteins. 2018;86(Suppl 1):283-291. |
| [49] | Azzaz F, Fantini J. The epigenetic dimension of protein structure. Biomol Concepts. 2022;13(1):55-60. |
| [50] | Cheng J, Hanada Y, Miura A, Tsuda S, Kondo H. Hydrophobic ice-binding sites confer hyperactivity of an antifreeze protein from a snow mold fungus. Biochem J. 2016;473(21):4011-4026. |
| [51] | Wang, Y, Miyazaki, R, Saitou, S, Hirasaka, K, Takeshita, S, Tachibana, K, Taniyama, S. The effect of ice crystals formations on the flesh quality of frozen horse mackerel (Trachurus japonicus). J Texture Stud. 2018;49(5):485-491. |
| [52] | Feng S, Yi J, Wu X, Ma Y, Bi J. Effects of cell morphology on the textural attributes of fruit cubes in freeze-drying: Apples, strawberries, and mangoes as examples. J Texture Stud. 2023;54(5):775-786. |
| [53] | Schaudinn C, Tautz C, Laue M. Thin polyester filters as versatile sample substrates for high-pressure freezing of bacterial biofilms, suspended microorganisms and adherent eukaryotic cells. J Microsc. 2019;274(2):92-101. |
| [54] | Lee J, Hitzenberger M, Rieger M, Kern NR, Zacharias M, Im W. CHARMM-GUI supports the Amber force fields. J Chem Phys. 2020;153(3):035103. |
| [55] | Kognole AA, Lee J, Park SJ, Jo S, Chatterjee P, Lemkul JA, Huang J, MacKerell AD, Jr Im W. CHARMM-GUI Drude prepper for molecular dynamics simulation using the classical Drude polarizable force field. J Comput Chem. 2022;43(5):359-375. |
| [56] | McMullen P, Fang L, Qiao Q, Shao Q, Jiang S. Impacts of a Zwitterionic Peptide on its Fusion Protein. Bioconjug Chem. 2022;33(8):1485-1493. |
| [57] | Li C, Xia Y, Liu C, Huang R, Qi W, He Z, Su R. Lubricin-Inspired Loop Zwitterionic Peptide for Fabrication of Superior Antifouling Surfaces. ACS Appl Mater Interfaces. 2021;13(35):41978-41986. |
| [58] | Prajakta N, Bappa G, Subhadip D, Sudip R, Rajnish K,Molecular dynamics study on growth of carbon dioxide and methane hydrate from a seed crystal, Chin. J. Chem. Eng, 2019; 27(9): 2074-2080. |
| [59] | Dalal P, Sönnichsen FD. Source of the ice-binding specificity of antifreeze protein type I. J Chem Inf Comput Sci. 2000;40(5):1276-1284. |
| [60] | Jung W, Campbell RL, Gwak Y, Kim JI, Davies PL, Jin E. New Cysteine-Rich Ice-Binding Protein Secreted from Antarctic Microalga, Chloromonas sp. PLoS One. 2016;11(4):e0154056. |
| [61] | Hudait A, Qiu Y, Odendahl N, Molinero V. Hydrogen-Bonding and Hydrophobic Groups Contribute Equally to the Binding of Hyperactive Antifreeze and Ice-Nucleating Proteins to Ice. J Am Chem Soc. 2019;141(19):7887-7898. |
| [62] | Hudait A, Moberg DR, Qiu Y, Odendahl N, Paesani F, Molinero V. Preordering of water is not needed for ice recognition by hyperactive antifreeze proteins. Proc Natl Acad Sci U S A. 2018;115(33):8266-8271. |
| [1] | LI Yi, LI Yinshuang, LI Shuang, LING Peixue, FANG Junqiang. Recent Progress on the in vitro Bio-transformation Synthesis of Human Milk Oligosaccharides [J]. Synthetic Biology Journal, 2025, (): 1-16. |
| [2] | Ding Yi, Su Min, Gao Xiaodong, Wang Ning. Advances in Research of N-Glycan Synthesis [J]. Synthetic Biology Journal, 2025, (): 1-16. |
| [3] | HUANG Yang, LI Yiye, NIE Guangjun. Synthetic Biology-Powered Cell Membrane-Derived Nanoparticles for Precision Theranostics [J]. Synthetic Biology Journal, 2025, (): 1-24. |
| [4] | LI Quanfei, CHEN Qian, YANG Kai, LIU Hao, LI Sha, LEI Peng, GU Yian, SUN Liang, WANG Rui, XU Hong. Biosynthesis and applications of high-adhesion protein materials [J]. Synthetic Biology Journal, 2025, (): 1-20. |
| [5] | ZHU Yueming, YANG Jiangang, ZENG Yan, DONG Qianzhen, SUN Yuanxia. Advances and applications of in vitro biosynthesis of carbohydrates [J]. Synthetic Biology Journal, 2025, (): 1-16. |
| [6] | HU Die, XU Daozhu, LU Zhiyi, TANG Wei, FAN Bo, HE Yucai. Biosynthesis of xylo-oligosaccharides from wheat straw xylan via the synergistic hydrolysis by xylanase Xyn11A and arabinofuranosidase Abf62A [J]. Synthetic Biology Journal, 2025, (): 1-15. |
| [7] | MA Muqing, WU Yan, QU Maohua, LU Xiafeng, CAO Min, DU Feng, JI Rongtao, DONG Leichi, LUO Zhibo. Extracellular Multi-enzyme Assembly and Biocatalytic Cascade: Advances and Prospects [J]. Synthetic Biology Journal, 2025, (): 1-20. |
| [8] | XIAO He, YAO Mingju, QIAO Xue. Advances in the biosynthesis of long sugar-chain saponins [J]. Synthetic Biology Journal, 2025, (): 1-7. |
| [9] | . [J]. Synthetic Biology Journal, 2025, 6(3): 493-496. |
| [10] | YANG Ying, LI Xia, LIU Lizhong. Applications of synthetic biology to stem-cell-derived modeling of early embryonic development [J]. Synthetic Biology Journal, 2025, 6(3): 669-684. |
| [11] | LI Mingchen, ZHONG Bozitao, YU Yuanxi, JIANG Fan, ZHANG Liang, TAN Yang, YU Huiqun, FAN Guisheng, HONG Liang. DeepSeek model analysis and its applications in AI-assistant protein engineering [J]. Synthetic Biology Journal, 2025, 6(3): 636-650. |
| [12] | HUANG Yi, SI Tong, LU Anjing. Standardization for biomanufacturing: global landscape, critical challenges, and pathways forward [J]. Synthetic Biology Journal, 2025, 6(3): 701-714. |
| [13] | SONG Chengzhi, LIN Yihan. AI-enabled directed evolution for protein engineering and optimization [J]. Synthetic Biology Journal, 2025, 6(3): 617-635. |
| [14] | WU Ke, LUO Jiahao, LI Feiran. Applications of machine learning in the reconstruction and curation of genome-scale metabolic models [J]. Synthetic Biology Journal, 2025, 6(3): 566-584. |
| [15] | TIAN Xiao-jun, ZHANG Rixin. “Economics Paradox” with cells in synthetic gene circuits [J]. Synthetic Biology Journal, 2025, 6(3): 532-546. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||