Single-cell transcriptome combined with biosensing: a new framework for development of molecular diagnostic methods for cervical cancer
Yan ZHEN1,2, ZHAO Chao3, AN Jiahui1,2, XIE Wenjing1,2, PENG Hanyong1,2, ZHANG Xiaobo4, LI Mingzhu3, CHEN Xin1,2, XU Li1,2, XIE Qunhui1,2, WEI Lihui3
- 1.State Key Laboratory of Environmental Chemistry and Toxicology,Research Center for Eco-Environmental Sciences,Chinese Academy of Sciences,Beijing,100085,China
2.University of Chinese Academy of Sciences,College of Resources and Environment,Beijing,100049,China
3.Department of Obstetrics and Gynecology,Peking University People’s Hospital,Beijing,100044,China
4.Department of Pathology,Peking University People’s Hospital,Beijing,100044,China
-
Received:2025-08-04Revised:2025-10-11Published:2025-10-14 -
Contact:XIE Qunhui, WEI Lihui
单细胞转录组联合生物传感——宫颈癌分子诊断方法研发新框架
鄢震1,2, 赵超3, 安佳慧1,2, 谢文菁1,2, 彭汉勇1,2, 张晓波4, 李明珠3, 陈新1,2, 徐丽1,2, 谢群慧1,2, 魏丽惠3
- 1.中国科学院生态环境研究中心,环境化学与环境毒理全国重点实验室,北京 100085
2.中国科学院大学资源与环境学院,北京 100044
3.北京大学人民医院妇产科,北京 100049
4.北京大学人民医院病理科,北京 100049
-
通讯作者:谢群慧,魏丽惠 -
作者简介:鄢震 (1998—),男,硕士研究生。研究方向环境分子毒理学,网络毒理学。 E-mail:zhenyan_st@rcees.ac.cn赵超 (1976—),女,北京大学人民医院妇科主任医师,研究方向为子宫颈癌前病变的筛查及诊断治疗,尤其是阴道镜检查、宫颈锥切手术治疗及妇科腹腔镜、宫腔镜手术治疗等。 E-mail:zhaochaocjr@163.com谢群慧 (1976—),女,副研究员,硕士生导师,研究方向为持久性有机污染物的神经毒理机制、环境健康问题相关的生物标志物、基于分子毒理机制的毒性评估方法与技术等。 E-mail:qhxie@rcees.ac.cn魏丽惠 (1944—),女,北京大学人民医院妇科主任医师,教授、博士生导师。研究方向为妇科疾病诊治及恶性肿瘤研究,擅长宫颈癌、子宫内膜癌、卵巢癌诊疗。 E-mail:weilhpku@163.com -
基金资助:科技部重点研发项目课题(E5L3030403);北京市通州区科学技术委员会项目(KJ2024CX060);中国科学院B 类先导专项项目(XDB0750300)
CLC Number:
Cite this article
Yan ZHEN, ZHAO Chao, AN Jiahui, XIE Wenjing, PENG Hanyong, ZHANG Xiaobo, LI Mingzhu, CHEN Xin, XU Li, XIE Qunhui, WEI Lihui. Single-cell transcriptome combined with biosensing: a new framework for development of molecular diagnostic methods for cervical cancer[J]. Synthetic Biology Journal, DOI: 10.12211/2096-8280.2025-084.
鄢震, 赵超, 安佳慧, 谢文菁, 彭汉勇, 张晓波, 李明珠, 陈新, 徐丽, 谢群慧, 魏丽惠. 单细胞转录组联合生物传感——宫颈癌分子诊断方法研发新框架[J]. 合成生物学, DOI: 10.12211/2096-8280.2025-084.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2025-084
| 类别 | 代表分子标志物 | 主要功能与意义 |
|---|---|---|
| HPV感染及癌变 | HPV DNA;E6/E7 mRNA; p16 | 持续性人乳头瘤病毒感染; 病毒致癌基因表达; 通过 E7-RB 通路破坏导致 p16 过表达 |
| 细胞周期与增殖 | p53;Ki-67 | p53:突变累积提示肿瘤进展。 Ki-67:经典的细胞增殖标志物,高表达是细胞增殖活跃的标志。 |
| 细胞凋亡 | BCL-2/BAX | 表达失衡反映肿瘤细胞逃逸凋亡的能力。 |
| 上皮分化 | CK7;CK8;CK17;CK19 | 表达模式的改变可反向指示肿瘤的恶性程度。 部分细胞角蛋白(如CK7; CK17)有鉴别病变类型及进展趋势的潜力。 |
| 侵袭和转移 | E-cadherin; CD44; MMP家族 | E-cadherin下调:指示上皮-间质转化。 CD44/MMP上调:指示基质降解增强与远端转移。 |
| 癌干细胞 | ALDH1A1; OCT4 | 与肿瘤干性、耐药及复发相关,可用于早期检测和疗效监测。 |
| 血管生成 | VEGF; TSP-1 | 共同构成抗血管生成治疗的评估靶点。 VEGF:诱导新生血管,促进肿瘤进展。 TSP-1:内源性抑制剂。 |
Table 1 Current classification of molecular markers for cervical cancer
| 类别 | 代表分子标志物 | 主要功能与意义 |
|---|---|---|
| HPV感染及癌变 | HPV DNA;E6/E7 mRNA; p16 | 持续性人乳头瘤病毒感染; 病毒致癌基因表达; 通过 E7-RB 通路破坏导致 p16 过表达 |
| 细胞周期与增殖 | p53;Ki-67 | p53:突变累积提示肿瘤进展。 Ki-67:经典的细胞增殖标志物,高表达是细胞增殖活跃的标志。 |
| 细胞凋亡 | BCL-2/BAX | 表达失衡反映肿瘤细胞逃逸凋亡的能力。 |
| 上皮分化 | CK7;CK8;CK17;CK19 | 表达模式的改变可反向指示肿瘤的恶性程度。 部分细胞角蛋白(如CK7; CK17)有鉴别病变类型及进展趋势的潜力。 |
| 侵袭和转移 | E-cadherin; CD44; MMP家族 | E-cadherin下调:指示上皮-间质转化。 CD44/MMP上调:指示基质降解增强与远端转移。 |
| 癌干细胞 | ALDH1A1; OCT4 | 与肿瘤干性、耐药及复发相关,可用于早期检测和疗效监测。 |
| 血管生成 | VEGF; TSP-1 | 共同构成抗血管生成治疗的评估靶点。 VEGF:诱导新生血管,促进肿瘤进展。 TSP-1:内源性抑制剂。 |
| 基因 | Ensembl ID | 调控 | 功能 | 引文 | |||
|---|---|---|---|---|---|---|---|
HPV感染 及癌变 | 细胞增殖、凋亡及分化 | 细胞黏附、 侵袭与转移 | 血管生成相关 | ||||
| APOC1 | ENSG00000130208 | 促进 | ✔ | ✔ | [ | ||
| E2F8 | ENSG00000129173 | 促进 | ✔ | ✔ | [ | ||
| FABP5 | ENSG00000164687 | 促进 | ✔ | ✔ | [ | ||
| KLF5 | ENSG00000102554 | 促进 | ✔ | ✔ | [ | ||
| HSPA2 | ENSG00000126803 | 促进 | ✔ | ✔ | [ | ||
| ITGA3 | ENSG00000005884 | 促进 | ✔ | ✔ | [ | ||
| LPCAT1 | ENSG00000153395 | 促进 | ✔ | ✔ | [ | ||
| MYO1B | ENSG00000128641 | 促进 | ✔ | ✔ | ✔ | [ | |
| NRP1 | ENSG00000099250 | 促进 | ✔ | [ | |||
| PAK5 | ENSG00000101349 | 促进 | ✔ | [ | |||
| POU5F1B | ENSG00000212993 | 促进 | ✔ | ✔ | [ | ||
| SEPTIN9 | ENSG00000184640 | 促进 | ✔ | ✔ | ✔ | [ | |
| SND1 | ENSG00000197157 | 促进 | ✔ | [ | |||
| STAT3 | ENSG00000168610 | 促进 | ✔ | ✔ | [ | ||
| FSTL1 | ENSG00000163430 | 抑制 | ✔ | ✔ | [ | ||
| RHCG | ENSG00000140519 | 抑制 | ✔ | ✔ | [ | ||
Table 2 Potential Molecular Markers Implicated in Cervical Cancer and Precancerous Lesions.
| 基因 | Ensembl ID | 调控 | 功能 | 引文 | |||
|---|---|---|---|---|---|---|---|
HPV感染 及癌变 | 细胞增殖、凋亡及分化 | 细胞黏附、 侵袭与转移 | 血管生成相关 | ||||
| APOC1 | ENSG00000130208 | 促进 | ✔ | ✔ | [ | ||
| E2F8 | ENSG00000129173 | 促进 | ✔ | ✔ | [ | ||
| FABP5 | ENSG00000164687 | 促进 | ✔ | ✔ | [ | ||
| KLF5 | ENSG00000102554 | 促进 | ✔ | ✔ | [ | ||
| HSPA2 | ENSG00000126803 | 促进 | ✔ | ✔ | [ | ||
| ITGA3 | ENSG00000005884 | 促进 | ✔ | ✔ | [ | ||
| LPCAT1 | ENSG00000153395 | 促进 | ✔ | ✔ | [ | ||
| MYO1B | ENSG00000128641 | 促进 | ✔ | ✔ | ✔ | [ | |
| NRP1 | ENSG00000099250 | 促进 | ✔ | [ | |||
| PAK5 | ENSG00000101349 | 促进 | ✔ | [ | |||
| POU5F1B | ENSG00000212993 | 促进 | ✔ | ✔ | [ | ||
| SEPTIN9 | ENSG00000184640 | 促进 | ✔ | ✔ | ✔ | [ | |
| SND1 | ENSG00000197157 | 促进 | ✔ | [ | |||
| STAT3 | ENSG00000168610 | 促进 | ✔ | ✔ | [ | ||
| FSTL1 | ENSG00000163430 | 抑制 | ✔ | ✔ | [ | ||
| RHCG | ENSG00000140519 | 抑制 | ✔ | ✔ | [ | ||
| [107] | CHEN L, PARK J E, PAA P, et al. Programmable C: G to G: C genome editing with CRISPR-Cas9-directed base excision repair proteins [J]. Nature communications, 2021, 12(1): 1384. |
| [108] | LI C, ZHANG R, MENG X, et al. Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors [J]. Nature biotechnology, 2020, 38(7): 875-82. |
| [109] | LU Y, TIAN Y, SHEN R, et al. Targeted, efficient sequence insertion and replacement in rice [J]. Nature Biotechnology, 2020, 38(12): 1402-7. |
| [110] | LIN Q, ZONG Y, XUE C, et al. Prime genome editing in rice and wheat [J]. Nature biotechnology, 2020, 38(5): 582-5. |
| [111] | PAN C, SRETENOVIC S, QI Y. CRISPR/dCas-mediated transcriptional and epigenetic regulation in plants [J]. Current Opinion in Plant Biology, 2021, 60: 101980. |
| [112] | NISSIM L, BAR‐ZIV R H. A tunable dual‐promoter integrator for targeting of cancer cells [J]. Molecular systems biology, 2010, 6(1): 444. |
| [113] | KYO S, TAKAKURA M, FUJIWARA T, et al. Understanding and exploiting hTERT promoter regulation for diagnosis and treatment of human cancers [J]. Cancer science, 2008, 99(8): 1528-38. |
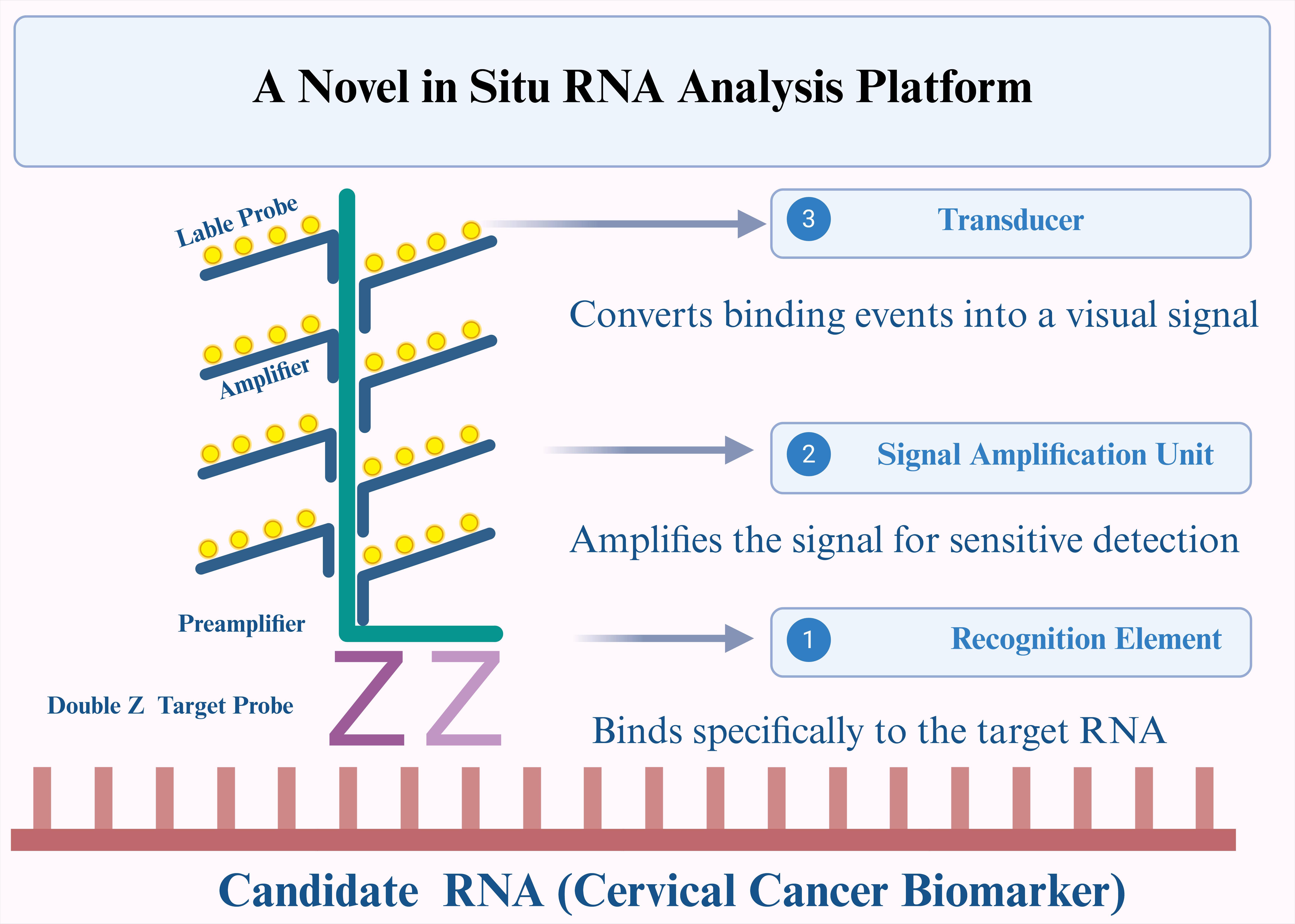
| [114] | KEMMER C, GITZINGER M, DAOUD-EL BABA M, et al. Self-sufficient control of urate homeostasis in mice by a synthetic circuit [J]. Nature biotechnology, 2010, 28(4): 355-60. |
| [1] | WU J, JIN Q, ZHANG Y, et al. Global burden of cervical cancer: current estimates, temporal trend and future projections based on the GLOBOCAN 2022 [J]. Journal of the National Cancer Center, 2025. |
| [2] | SHEN X, CHENG Y, REN F, et al. The burden of cervical cancer in China [J]. Frontiers in Oncology, 2022, 12: 979809. |
| [3] | CROSBIE E J, EINSTEIN M H, FRANCESCHI S, et al. Human papillomavirus and cervical cancer [J]. The Lancet, 2013, 382(9895): 889-99. |
| [4] | BANSAL A, SINGH M P, RAI B. Human papillomavirus-associated cancers: A growing global problem [J]. International Journal of Applied and Basic Medical Research, 2016, 6(2): 84-9. |
| [5] | VOELKER R A. Cervical cancer screening [J]. Jama, 2023, 330(20): 2030-. |
| [6] | KOMDEUR F L, PRINS T M, VAN DE WALL S, et al. CD103+ tumor-infiltrating lymphocytes are tumor-reactive intraepithelial CD8+ T cells associated with prognostic benefit and therapy response in cervical cancer [J]. Oncoimmunology, 2017, 6(9): e1338230. |
| [7] | MEDDA A, DUCA D, CHIOCCA S. Human papillomavirus and cellular pathways: hits and targets [J]. Pathogens, 2021, 10(3): 262. |
| [8] | GüZEL C, VAN STEN-VAN'T HOFF J, DE KOK I M, et al. Molecular markers for cervical cancer screening [J]. Expert review of proteomics, 2021, 18(8): 675-91. |
| [9] | BOUVARD V, WENTZENSEN N, MACKIE A, et al. The IARC perspective on cervical cancer screening [J]. New England Journal of Medicine, 2021, 385(20): 1908-18. |
| [10] | FONTHAM E T, WOLF A M, CHURCH T R, et al. Cervical cancer screening for individuals at average risk: 2020 guideline update from the American Cancer Society [J]. CA: a cancer journal for clinicians, 2020, 70(5): 321-46. |
| [11] | 胡尚英, 赵雪莲, 张勇. 预防宫颈癌: WHO 宫颈癌前病变筛查和治疗指南 (第二版)[J]. 解读, 2021, 101(34): 2653-2657. |
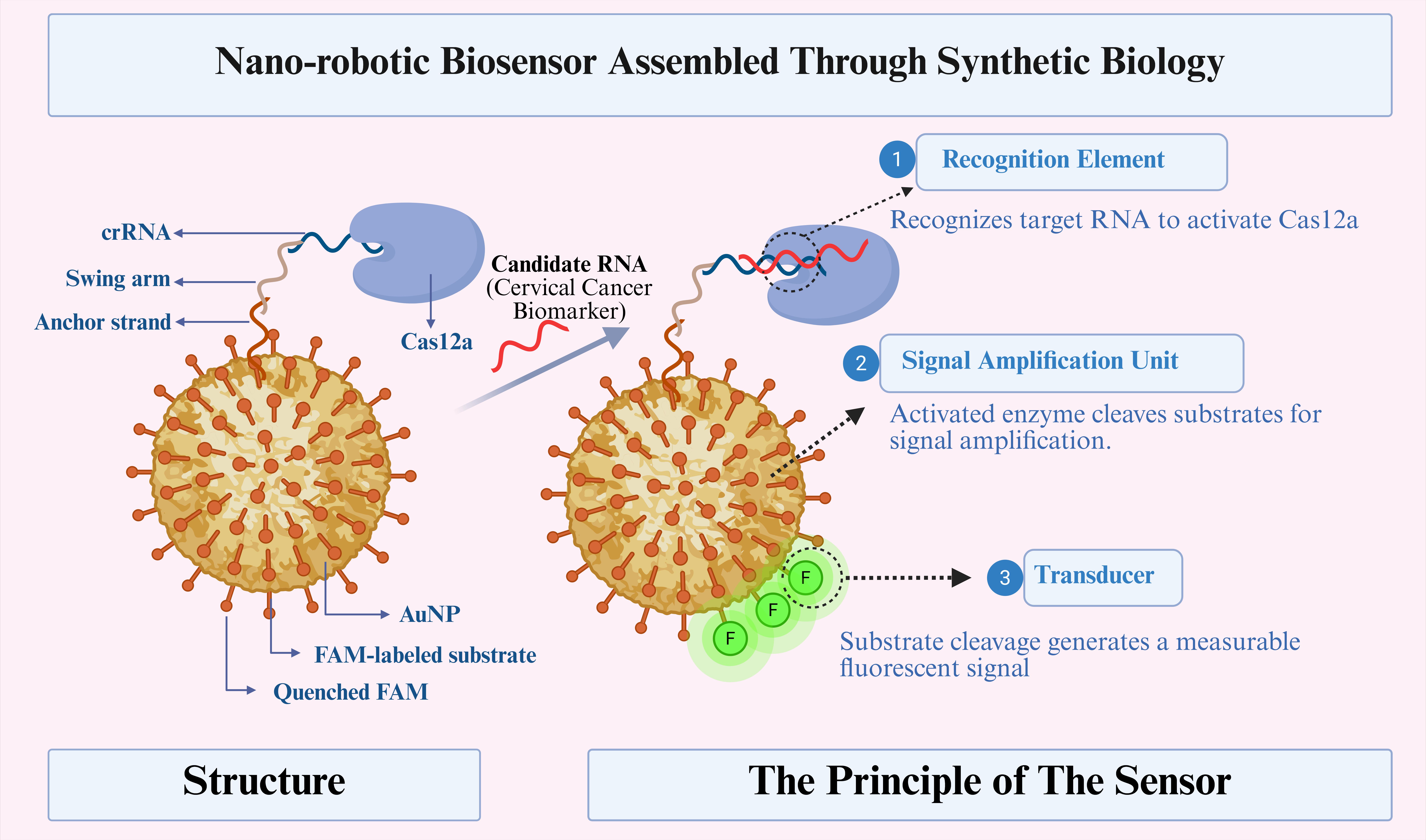
| [12] | SARHADI V K, ARMENGOL G. Molecular biomarkers in cancer [J]. Biomolecules, 2022, 12(8): 1021. |
| [13] | VOLKOVA L V, PASHOV A I, OMELCHUK N N. Cervical carcinoma: oncobiology and biomarkers [J]. International journal of molecular sciences, 2021, 22(22): 12571. |
| [14] | BALASUBRAMANIAM S D, BALAKRISHNAN V, OON C E, et al. Key molecular events in cervical cancer development [J]. Medicina, 2019, 55(7): 384. |
| [15] | NAKAMURA M, OBATA T, DAIKOKU T, et al. The association and significance of p53 in gynecologic cancers: the potential of targeted therapy [J]. International Journal of Molecular Sciences, 2019, 20(21): 5482. |
| [16] | BABICHENKO I. New methods of immunohistochemical diagnostic of tumor grows [J]. RUDN Journal of Medicine, 2008, (4): 94-9. |
| [17] | VAN ZUMMEREN M, LEEMAN A, KREMER W W, et al. Three-tiered score for Ki-67 and p16ink4a improves accuracy and reproducibility of grading CIN lesions [J]. Journal of clinical pathology, 2018, 71(11): 981-8. |
| [18] | SILVA D C, GONCALVES A K, COBUCCI R N, et al. Immunohistochemical expression of p16, Ki-67 and p53 in cervical lesions-A systematic review [J]. Pathology-Research and Practice, 2017, 213(7): 723-9. |
| [19] | MITILDZANS A, ARECHVO A, REZEBERGA D, et al. Expression of p63, p53 and Ki-67 in Patients with Cervical Intraepithelial Neoplasia [J]. Turkish Journal of Pathology, 2017, 33(1). |
| [20] | CHEN C-C, HUANG L-W, BAI C-H, et al. Predictive value of p16/Ki-67 immunocytochemistry for triage of women with abnormal Papanicolaou test in cervical cancer screening: a systematic review and meta-analysis [J]. Annals of Saudi medicine, 2016, 36(4): 245-51. |
| [21] | PIRI R, GHAFFARI A, GHOLAMI N, et al. Ki-67/MIB-1 as a prognostic marker in cervical cancer-a systematic review with meta-analysis [J]. Asian Pacific Journal of Cancer Prevention, 2015, 16(16): 6997-7002. |
| [22] | SOOD S, PATEL F D, SRINIVASAN R, et al. Chemoradiation therapy induces in vivo changes in gene promoter methylation & gene transcript expression in patients with invasive cervical cancer [J]. Indian Journal of Medical Research, 2018, 147(2): 151-7. |
| [23] | KILIC S, CRACCHIOLO B, GABEL M, et al. The relevance of molecular biomarkers in cervical cancer patients treated with radiotherapy [J]. Annals of translational medicine, 2015, 3(18): 261. |
| [24] | QIN C, CHEN X, BAI Q, et al. Factors associated with radiosensitivity of cervical cancer [J]. Anticancer research, 2014, 34(9): 4649-56. |
| [25] | YAO T, LU R, ZHANG Y, et al. Cervical cancer stem cells [J]. Cell proliferation, 2015, 48(6): 611-25. |
| [26] | IMMUNOHISTOCHEMISTRY D. Theranostic and Genomic Applications, ; Dabbs, DJ, Ed [Z]. Elsevier: Amsterdam, Netherlands The. 2017 |
| [27] | LI B, SHI H, WANG F, et al. Expression of E-, P-and N-cadherin and its clinical significance in cervical squamous cell carcinoma and precancerous lesions [J]. PloS one, 2016, 11(5): e0155910. |
| [28] | SOLOVУEVA N, TIMOSHENKO O, GUREEVA T, et al. Matrix metalloproteinases and their endogenous regulators in squamous cervical carcinoma (review of the own data) [J]. Biomeditsinskaia Khimiia, 2015, 61(6): 694-704. |
| [29] | TIMOSHENKO O, GUREEVA T, KUGAEVSKAIA E, et al. Membrane type 1 matrix metalloproteinase (MT1-MMP) and the regulators of its activity as invasive factors in squamous cell cervical carcinomas [J]. Biomeditsinskaia Khimiia, 2014, 60(6): 683-8. |
| [30] | TULAKE W, YUEMAIER R, SHENG L, et al. Upregulation of stem cell markers ALDH1A1 and OCT4 as potential biomarkers for the early detection of cervical carcinoma [J]. Oncology Letters, 2018, 16(5): 5525-34. |
| [31] | NAGY V M, BUIGA R, BRIE I, et al. Expression of VEGF, VEGFR, EGFR, COX-2 and MVD in cervical carcinoma, in relation with the response to radio-chemotherapy [J]. Rom J Morphol Embryol, 2011, 52(1): 53-9. |
| [32] | LI Y, ZHAO S. The expression and underlying angiogenesis effect of DPC4 and VEGF on the progression of cervical carcinoma [J]. Oncology Letters, 2018, 15(2): 2534-40. |
| [33] | VAN TRAPPEN P O, STEELE D, LOWE D G, et al. Expression of vascular endothelial growth factor (VEGF)‐C and VEGF‐D, and their receptor VEGFR‐3, during different stages of cervical carcinogenesis [J]. The Journal of Pathology: A Journal of the Pathological Society of Great Britain and Ireland, 2003, 201(4): 544-54. |
| [34] | BELFORT-MATTOS P N, FOCCHI G R D A, RIBALTA J C L, et al. Immunohistochemical expression of VEGF and podoplanin in uterine cervical squamous intraepithelial lesions [J]. Disease Markers, 2016, 2016(1): 8293196. |
| [35] | 李晓, 汪辉, 李静然, et al. p16/Ki-67双染检测用于子宫颈癌筛查异常人群分流的专家共识 [J]. 中国妇产科临床杂志, 2025, 26(02): 188-92. |
| [36] | 李明珠, 魏丽惠. 高危型HPV持续阳性人群的分流和管理 [J]. 中国妇产科临床杂志, 2025, 26(01): 1-2. |
| [37] | SARWATH H, BANSAL D, HUSAIN N E, et al. Introduction of p16(INK4a) as a surrogate biomarker for HPV in women with invasive cervical cancer in Sudan [J]. Infectious agents and cancer, 2017, 12: 50. |
| [38] | VON KNEBEL DOEBERITZ M, REUSCHENBACH M, SCHMIDT D, et al. Biomarkers for cervical cancer screening: the role of p16INK4a to highlight transforming HPV infections [J]. Expert review of proteomics, 2012, 9(2): 149-63. |
| [39] | IKENBERG H, BERGERON C, SCHMIDT D, et al. Screening for cervical cancer precursors with p16/Ki-67 dual-stained cytology: results of the PALMS study [J]. Journal of the National Cancer Institute, 2013, 105(20): 1550-7. |
| [40] | ZHANG R, GE X, YOU K, et al. p16/Ki67 dual staining improves the detection specificity of high-grade cervical lesions [J]. Journal of Obstetrics and Gynaecology Research, 2018, 44(11): 2077-84. |
| [41] | CHARAKORN C, THADANIPON K, CHAIJINDARATANA S, et al. The association between serum squamous cell carcinoma antigen and recurrence and survival of patients with cervical squamous cell carcinoma: A systematic review and meta-analysis [J]. Gynecologic oncology, 2018, 150(1): 190-200. |
| [42] | ZHOU Z, LI W, ZHANG F, et al. The value of squamous cell carcinoma antigen (SCCa) to determine the lymph nodal metastasis in cervical cancer: A meta-analysis and literature review [J]. PloS one, 2017, 12(12): e0186165. |
| [43] | SHI X, WANG J, DAI S, et al. Apolipoprotein C1 (APOC1): a novel diagnostic and prognostic biomarker for cervical cancer [J]. OncoTargets and therapy, 2020: 12881-91. |
| [44] | KIM L K, S-A PARK, EOH K J, et al. E2F8 regulates the proliferation and invasion through epithelial-mesenchymal transition in cervical cancer [J]. International Journal of Biological Sciences, 2020, 16(2): 320. |
| [45] | WANG W, CHU H-J, LIANG Y-C, et al. FABP5 correlates with poor prognosis and promotes tumor cell growth and metastasis in cervical cancer [J]. Tumor Biology, 2016, 37(11): 14873-83. |
| [46] | MA D, CHANG L-Y, ZHAO S, et al. KLF5 promotes cervical cancer proliferation, migration and invasion in a manner partly dependent on TNFRSF11a expression [J]. Scientific reports, 2017, 7(1): 15683. |
| [47] | GARG M, KANOJIA D, SAINI S, et al. Germ cell‐specific heat shock protein 70‐2 is expressed in cervical carcinoma and is involved in the growth, migration, and invasion of cervical cells [J]. Cancer, 2010, 116(16): 3785-96. |
| [48] | DU Q, WANG W, LIU T, et al. High expression of integrin α3 predicts poor prognosis and promotes tumor metastasis and angiogenesis by activating the c-Src/extracellular signal-regulated protein kinase/focal adhesion kinase signaling pathway in cervical cancer [J]. Frontiers in Oncology, 2020, 10: 36. |
| [49] | GAO F, CHEN J, ZHANG T, et al. LPCAT1 functions as an oncogene in cervical cancer through mediating JAK2/STAT3 signaling [J]. Experimental Cell Research, 2022, 421(1): 113360. |
| [50] | ZHANG H-R, LAI S-Y, HUANG L-J, et al. Myosin 1b promotes cell proliferation, migration, and invasion in cervical cancer [J]. Gynecologic oncology, 2018, 149(1): 188-97. |
| [51] | YANG L, LIU L, ZHU Y-H, et al. Neuropilin-1 is associated with the prognosis of cervical cancer in Henan Chinese population [J]. OncoTargets and therapy, 2019: 2911-20. |
| [52] | HUO F-C, PAN Y-J, LI T-T, et al. PAK5 promotes the migration and invasion of cervical cancer cells by phosphorylating SATB1 [J]. Cell Death & Differentiation, 2019, 26(6): 994-1006. |
| [53] | YU J, ZHANG J, ZHOU L, et al. The octamer-binding transcription factor 4 (OCT4) pseudogene, POU domain class 5 transcription factor 1B (POU5F1B), is upregulated in cervical cancer and down-regulation inhibits cell proliferation and migration and induces apoptosis in cervical cancer cell lines [J]. Medical science monitor: international medical journal of experimental and clinical research, 2019, 25: 1204. |
| [54] | JIAO X, ZHANG S, JIAO J, et al. Promoter methylation of SEPT9 as a potential biomarker for early detection of cervical cancer and its overexpression predicts radioresistance [J]. Clinical Epigenetics, 2019, 11(1): 120. |
| [55] | ZHAN F, ZHONG Y, QIN Y, et al. SND1 facilitates the invasion and migration of cervical cancer cells by Smurf1-mediated degradation of FOXA2 [J]. Experimental cell research, 2020, 388(1): 111809. |
| [56] | WU L, SHEN B, LI J, et al. STAT3 exerts pro-tumor and anti-autophagy roles in cervical cancer [J]. Diagnostic Pathology, 2022, 17(1): 13. |
| [57] | LIU Z, ZHANG H, HU X. Analysis of the expression and mechanism of follistatin‑like protein 1 in cervical cancer [J]. Oncology Reports, 2023, 50(6): 215. |
| [58] | WANG D-G, LI T-M, LIU X. RHCG suppresses cervical cancer progression through inhibiting migration and inducing apoptosis regulated by TGF-β1 [J]. Biochemical and Biophysical Research Communications, 2018, 503(1): 86-93. |
| [59] | DU N, LI D, ZHAO W, et al. Stratifin (SFN) regulates cervical cancer cell proliferation, apoptosis, and cytoskeletal remodeling and metastasis progression through LIMK2/Cofilin signaling [J]. Molecular Biotechnology, 2024, 66(11): 3369-81. |
| [60] | ZHAO Y-C, WANG T-J, SHE L-Z, et al. S100A10 overexpression correlates with adverse prognosis, tumor microenvironment, and aggressive behavior in vitro and in vivo of cervical cancer [J]. Journal of Cancer, 2023, 14(15): 2931. |
| [61] | WANG S, CHEN X. Identification of potential biomarkers in cervical cancer with combined public mRNA and miRNA expression microarray data analysis [J]. Oncology letters, 2018, 16(4): 5200-8. |
| [62] | ESCOBAR-HOYOS L F, YANG J, ZHU J, et al. Keratin 17 in premalignant and malignant squamous lesions of the cervix: proteomic discovery and immunohistochemical validation as a diagnostic and prognostic biomarker [J]. Modern Pathology, 2014, 27(4): 621-30. |
| [63] | LI Z, CHEN J, ZHAO S, et al. Discovery and validation of novel biomarkers for detection of cervical cancer [J]. Cancer medicine, 2021, 10(6): 2063-74. |
| [64] | AN R, MENG S, QIAN H. Identification of Key Pathways and Establishment of a Seven‐Gene Prognostic Signature in Cervical Cancer [J]. Journal of oncology, 2022, 2022(1): 4748796. |
| [65] | LAI Y, ZHOU B, TAN Q, et al. LINC00116 enhances cervical cancer tumorigenesis through miR-106a/c-Jun pathway [J]. Journal of Cellular Biochemistry, 2020, 121(3): 2247-57. |
| [66] | LIU J, HAN B, HU X, et al. Identification of N6-methyladenosine-associated ferroptosis biomarkers in cervical cancer [J]. Hereditas, 2025, 162(1): 53. |
| [67] | BODZEK P, SZYMALA B, DAMASIEWICZ-BODZEK A, et al. Are IgG antibodies to heat shock proteins HSP27 and HSP60 useful markers in endometrial cancer and cervical cancer? [J]. Ginekologia Polska, 2021, 92(11): 760-6. |
| [68] | WU X, PENG L, ZHANG Y, et al. Identification of key genes and pathways in cervical cancer by bioinformatics analysis [J]. International journal of medical sciences, 2019, 16(6): 800. |
| [69] | WANG T, ZHANG L, MEI S, et al. Single-cell RNA sequencing highlights the unique tumor microenvironment of small cell neuroendocrine cervical carcinoma [J]. Journal of Translational Medicine, 2025, 23(1): 19. |
| [70] | ZHAO J, LI H, YUAN M. EGR1 promotes stemness and predicts a poor outcome of uterine cervical cancer by inducing SOX9 expression [J]. Genes & Genomics, 2021, 43(5): 459-70. |
| [71] | BHATTACHARJEE R, DAS S S, BISWAL S S, et al. Mechanistic role of HPV-associated early proteins in cervical cancer: Molecular pathways and targeted therapeutic strategies [J]. Critical reviews in oncology/hematology, 2022, 174: 103675. |
| [72] | 狄晨红,金帆.密封蛋白4与高危型人乳头瘤病毒联合检测对于高级别鳞状上皮内病变及宫颈鳞癌的诊断价值[J].浙江大学学报(医学版),2018,47(04):344-350. |
| [73] | ROYCHOWDHURY A, PAL D, BASU M, et al. Promoter methylation and enhanced SKP2 are associated with the downregulation of CDKN1C in cervical squamous cell carcinoma [J]. Cellular Signalling, 2023, 109: 110735. |
| [74] | LIZANO M, CARRILLO-GARCíA A, DE LA CRUZ-HERNáNDEZ E, et al. Promising predictive molecular biomarkers for cervical cancer [J]. International Journal of Molecular Medicine, 2024, 53(6): 50. |
| [75] | NADERZADEH E, KARGAR M, MOKHTARI M J, et al. Activating transcription factor 3 induces oxidative stress and genotoxicity, transcriptionally modulating metastasis-related gene expression in human papillomavirus-infected cervical cancer [J]. Virology Journal, 2025, 22(1): 46. |
| [76] | WANG J, SU Y, TIAN Y, et al. Characterization of DNA hydroxymethylation profile in cervical cancer [J]. Artificial Cells, Nanomedicine, and Biotechnology, 2019, 47(1): 2706-14. |
| [77] | DAI F, CHEN G, WANG Y, et al. Identification of candidate biomarkers correlated with the diagnosis and prognosis of cervical cancer via integrated bioinformatics analysis [J]. OncoTargets and therapy, 2019: 4517-32. |
| [78] | TANIM M T H, NATH S D, KHAN S F, et al. Transcriptomes of cervical cancer provide novel insights into dysregulated pathways, potential therapeutic targets, and repurposed drugs [J]. Cancer Treatment and Research Communications, 2024, 39: 100808. |
| [79] | LIU C, ZHANG M, YAN X, et al. Single-cell dissection of cellular and molecular features underlying human cervical squamous cell carcinoma initiation and progression [J]. Science advances, 2023, 9(4): eadd8977. |
| [80] | QIU J, QU X, WANG Y, et al. Single‐cell landscape highlights heterogenous microenvironment, novel immune reaction patterns, potential biomarkers and unique therapeutic strategies of cervical squamous carcinoma, Human Papillomavirus-Associated (HPVA) and Non-HPVA Adenocarcinoma [J]. Advanced Science, 2023, 10(10): 2204951. |
| [81] | GUO C, QU X, TANG X, et al. Spatiotemporally deciphering the mysterious mechanism of persistent HPV-induced malignant transition and immune remodelling from HPV-infected normal cervix, precancer to cervical cancer: Integrating single-cell RNA-sequencing and spatial transcriptome [J]. Clinical and translational medicine, 2023, 13(3): e1219. |
| [82] | PENG Y, YANG J, AO J, et al. Single-cell profiling reveals the intratumor heterogeneity and immunosuppressive microenvironment in cervical adenocarcinoma [J]. eLife, 2025, 13: RP97335. |
| [83] | DING H, MEI X, LI L, et al. RUNX1 ameliorates rheumatoid arthritis progression through epigenetic inhibition of LRRC15 [J]. Molecules and cells, 2023, 46(4): 231-44. |
| [84] | ARTIKA I M, DEWI Y P, NAINGGOLAN I M, et al. Real-time polymerase chain reaction: current techniques, applications, and role in COVID-19 diagnosis [J]. Genes, 2022, 13(12): 2387. |
| [85] | GINZINGER D G. Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream [J]. Experimental hematology, 2002, 30(6): 503-12. |
| [86] | KIM K, RYU T Y, LEE J, et al. Epigenetic silencing of CHOP expression by the histone methyltransferase EHMT1 regulates apoptosis in colorectal cancer cells [J]. Molecules and cells, 2022, 45(9): 622-30. |
| [87] | WAGNER E M. Monitoring gene expression: quantitative real-time rt-PCR [M]. Lipoproteins and Cardiovascular Disease: Methods and Protocols. Springer. 2013: 19-45. |
| [88] | HARSHITHA R, ARUNRAJ D R. Real-time quantitative PCR: A tool for absolute and relative quantification [J]. Biochemistry and Molecular Biology Education, 2021, 49(5): 800-12. |
| [89] | WALTS A E, LECHAGO J, BOSE S. P16 and Ki67 immunostaining is a useful adjunct in the assessment of biopsies for HPV-associated anal intraepithelial neoplasia [J]. The American journal of surgical pathology, 2006, 30(7): 795-801. |
| [90] | MOTER A, GöBEL U B. Fluorescence in situ hybridization (FISH) for direct visualization of microorganisms [J]. Journal of microbiological methods, 2000, 41(2): 85-112. |
| [91] | WANG F, FLANAGAN J, SU N, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues [J]. The Journal of molecular diagnostics, 2012, 14(1): 22-9. |
| [92] | DAVID F, DAVIS A M, GOSSING M, et al. A perspective on synthetic biology in drug discovery and development—current impact and future opportunities [J]. SLAS DISCOVERY: Advancing the Science of Drug Discovery, 2021, 26(5): 581-603. |
| [93] | ABDEEN A A, COSGROVE B D, GERSBACH C A, et al. Integrating biomaterials and genome editing approaches to advance biomedical science [J]. Annual Review of Biomedical Engineering, 2021, 23(1): 493-516. |
| [94] | BLACK J B, PEREZ-PINERA P, GERSBACH C A. Mammalian synthetic biology: engineering biological systems [J]. Annual Review of Biomedical Engineering, 2017, 19(1): 249-77. |
| [95] | BHAYA D, DAVISON M, BARRANGOU R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation [J]. Annual review of genetics, 2011, 45(1): 273-97. |
| [96] | ISHINO Y, SHINAGAWA H, MAKINO K, et al. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product [J]. Journal of bacteriology, 1987, 169(12): 5429-33. |
| [97] | HILLE F, RICHTER H, WONG S P, et al. The biology of CRISPR-Cas: backward and forward [J]. Cell, 2018, 172(6): 1239-59. |
| [98] | CONG L, RAN F A, COX D, et al. Multiplex genome engineering using CRISPR/Cas systems [J]. Science, 2013, 339(6121): 819-23. |
| [99] | ZETSCHE B, GOOTENBERG J S, ABUDAYYEH O O, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system [J]. cell, 2015, 163(3): 759-71. |
| [100] | TSUCHIDA C A, ZHANG S, DOOST M S, et al. Chimeric CRISPR-CasX enzymes and guide RNAs for improved genome editing activity [J]. Molecular cell, 2022, 82(6): 1199-209. e6. |
| [101] | GHOUNEIMY A, MAHAS A, MARSIC T, et al. CRISPR-Based diagnostics: challenges and potential solutions toward point-of-care applications [J]. ACS synthetic biology, 2022, 12(1): 1-16. |
| [102] | FENG W, PENG H, XU J, et al. Integrating reverse transcription recombinase polymerase amplification with CRISPR technology for the one-tube assay of RNA [J]. Analytical Chemistry, 2021, 93(37): 12808-16. |
| [103] | YUAN A, SUN T, CHEN L, et al. CRISPR/Cas12a corona nanomachine for detecting circulating tumor nucleic acids in serum [J]. Analytical Chemistry, 2024, 96(50): 20074-81. |
| [104] | YUAN A, SHA R, XIE W, et al. RNA-activated CRISPR/Cas12a nanorobots operating in living cells [J]. Journal of the American Chemical Society, 2024, 146(39): 26657-66. |
| [105] | LI C, ZONG Y, WANG Y, et al. Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion [J]. Genome biology, 2018, 19(1): 59. |
| [106] | TAN J, ZENG D, ZHAO Y, et al. PhieABEs: a PAM‐less/free high‐efficiency adenine base editor toolbox with wide target scope in plants [J]. Plant Biotechnology Journal, 2022, 20(5): 934-43. |
| [1] | LI Jian, CHEN Yun, LIU Haiyan, TAN Zaigao. Advances in the biological utilization of one-carbon compounds [J]. Synthetic Biology Journal, 2025, (): 1-18. |
| [2] | HAN Lin, GUO Yuman, LI Yan, CAO Hengheng, LI Jiajing, YANG Minghao, WANG Mengmeng, LI Jinping, LV Yongqin. Advances in electro-microbial synergistic systems for value-added conversion of carbon dioxide [J]. Synthetic Biology Journal, 2025, (): 1-30. |
| [3] | PU Ya, JIAO Yuling. Plant artificial chromosomes: current research progress and future application perspectives [J]. Synthetic Biology Journal, 2025, (): 1-21. |
| [4] | WEI Jiaxiu, JI Peiyun, JIE Qingyu, HUANG Qiuyan, YE Hao, DAI Junbiao. Construction and application of plant artificial chromosomes [J]. Synthetic Biology Journal, 2025, (): 1-14. |
| [5] | LIU Dan, WANG Jianyu, JIANG Zhengqiang. Research progress and development trends in the biosynthesis of neutral core human milk oligosaccharides [J]. Synthetic Biology Journal, 2025, (): 1-18. |
| [6] | LIU Jie, GAO Yu, MA Yongshuo, SHANG Yi. Progress and challenges of synthetic biology in agriculture [J]. Synthetic Biology Journal, 2025, (): 1-27. |
| [7] | LIU Jiakun, YOU Di, XIANYU Yunlei, QU Qiang. Multi-party collaborative secured DNA data storage and access: toward a hybrid dna-silicon storage facility [J]. Synthetic Biology Journal, 2025, (): 1-11. |
| [8] | LAI Xia, ZHANG Yanmei, ZHANG Hongtao, DU Yuguang, ZHAN Xiaobei, CHAI Wengang. Progress on synthetic methods and applications of sialyllactose based on synthetic biology [J]. Synthetic Biology Journal, 2025, (): 1-16. |
| [9] | LI Yicheng, LUO Huiying, YAO Bin, TU Tao. Agricultural Synthetic Biology Driving Innovation in Animal Nutrition: Advances and Prospects [J]. Synthetic Biology Journal, 2025, (): 1-21. |
| [10] | ZHONG Naicai, CHEN Yuan, PAN Wenfeng, SU Xiaofeng, LIAO Jingwen, ZHAI Yinglei, ZHONG Jinyi. Application of plasma microbial breeding technology in biofabrication [J]. Synthetic Biology Journal, 2025, 6(4): 789-805. |
| [11] | FANG Xinyi, SUN Lichao, HUO Yixin, WANG Ying, YUE Haitao. Trends and challenges in microbial synthesis of higher alcohols [J]. Synthetic Biology Journal, 2025, 6(4): 873-898. |
| [12] | ZHANG Yi-Heng P. Job, CHEN Xuemei, HAN Pingping. PE and PX values in biomanufacturing: definitions and applications [J]. Synthetic Biology Journal, 2025, 6(4): 715-727. |
| [13] | WU Xiaoyan, SONG Qi, XU Rui, DING Chenjun, CHEN Fang, GUO Qing, ZHANG Bo. A comparative analysis of global research and development competition in synthetic biology [J]. Synthetic Biology Journal, 2025, 6(4): 940-955. |
| [14] | WANG Hong, LU Kongyong, ZHENG Yangyang, CHEN Tao, WANG Zhiwen. Construction and advances in the applications of transcription factor-based biosensors [J]. Synthetic Biology Journal, 2025, 6(4): 829-845. |
| [15] | SONG Xinyu, PAN Weisong, WU Tairu, PAN Jiahao, WU Chuan, LI Waichin. Research progress in plant-derived vaccines [J]. Synthetic Biology Journal, 2025, 6(4): 846-872. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||