Advances in the biological utilization of one-carbon compounds
LI Jian1,2, CHEN Yun1,2, LIU Haiyan1,2, TAN Zaigao1,2
- 1.State Key Laboratory of Microbial Metabolism,Shanghai Jiao Tong University,Shanghai 200240,China
2.Department of Bioengineering,School of Life Sciences and Biotechnology,Shanghai Jiao Tong University,Shanghai 200240,China
-
Received:2025-07-29Revised:2025-10-10Published:2025-10-13 -
Contact:TAN Zaigao
一碳化合物生物利用的合成生物学研究进展
李健1,2, 陈云1,2, 刘海艳1,2, 谭在高1,2
- 1.上海交通大学微生物代谢全国重点实验室,上海 200240
2.上海交通大学生命科学技术学院生物工程系,上海 200240
-
通讯作者:谭在高 -
作者简介:李健 (1992—),男,博士后,研究方向为植物源药物分子的合成生物学研究。E-mail:lij0813@163.com谭在高 (1987—),男,上海交通大学生命科学技术学院研究员,博士生导师,国家自然科学基金优秀青年基金获得者。研究方向为微生物细胞工厂的人工创制。以通讯作者在Nature Catalysis, Biotechnology Advances, Fundamental Research, Chemical Engineering Journal, Biotechnology for Biofuels, Synthetic and Systems Biotechnology, ACS Synthetic Biology等发表多篇论文。目前担任工业生物技术领域期刊J Ind Microbiol Biotechnol的编委。E-mail:ZTAN0918@sjtu.edu.cn -
基金资助:国家自然科学基金优秀青年基金项目(32422047);国家自然科学基金面上项目(32371482)
CLC Number:
Cite this article
LI Jian, CHEN Yun, LIU Haiyan, TAN Zaigao. Advances in the biological utilization of one-carbon compounds[J]. Synthetic Biology Journal, DOI: 10.12211/2096-8280.2025-077.
李健, 陈云, 刘海艳, 谭在高. 一碳化合物生物利用的合成生物学研究进展[J]. 合成生物学, DOI: 10.12211/2096-8280.2025-077.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2025-077
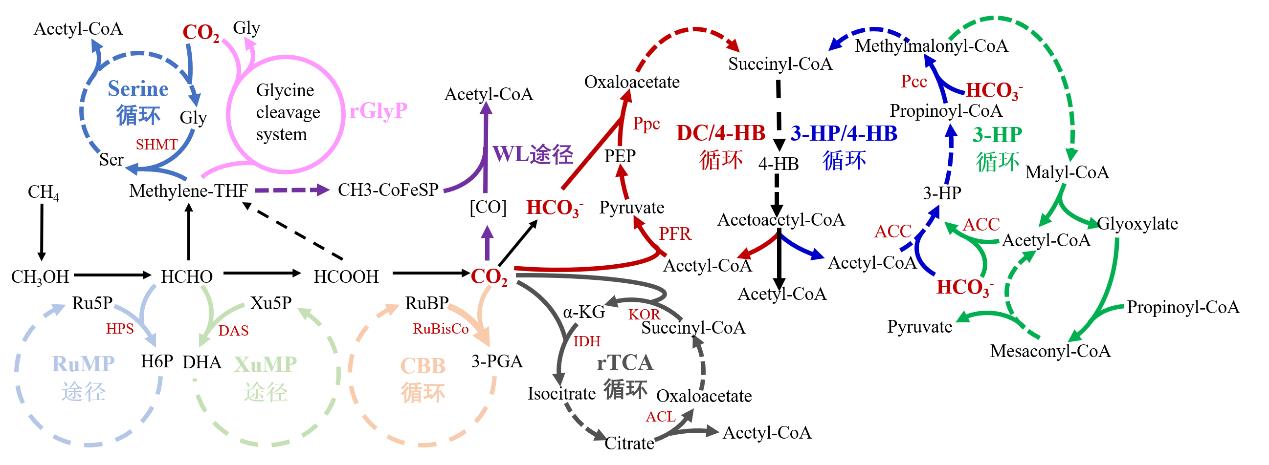
Fig. 1 Natural metabolic pathways of one-carbon compounds(Dotted lines indicate multi-step reactions. Ser, serine; Gly, glycine; PEP, phosphoenolpyruvate; DC, dicarboxylate; 4-HB, 4-hydroxybutanoate; 3-HP, 3-hydroxypropanoate; H6P, hexulose 6-phosphate; Xu5P, xylulose 5-phosphate; DHA, dihydroxyacetone; RuBP, ribulose-1,5-bisphosphate; PGA, 3-phosphoglycerate; α-KG, α-ketoglutarate; SHMT, serine hydroxymethyltransferase; Ppc, PEP carboxylase; PFR, pyruvate-ferredoxin oxidoreductase; Pcc, propionyl-CoA carboxylase; ACC, acetyl-CoA carboxylase; HPS, hexulose-6-phosphate synthase; DAS, dihydroxyacetone synthase; RuBisCo, ribulose-1, 5-bisphosphate carboxylase; IDH, isocitrate dehydrogenase; KOR, α-ketoglutarate synthase; ACL, ATP-depentent citrate lyase.)
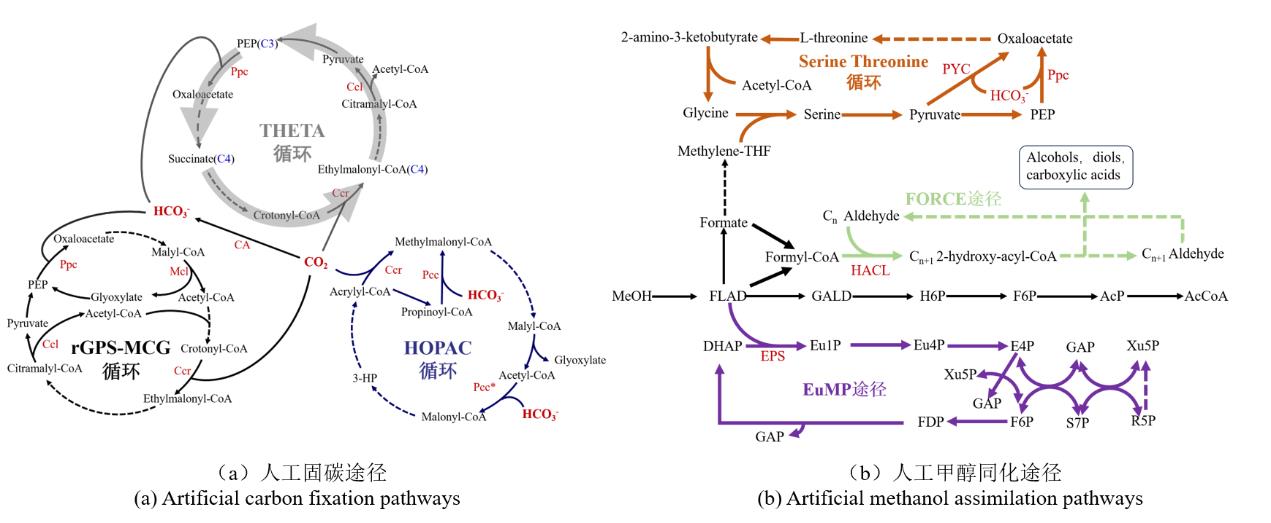
Fig. 2 Artificial metabolic pathways of one-carbon compounds((A) Artificial carbon fixation pathways. Dotted lines indicate multi-step reactions. PEP, phosphoenolpyruvate; 3-HP, 3-hydroxypropanoate; Ppc, PEP carboxylase; Ccl, citramalyl-CoA lyase; Pcc, propionyl-CoA carboxylase; Ccr, crotonyl-CoA carboxylase; Mcl, malyl-CoA lyase; CA, carbonic anhydras. (B) Artificial methanol assimilation pathways. Dotted lines indicate multi-step reactions. MeOH, methanol; FLAD, Formaldehyde; GALD, glycolaldehyde; H6P, hexose-6-phosphate; F6P, fructose 6-phosphate; AcP, acetyl-phosphate; AcCoA, acetyl-CoA; DHAP, dihydroxyacetone phosphate; Eu1P, erythrulose 1-phosphate; Eu4P, erythrulose 4-phosphate; E4P, erythrose 4-phosphate; GAP, glyceraldehyde 3-phosphate; Ru5P, ribulose 5-phosphate; Xu5P, xylulose 5-phosphate; R5P, ribose 5-phosphate; S7P, sedoheptulose 7-phosphate; FDP, fructose-1,6-bisphosphate; PYC, pyruvate carboxylase; HACL, 2-hydroxyacyl-CoA lyase; EPS, erythrulose 1-phosphate synthase.)
| Pathways | C1 substrates | Products | ATP consumption | NAD(P)H consumption | steps | others |
|---|---|---|---|---|---|---|
| XuMP | Methanol | GAP | 1 | 0 | 10 | - |
| RuMP | Methanol | GAP | 1 | 0 | 8 | - |
| rGlyP | Formate、CO2 | Acetyl-CoA | 1 | 3 | 7 | - |
| SC | Formate、CO2 | Acetyl-CoA | 3 | 3 | 12 | - |
| STC | Formate、CO2 | Acetyl-CoA | 4 | 4 | 14 | Adaptability to low CO2 concentration |
| FORCE | Methanol | Organic acid | 0 | - | - | Wide product spectrum |
| EuMP | Methanol | GAP | 1 | 0 | 11 | - |
| SMA | Methanol | Acetyl-CoA | 0 | 0 | 6 | No ATP consumed |
Table 1 Comparison between artificial and natural pathways of methylotrophy
| Pathways | C1 substrates | Products | ATP consumption | NAD(P)H consumption | steps | others |
|---|---|---|---|---|---|---|
| XuMP | Methanol | GAP | 1 | 0 | 10 | - |
| RuMP | Methanol | GAP | 1 | 0 | 8 | - |
| rGlyP | Formate、CO2 | Acetyl-CoA | 1 | 3 | 7 | - |
| SC | Formate、CO2 | Acetyl-CoA | 3 | 3 | 12 | - |
| STC | Formate、CO2 | Acetyl-CoA | 4 | 4 | 14 | Adaptability to low CO2 concentration |
| FORCE | Methanol | Organic acid | 0 | - | - | Wide product spectrum |
| EuMP | Methanol | GAP | 1 | 0 | 11 | - |
| SMA | Methanol | Acetyl-CoA | 0 | 0 | 6 | No ATP consumed |
| Hosts | C1 pathway | Safety | Doubling Time | Features |
|---|---|---|---|---|
| S. elongatus | CBB | To be evaluated | 6-12 h | Photoautotrophic, salt-tolerant |
| C. reinhardtii | CBB | GRAS | 6-8 h | Photoautotrophic, eukaryotic expression system |
| C. necator | CBB | Industrial safety | 4-6 h | facultative autotrophy, high metabolic flexibility, clear genetic background |
| K. phaffii | XuMP | GRAS | 2-3 h | Efficient expression of proteins, high density fermentation, natural methylotroph |
| M. extorquens | SC | Industrial safety | 3-5 h | Natural methylotroph, clear genetic background |
| B. methanolicus | RuMP | Industrial safety | 1-1.5 h | Natural methylotroph, high temperature resistance |
| O. polymorpha | XuMP | GRAS | 1.5-2 h | Wide substrate spectrum, natural methylotroph, high robustness |
| E. coli | RuMP、FORCE、SMA、rGlyP、EuMP、STC | Industrial safety | 20-30 min | Clear genetic background, rapid growth, wide substrates spectrum |
| S. cerevisiae | XuMP、RuMP、rGlyP | GRAS | 1.5-2 h | Clear genetic background, eukaryotic expression system, wide products spectrum, |
| Y. lipolytica | XuMP | GRAS | 1.5-2 h | high lipid synthesis flux |
| S. marcescens | XuMP | Opportunistic infection | 0.5-1 h | High robustness |
| Cell free | THETA、ACSP | - | - | High orthogonality, high efficiency |
Table 2 The hosts of C1 utilization pathways mentioned in this paper
| Hosts | C1 pathway | Safety | Doubling Time | Features |
|---|---|---|---|---|
| S. elongatus | CBB | To be evaluated | 6-12 h | Photoautotrophic, salt-tolerant |
| C. reinhardtii | CBB | GRAS | 6-8 h | Photoautotrophic, eukaryotic expression system |
| C. necator | CBB | Industrial safety | 4-6 h | facultative autotrophy, high metabolic flexibility, clear genetic background |
| K. phaffii | XuMP | GRAS | 2-3 h | Efficient expression of proteins, high density fermentation, natural methylotroph |
| M. extorquens | SC | Industrial safety | 3-5 h | Natural methylotroph, clear genetic background |
| B. methanolicus | RuMP | Industrial safety | 1-1.5 h | Natural methylotroph, high temperature resistance |
| O. polymorpha | XuMP | GRAS | 1.5-2 h | Wide substrate spectrum, natural methylotroph, high robustness |
| E. coli | RuMP、FORCE、SMA、rGlyP、EuMP、STC | Industrial safety | 20-30 min | Clear genetic background, rapid growth, wide substrates spectrum |
| S. cerevisiae | XuMP、RuMP、rGlyP | GRAS | 1.5-2 h | Clear genetic background, eukaryotic expression system, wide products spectrum, |
| Y. lipolytica | XuMP | GRAS | 1.5-2 h | high lipid synthesis flux |
| S. marcescens | XuMP | Opportunistic infection | 0.5-1 h | High robustness |
| Cell free | THETA、ACSP | - | - | High orthogonality, high efficiency |
| Hosts | C1 pathway | Carbon source | Products | Titer | Ref. |
|---|---|---|---|---|---|
| S. elongatus | CBB | CO2 | Mannitol | 701 mg/L | [ |
| CBB | CO2 | α-Farnesene | 12.87 mg/L | [ | |
| CBB | CO2 | Sucrose | 3.8 g/L | [ | |
| C. reinhardtii | CBB | CO2 | Limonene | 117 µg/L | [ |
| CBB | CO2 | Astaxanthin | 23.5 mg/L | [ | |
| CBB | CO2 | Lipid | 672 mg/L | [ | |
| C. reinhardtii + E.coli | CBB | CO2 | Lycopene | 1.48 mg/L | [ |
| C. necator | CBB | CO2 | N-acetylglucosamine | 75.3 mg/L | [ |
| CBB | CO2 | Myoinositol | 1054.8 mg/L | [ | |
| CBB | CO2 | L-isoleucine | 105 mg/L | [ | |
| CBB | CO2 | Valine | 319 mg/L | [ | |
| S. elongatus + V. natriegens | CBB | CO2 | Lactate | 472.1 mg/L | [ |
| S. elongatus + E. coli | CBB | CO2 | 3-HP | 120.3 mg/L | [ |
| S. elongatus + E. coli | CBB | CO2 | Pinocembrin | 152.7 mg/L | [ |
| B. methylotrophicum | WL | Methanol, HCO3- | Butyric acid | 3.69 g/L | [ |
| K. phaffii | XuMP | Methanol | α-Bisabolene | 1.1 g/L | [ |
| XuMP | Methanol | Zealexin A1 | 102.5 mg/L | [ | |
| XuMP | Methanol | Lactate | 5.18 g/L | [ | |
| XuMP | Methanol | Itaconic acid | 28 g/L | [ | |
| XuMP | Methanol | Fatty alcohol | 5.6 g/L | [ | |
| XuMP | Methanol | Fatty acid | 23.4 g/L | [ | |
| rGlyP | Methanol, HCO3- | Fatty alcohol | 0.21 g/L | [ | |
| XuMP-RuMP | Methanol | Erythritol | 31.5 g/L | [ | |
| XuMP | Methanol | Cordycepin | 8.11 g/L | [ | |
| XuMP | Methanol | 3-HP | 27 g/L | [ | |
| XuMP | Methanol | single cell protein | 0.506 g/g DCW | [ | |
| M. extorquens | SC | Methanol | 3-HP | 1.75 g/L | [ |
| SC | Methanol | Polyhydroxyalkanoate | 11.07 g/L | [ | |
| RuMP | Methanol | Riboflavin | 2579 mg/L | [ | |
| E. coli | RuMP | Methanol | Itaconic acid | 1 g/L | [ |
| RuMP | Methanol, xylose | D-allulose | 98 mM | [ | |
| rGlyP | Formate, CO2 | Lactate | 1.2 mM | [ | |
| RuMP | Methanol, xylose | 3-HP | 437 mg/L | [ | |
| RuMP | Methanol, xylose | D-glucaric acid | 3.0 g/L | [ | |
| FORCE | Methanol | Glycolate | 5.2 g/L | [ | |
| O. polymorpha | XuMP | Methanol | Succinate | 0.35 g/L | [ |
| XuMP | Methanol | Malate | 13 g/L | [ | |
| XuMP | Methanol | 3-HP | 7.10 g/L | [ | |
| XuMP | Methanol | Fatty alcohol | 3.6 g/L | [ | |
| XuMP | Methanol | Fatty acid | 15.9 g/L | [ | |
| XuMP | Methanol | Lactate | 3.8 g/L | [ | |
| S. cerevisiae | XuMP | Methanol, CO2, 0.1% yeast extract | Cannabigerolic acid | 18 μg/L | [ |
| rGlyP | Methanol, HCO3- | 5-aminolevulinic acid | 1.67 mg/L | [ | |
| S. marcescens | XuMP | Methanol, xylose | Bisabolol | 1256.41 mg/L | [ |
| Cell free | THETA | CO2, HCO3- | Glyoxylate | 760.3 μM | [ |
| ACSP | CO2 | Glucose | 11.4 g/L | [ | |
| ACSP | CO2 | sucrose | 14 g/L | [ |
Table 3 Progress in biomanufacturing research based on C1
| Hosts | C1 pathway | Carbon source | Products | Titer | Ref. |
|---|---|---|---|---|---|
| S. elongatus | CBB | CO2 | Mannitol | 701 mg/L | [ |
| CBB | CO2 | α-Farnesene | 12.87 mg/L | [ | |
| CBB | CO2 | Sucrose | 3.8 g/L | [ | |
| C. reinhardtii | CBB | CO2 | Limonene | 117 µg/L | [ |
| CBB | CO2 | Astaxanthin | 23.5 mg/L | [ | |
| CBB | CO2 | Lipid | 672 mg/L | [ | |
| C. reinhardtii + E.coli | CBB | CO2 | Lycopene | 1.48 mg/L | [ |
| C. necator | CBB | CO2 | N-acetylglucosamine | 75.3 mg/L | [ |
| CBB | CO2 | Myoinositol | 1054.8 mg/L | [ | |
| CBB | CO2 | L-isoleucine | 105 mg/L | [ | |
| CBB | CO2 | Valine | 319 mg/L | [ | |
| S. elongatus + V. natriegens | CBB | CO2 | Lactate | 472.1 mg/L | [ |
| S. elongatus + E. coli | CBB | CO2 | 3-HP | 120.3 mg/L | [ |
| S. elongatus + E. coli | CBB | CO2 | Pinocembrin | 152.7 mg/L | [ |
| B. methylotrophicum | WL | Methanol, HCO3- | Butyric acid | 3.69 g/L | [ |
| K. phaffii | XuMP | Methanol | α-Bisabolene | 1.1 g/L | [ |
| XuMP | Methanol | Zealexin A1 | 102.5 mg/L | [ | |
| XuMP | Methanol | Lactate | 5.18 g/L | [ | |
| XuMP | Methanol | Itaconic acid | 28 g/L | [ | |
| XuMP | Methanol | Fatty alcohol | 5.6 g/L | [ | |
| XuMP | Methanol | Fatty acid | 23.4 g/L | [ | |
| rGlyP | Methanol, HCO3- | Fatty alcohol | 0.21 g/L | [ | |
| XuMP-RuMP | Methanol | Erythritol | 31.5 g/L | [ | |
| XuMP | Methanol | Cordycepin | 8.11 g/L | [ | |
| XuMP | Methanol | 3-HP | 27 g/L | [ | |
| XuMP | Methanol | single cell protein | 0.506 g/g DCW | [ | |
| M. extorquens | SC | Methanol | 3-HP | 1.75 g/L | [ |
| SC | Methanol | Polyhydroxyalkanoate | 11.07 g/L | [ | |
| RuMP | Methanol | Riboflavin | 2579 mg/L | [ | |
| E. coli | RuMP | Methanol | Itaconic acid | 1 g/L | [ |
| RuMP | Methanol, xylose | D-allulose | 98 mM | [ | |
| rGlyP | Formate, CO2 | Lactate | 1.2 mM | [ | |
| RuMP | Methanol, xylose | 3-HP | 437 mg/L | [ | |
| RuMP | Methanol, xylose | D-glucaric acid | 3.0 g/L | [ | |
| FORCE | Methanol | Glycolate | 5.2 g/L | [ | |
| O. polymorpha | XuMP | Methanol | Succinate | 0.35 g/L | [ |
| XuMP | Methanol | Malate | 13 g/L | [ | |
| XuMP | Methanol | 3-HP | 7.10 g/L | [ | |
| XuMP | Methanol | Fatty alcohol | 3.6 g/L | [ | |
| XuMP | Methanol | Fatty acid | 15.9 g/L | [ | |
| XuMP | Methanol | Lactate | 3.8 g/L | [ | |
| S. cerevisiae | XuMP | Methanol, CO2, 0.1% yeast extract | Cannabigerolic acid | 18 μg/L | [ |
| rGlyP | Methanol, HCO3- | 5-aminolevulinic acid | 1.67 mg/L | [ | |
| S. marcescens | XuMP | Methanol, xylose | Bisabolol | 1256.41 mg/L | [ |
| Cell free | THETA | CO2, HCO3- | Glyoxylate | 760.3 μM | [ |
| ACSP | CO2 | Glucose | 11.4 g/L | [ | |
| ACSP | CO2 | sucrose | 14 g/L | [ |
| [1] | O'KEEFFE S, GARCIA L, CHEN Y, et al. Bringing carbon to life via one-carbon metabolism [J]. Trends in Biotechnology, 2025, 43(3): 572-585. |
| [2] | 高教琪, 周雍进. 甲醇生物转化的机遇与挑战 [J]. 合成生物学, 2020, 1: 158-173. |
| GAO J Q, ZHOU Y J. Advances in methanol bio-transformation [J]. Synthetic biology Journal, 2020, 1: 572-585. | |
| [3] | MENG X, HU G, LI X, et al. A synthetic methylotroph achieves accelerated cell growth by alleviating transcription-replication conflicts [J]. Nature Communications, 2025, 16(1): 31 |
| [4] | MENG Q, WANG D, FU X, et al. Converting Bacillus subtilis 168 to a synthetic methylotroph by combinatorial metabolic regulation strategies [J]. Journal of Agricultural and Food Chemistry, 2025, 73(8): 4755-4763. |
| [5] | NIEH L Y, CHEN F Y H, JUNG H W, et al. Evolutionary engineering of methylotrophic E. coli enables fast growth on methanol [J]. Nature Communications, 2024, 15(1): 8840. |
| [6] | ZHAN C J, LI X, LAN G, et al. Reprogramming methanol utilization pathways to convert Saccharomyces cerevisiae to a synthetic methylotroph [J]. Nature Catalysis, 2023, 6(5): 435-450. |
| [7] | REITER M A, BRADLEY T, BüCHEL L A, et al. A synthetic methylotrophic Escherichia coli as a chassis for bioproduction from methanol [J]. Nature Catalysis, 2024, 7(5): 560-573. |
| [8] | KRÜSEMANN J L, LINDNER S N. Bioproduction from methanol [J]. Nature Catalysis, 2024, 7(5): 472-474. |
| [9] | CAI P, WU X Y, DENG J, et al. Methanol biotransformation toward high-level production of fatty acid derivatives by engineering the industrial yeast Pichia pastoris [J]. Proceedings of the National Academy of Sciences, 2022, 119: e2201711119. |
| [10] | MENG J, LIU S, GAO L, et al. Economical production of Pichia pastoris single cell protein from methanol at industrial pilot scale [J]. Microbial Cell Factories, 2023, 22(1): 198. |
| [11] | WU X Y, REN Y Y, CHEN S S, et al. Production of L-lactic acid from methanol by engineered yeast Pichia pastoris [J]. Bioresource Technology, 2025, 415: 131730. |
| [12] | SHEN Y W, CAI P, GAO L H, et al. Engineering high production of fatty alcohols from methanol by constructing coordinated dual biosynthetic pathways [J]. Bioresource Technology, 2024, 412: 131396. |
| [13] | ZHAI X X, GAO J Q, LI X Y, et al. Peroxisomal metabolic coupling improves fatty alcohol production from sole methanol in yeast [J]. Proceedings of the National Academy of Sciences, 2023, 120: e2220816120. |
| [14] | WEFELMEIER K, SCHMITZ S, KöSTERS B J, et al. Methanol bioconversion into C3, C4, and C5 platform chemicals by the yeast Ogataea polymorpha [J]. Microbial Cell Factories, 2024, 23(1): 8. |
| [15] | KANG N K, CHAU T H T, LEE E Y. Engineered methane biocatalysis: strategies to assimilate methane for chemical production [J]. Current Opinion in Biotechnology, 2024, 85: 103031. |
| [16] | KELLER P, REITER M A, KIEFER P, et al. Generation of an Escherichia coli strain growing on methanol via the ribulose monophosphate cycle [J]. Nature Communications, 2022, 13: 5423. |
| [17] | WENK S, RAINALDI V, SCHANN K, et al. Evolution-assisted engineering of E. coli enables growth on formic acid at ambient CO2 via the serine threonine cycle [J]. Metabolic Engineering, 2025, 88: 14-24. |
| [18] | GREGORY G J, BENNETT R K, PAPOUTSAKIS E T. Recent advances toward the bioconversion of methane and methanol in synthetic methylotrophs [J]. Metabolic Engineering, 2022, 71: 99-116. |
| [19] | LIU L N. Advances in the bacterial organelles for CO2 fixation [J]. Trends in Microbiology, 2022, 30(6): 567-580. |
| [20] | LUO S S, DIEHL C, HE H, et al. Construction and modular implementation of the THETA cycle for synthetic CO2 fixation [J]. Nature Catalysis, 2023, 6(12): 1228-1240. |
| [21] | SATANOWSKI A, MARCHAL D G, PERRET A, et al. Design and implementation of aerobic and ambient CO2-reduction as an entry-point for enhanced carbon fixation [J]. Nature Communications, 2025, 16(1): 3134. |
| [22] | MCLEAN R, Schwander T, CHRISTOPH DIEHL, et al. Exploring alternative pathways for the in vitro establishment of the HOPAC cycle for synthetic CO2 fixation [J]. Science Advances, 2023, 9(24): eadh4299. |
| [23] | LUO S S, LIN P P, NIEH LY, et al. A cell-free self-replenishing CO2-fixing system [J]. Nature Catalysis, 2022, 5(2): 154-162. |
| [24] | FENG J, LI X, TENG X, et al. Harnessing CO2 fixation and reducing power recycling for enhanced polyhydroxyalkanoates industrial bioproduction [J]. Metabolic Engineering, 2025, 91: 204-216. |
| [25] | ZHU P, CHEN X. Converting heterotrophic Escherichia coli into synthetic C1-trophic modes [J]. Trends in Chemistry, 2022, 4(10): 860-862. |
| [26] | LI C, ZHENG H, LI Y, et al. Facilitated channeling of fixed carbon and energy into chemicals in artificial phototrophic communities [J]. Journal of the American Chemical Society, 2025, 147(6): 4707-4713. |
| [27] | FENG J, MA D, GAO S, et al. Recent advances in engineering heterotrophic microorganisms for reinforcing CO2 fixation based on Calvin-Benson-Bassham Cycle [J]. ACS Sustainable Chemistry & Engineering, 2023, 11(26): 9509-9522. |
| [28] | PENG J H, LO S C, YU Y N, et al. Carbon fluxes rewiring in engineered E. coli via reverse tricarboxylic acid cycle pathway under chemolithotrophic condition [J]. Journal of Biological Engineering, 2025, 19(1): 20. |
| [29] | CUI Z Y, ZHONG Y T, SUN Z J, et al. Reconfiguration of the reductive TCA cycle enables high-level succinic acid production by Yarrowia lipolytica [J]. Nature Communications, 2023, 14: 8480. |
| [30] | PAN J, ZHANG X X, XU W, et al. Wood–Ljungdahl pathway found in novel marine Korarchaeota groups illuminates their evolutionary history [J]. Msystems, 2023, 8(4): e00305-23. |
| [31] | MATTOZZI M D, ZIESACK M, VOGES M J, et al. Expression of the sub-pathways of the Chloroflexus aurantiacus 3-hydroxypropionate carbon fixation bicycle in E. coli: Toward horizontal transfer of autotrophic growth [J]. Metabolic Engineering, 2013, 16: 130-139. |
| [32] | QIN N, LI L Y, WAN X Z, et al. Increased CO2 fixation enables high carbon-yield production of 3-hydroxypropionic acid in yeast [J]. Nature Communications, 2024, 15: 1591. |
| [33] | SIMONE GIAVERI, NITIN BOHRA, CHRISTOPH DIEHL, et al. Integrated translation and metabolism in a partially self-synthesizing biochemical network [J]. Science, 385(6705): 174-178. |
| [34] | HARALD H, Martin G, ULRIKE J, et al. A dicarboxylate/4-hydroxybutyrate autotrophic carbon assimilation cycle in the hyperthermophilic Archaeum Ignicoccus hospitalis [J]. Proceedings of the National Academy of Sciences, 2008, 105(22): 7851-7856. |
| [35] | SARWAR A, LEE E Y. Methanol-based biomanufacturing of fuels and chemicals using native and synthetic methylotrophs [J]. Synthetic and Systems Biotechnology, 2023, 8(3): 396-415. |
| [36] | YISHAI O, LINDNER S N, GONZALEZ DE LA CRUZ J, et al. The formate bio-economy [J]. Current Opinion in Chemical Biology, 2016, 35: 1-9. |
| [37] | JIA M, LIU M, LI J, et al. Formaldehyde: an essential intermediate for C1 metabolism and bioconversion [J]. ACS Synthetic Biology, 2024, 13(11): 3507-3522. |
| [38] | WANG Y Y, LI R S, ZHAO F G, et al. Metabolic engineering of Komagataella phaffii for the efficient utilization of methanol [J]. Microbial Cell Factories, 2024, 23(1): 198. |
| [39] | DRONSELLA B, ORSI E, SCHULZ-MIRBACH H, et al. One-carbon fixation via the synthetic reductive glycine pathway exceeds yield of the Calvin cycle [J]. Nature Microbiology, 2025, 10(3): 646-653. |
| [40] | BYSANI V R, ALAM A S, BAR-EVEN A, et al. Engineering and evolution of the complete reductive glycine pathway in Saccharomyces cerevisiae for formate and CO2 assimilation [J]. Metabolic Engineering, 2024, 81: 167-181. |
| [41] | CHOU A, LEE S H, ZHU F, et al. An orthogonal metabolic framework for one-carbon utilization [J]. Nature Metabolism, 2021, 3(10): 1385-1399. |
| [42] | SEUNG HWAN LEE A C, MAREN NATTERMANN, FAYIN ZHU, CLOMBURG JAMES M., NICOLE PACZIA, TOBIAS J. ERB, RAMON GONZALEZ. Identification of 2-hydroxyacyl-CoA synthases with high acyloin condensation activity for orthogonal one-carbon bioconversion [J]. ACS Catalysis, 2023, 13: 12007-12020. |
| [43] | WU T, GÓMEZ-CORONADO P A, KUBIS A, et al. Engineering a synthetic energy-efficient formaldehyde assimilation cycle in Escherichia coli [J]. Nature Communications, 2023, 14(1): 8490. |
| [44] | LU X Y, LIU Y W, YANG Y Y, et al. Constructing a synthetic pathway for acetyl-coenzyme A from one-carbon through enzyme design [J]. Nature Communications, 2019, 10(1): 1378. |
| [45] | DANG B T, BUI X T, TRAN D P H, et al. Current application of algae derivatives for bioplastic production: A review [J]. Bioresource Technology, 2022, 347: 126698. |
| [46] | MEHDIZADEH ALLAF M, PEERHOSSAINI H. Cyanobacteria: model microorganisms and beyond [J]. Microorganisms, 2022, 10(4): 696. |
| [47] | PESSOA J D S, DE OLIVEIRA C F M, MENA‐CHALCO J P, et al. Trends on Chlamydomonas reinhardtii growth regimes and bioproducts [J]. Biotechnology and Applied Biochemistry, 2023, 70(6): 1830-1842. |
| [48] | DONATI S, JOHNSON C W. Optimizing Cupriavidus necator H16 as a host for aerobic C1 conversion [J]. Current Opinion in Biotechnology, 2025, 93. |
| [49] | JIANG J, LI X, YANG K, et al. Photosynthetic cultivation of Chlamydomonas reinhardtii with formate as a novel carbon source to the protein production [J]. Chemical Engineering Journal, 2024, 493: 152518. |
| [50] | MORLINO M S, SERNA GARCíA R, SAVIO F, et al. Cupriavidus necator as a platform for polyhydroxyalkanoate production: An overview of strains, metabolism, and modeling approaches [J]. Biotechnology Advances, 2023, 69: 108264. |
| [51] | MADHU S, SENGUPTA A, SARNAIK A P, et al. Expanding the synthetic biology repertoire of a fast‐growing cyanobacterium Synechococcus elongatus PCC 11801 [J]. Biotechnology and Bioengineering, 2024, 121(9): 2974-2980. |
| [52] | LI Z, LI S, CHEN L, et al. Fast-growing cyanobacterial chassis for synthetic biology application [J]. Critical Reviews in Biotechnology, 2023, 44(3): 414-428. |
| [53] | SANTOS-MERINO M, GUTIéRREZ-LANZA R, NOGALES J, et al. Synechococcus elongatus PCC 7942 as a platform for bioproduction of omega-3 fatty acids [J]. Life, 2022, 12(6): 810. |
| [54] | PRITAM P, SARNAIK A P, WANGIKAR P P. Metabolic engineering of Synechococcus elongatus for photoautotrophic production of mannitol [J]. Biotechnology and Bioengineering, 2023, 120(8): 2363-2370. |
| [55] | WANG B, Zuniga Cristal, GUARNIERI M T, et al. Metabolic engineering of Synechococcus elongatus 7942 for enhanced sucrose biosynthesis [J]. Metabolic Engineering, 2023, 80: 12-24. |
| [56] | RAUTELA A, YADAV I, GANGWAR A, et al. Photosynthetic production of α-farnesene by engineered Synechococcus elongatus UTEX 2973 from carbon dioxide [J]. Bioresource Technology, 2024, 396: 130432. |
| [57] | MA K, DENG L, WU H Z, et al. Towards green biomanufacturing of high-value recombinant proteins using promising cell factory: Chlamydomonas reinhardtii chloroplast [J]. Bioresources and Bioprocessing, 2022, 9(1): 1-14. |
| [58] | MASI A, LEONELLI F, SCOGNAMIGLIO V, et al. Chlamydomonas reinhardtii: a factory of nutraceutical and food supplements for human health [J]. Molecules, 2023, 28(3): 1185. |
| [59] | DELLA VALLE S, ORSI E, CREUTZBURG S C A, et al. Streamlined and efficient genome editing in Cupriavidus necator H16 using an optimised SIBR-Cas system [J]. Trends in Biotechnology, 2025, 43: 1470-1491. |
| [60] | YANG X T, ZHENG Z J, WANG Y. Bacillus methanolicus: an emerging chassis for low-carbon biomanufacturing [J]. Trends in Biotechnology, 2025, 43(2): 274-277. |
| [61] | XIE L F, YU W, GAO J Q, et al. Ogataea polymorpha as a next-generation chassis for industrial biotechnology [J]. Trends in Biotechnology, 2024, 42(11): 1363-1378. |
| [62] | GUO F, QIAO Y, XIN F, et al. Bioconversion of C1 feedstocks for chemical production using Pichia pastoris [J]. Trends in Biotechnology, 2023, 41(8): 1066-1079. |
| [63] | WU X Y, CAI P, YAO L, et al. Genetic tools for metabolic engineering of Pichia pastoris [J]. Engineering Microbiology, 2023, 3(4): 100094. |
| [64] | OCHSNER A M, SONNTAG F, BUCHHAUPT M, et al. Methylobacterium extorquens: methylotrophy and biotechnological applications [J]. Applied Microbiology and Biotechnology, 2014, 99(2): 517-534. |
| [65] | MüLLER J E N, HEGGESET T M B, WENDISCH V F, et al. Methylotrophy in the thermophilic Bacillus methanolicus, basic insights and application for commodity production from methanol [J]. Applied Microbiology and Biotechnology, 2014, 99(2): 535-551. |
| [66] | NI X, ZHAI X X, YU W, et al. Dynamically regulating homologous recombination enables precise genome editing in Ogataea polymorpha [J]. ACS Synthetic Biology, 2024, 13(9): 2938-2947. |
| [67] | JIA W, POUVREAU L, VAN DER GOOT A J, et al. Renewable methanol utilizing bacteria as future meat analogue: An explorative study on the physicochemical and texturing properties of Methylobacillus flagellatus biomass and fractions [J]. Food Hydrocolloids, 2024, 151: 109832. |
| [68] | XU D Y, LEUNG K M, LAI G K K, et al. Complete genome sequence of Klebsiella pneumoniae RX.G5M15, a methanol-metabolizing strain recovered from the sole of a shoe [J]. Microbiology Resource Announcements, 2023, 13(9): e00451-24. |
| [69] | PONTRELLI S, CHIU T Y, LAN E I, et al. Escherichia coli as a host for metabolic engineering [J]. Metabolic Engineering, 2018, 50: 16-46. |
| [70] | SANFORD P A, WOOLSTON B M. Synthetic or natural? Metabolic engineering for assimilation and valorization of methanol [J]. Current Opinion in Biotechnology, 2022, 74: 171-179. |
| [71] | WEGAT V, FABARIUS J T, SIEBER V. Synthetic methylotrophic yeasts for the sustainable fuel and chemical production [J]. Biotechnology for Biofuels and Bioproducts, 2022, 15: 113. |
| [72] | DELMAS V A, PERCHAT N, MONET O, et al. Genetic and biocatalytic basis of formate dependent growth of Escherichia coli strains evolved in continuous culture [J]. Metabolic Engineering, 2022, 72: 200-214. |
| [73] | SUN Q, LIU D, CHEN Z. Engineering and adaptive laboratory evolution of Escherichia coli for improving methanol utilization based on a hybrid methanol assimilation pathway [J]. Frontiers in Bioengineering and Biotechnology, 2023, 10: 1089639. |
| [74] | GUO Y, ZHANG R, WANG J, et al. Engineering yeasts to Co-utilize methanol or formate coupled with CO2 fixation [J]. Metabolic Engineering, 2024, 84: 1-12. |
| [75] | QI M, ZHU C, CHENG C, et al. Rewiring methanol assimilation and reductive glycine pathways in Saccharomyces cerevisiae to increase one-carbon recovery [J]. Green Chemistry, 2025, 27(12): 3261-3271. |
| [76] | LIU D, WANG L, Gou L B, et al. Hybrid Methylotrophic pathway in Serratia marcescens for sustainable terpenoid biosynthesis [J]. ACS Synthetic Biology, 2025, 14(5): 1766-1776. |
| [77] | ZHANG S, GUO F, YANG Q, et al. Improving methanol assimilation in Yarrowia lipolytica via systematic metabolic engineering combined with compartmentalization [J]. Green Chemistry, 2023, 25(1): 183-195. |
| [78] | GAO B, ZHAO N, DENG J, et al. Constructing a methanol-dependent Bacillus subtilis by engineering the methanol metabolism [J]. Journal of Biotechnology, 2022, 343: 128-137. |
| [79] | YANG J G, SONG W, CAI T, et al. De novo artificial synthesis of hexoses from carbon dioxide [J]. Science Bulletin, 2023, 68(20): 2370-2381. |
| [80] | WANG Y Y, CHEN P, LI W W, et al. Cell-free synthesis of high-order carbohydrates from low-carbon molecules [J]. Science Bulletin, 2025, 70(14): 2266-2276. |
| [81] | YUN L, ZEGARAC R, DUCAT D C. Impact of irradiance and inorganic carbon availability on heterologous sucrose production in Synechococcus elongatus PCC 7942 [J]. Frontiers in Plant Science, 2024, 15: 1378573. |
| [82] | ZHAO M L, CAI W S, ZHENG S Q, et al. Metabolic engineering of the isopentenol utilization pathway enhanced the production of terpenoids in Chlamydomonas reinhardtii [J]. Marine Drugs, 2022, 20(9): 577. |
| [83] | AMENDOLA S, KNEIP J S, MEYER F, et al. Metabolic engineering for efficient ketocarotenoid accumulation in the green microalga Chlamydomonas reinhardtii [J]. ACS Synthetic Biology, 2023, 12(3): 820-831. |
| [84] | LIN J Y, WAHYU EFFENDI S SRI, NG I S. Enhanced carbon capture and utilization (CCU) using heterologous carbonic anhydrase in Chlamydomonas reinhardtii for lutein and lipid production [J]. Bioresource Technology, 2022, 351: 127009. |
| [85] | KANG N K, KOH H G, CHOI Y, et al. Bioconversion of CO2 into valuable bioproducts via synthetic modular co-culture of engineered Chlamydomonas reinhardtii and Escherichia coli [J]. Metabolic Engineering, 2025, 90: 57-66. |
| [86] | WANG X L, CHANG F F, WANG T T, et al. Production of N-acetylglucosamine from carbon dioxide by engineering Cupriavidus necator H16 [J]. Bioresource Technology, 2023, 379: 129024. |
| [87] | WANG X L, WANG K K, WANG L, et al. Engineering Cupriavidus necator H16 for heterotrophic and autotrophic production of myo-inositol [J]. Bioresource Technology, 2023, 368: 128321. |
| [88] | WANG L, YAO J H, TU T, et al. Heterotrophic and autotrophic production of L-isoleucine and L-valine by engineered Cupriavidus necator H16 [J]. Bioresource Technology, 2024, 398: 130538. |
| [89] | LI C F, WANG R Y, WANG J W, et al. A highly compatible phototrophic community for carbon‐negative biosynthesis [J]. Angewandte Chemie International Edition, 2022. |
| [90] | LI C F, YIN L, WANG J W, et al. Light-driven biosynthesis of volatile, unstable and photosensitive chemicals from CO2 [J]. Nature Synthesis, 2023, 2(10): 960-971. |
| [91] | WANG J, LIAO Y, QIN J, et al. Increasing lysine level improved methanol assimilation toward butyric acid production in Butyribacterium methylotrophicum [J]. Biotechnology for Biofuels and Bioproducts, 2023, 16: 10. |
| [92] | GAO L, HOU R, CAI P, et al. Engineering yeast peroxisomes for α-bisabolene production from sole methanol with the aid of proteomic analysis [J]. JACS Au, 2024, 4(7): 2474-2483. |
| [93] | NIU T, YAN X, WANG J, et al. Engineering of Pichia pastoris for the de novo synthesis of the sesquiterpene zealexin a1 from methanol [J]. ACS Sustainable Chemistry & Engineering, 2024, 12(34): 12786-12794. |
| [94] | INOUE Y, YAMADA R, MATSUMOTO T, et al. Enhancing D-lactic acid production by optimizing the expression of D-LDH gene in methylotrophic yeast Komagataella phaffii [J]. Biotechnology for Biofuels and Bioproducts, 2024, 17(1): 149. |
| [95] | SEVERINSEN M M, BACHLEITNER S, MODENESE V, et al. Efficient production of itaconic acid from the single-carbon substrate methanol with engineered Komagataella phaffii [J]. Biotechnology for Biofuels and Bioproducts, 2024, 17(1): 98. |
| [96] | WANG S X, FANG J Y, WANG M Y, et al. Rewiring the methanol assimilation pathway in the methylotrophic yeast Pichia pastoris for high-level production of erythritol [J]. Bioresource Technology, 2025, 427: 132430. |
| [97] | ZHAO B J, LI Y, ZHANG Y, et al. Low-carbon and overproduction of cordycepin from methanol using engineered Pichia pastoris cell factory [J]. Bioresource Technology, 2024, 413: 131446. |
| [98] | ÀVILA-CABRé S, ALBIOL J, FERRER P. Metabolic engineering of Komagataella phaffii for enhanced 3-hydroxypropionic acid (3-HP) production from methanol [J]. Journal of Biological Engineering, 2025, 19: 19. |
| [99] | MA Z X, FENG C X, SONG Y Z, et al. Engineering photo-methylotrophic Methylobacterium for enhanced 3-hydroxypropionic acid production during non-growth stage fermentation [J]. Bioresource Technology, 2024, 393: 130104. |
| [100] | CHANG W, YOON J, M-K OH. Production of polyhydroxyalkanoates with the fermentation of Methylorubrum extorquens using formate as a carbon substrate [J]. Biotechnology and Bioprocess Engineering, 2022, 27(2): 268-275. |
| [101] | LI B, YANG Z, LI Z, et al. Enabling genetic manipulation and robustness of Bacillus methanolicus for methanol-based bio-manufacturing [J]. Metabolic Engineering, 2025, 89: 121-134. |
| [102] | GUO Q, ZHENG L J, ZHENG S H, et al. Enhanced biosynthesis of D-allulose from a D-xylose-methanol mixture and its self-inductive detoxification by using antisense RNAs in Escherichia coli [J]. Journal of Agricultural and Food Chemistry, 2024, 72(26): 14821-14829. |
| [103] | KIM S, GIRALDO N, RAINALDI V, et al. Optimizing E. coli as a formatotrophic platform for bioproduction via the reductive glycine pathway [J]. Frontiers in Bioengineering and Biotechnology, 2023, 11: 1091899. |
| [104] | LI M K, SUN W J, WANG X, et al. A eukaryote-featured membrane phospholipid enhances bacterial formaldehyde tolerance and assimilation of one-carbon feedstocks [J]. ACS Synthetic Biology, 2024, 13(12): 4074-4084. |
| [105] | CHEN W X, ZHWNF L J, LUO X,et al. Metabolic engineering and adaptive evolution of Escherichia coli for enhanced conversion of d‑xylose to d-glucaric acid mediated by methanol [J]. Biotechnology and Bioengineering, 2025, 122: 1472-1483. |
| [106] | YU W, GAO J Q, YAO L, et al. Bioconversion of methanol to 3-hydroxypropionate by engineering Ogataea polymorpha [J]. Chinese Journal of Catalysis, 2023, 46: 84-90. |
| [107] | GAO J Q, LI Y, YU W, et al. Rescuing yeast from cell death enables overproduction of fatty acids from sole methanol [J]. Nature Metabolism, 2022, 4(7): 932-943. |
| [108] | WEFELMEIER K, SCHMITZ S, HAUT A M, et al. Engineering the methylotrophic yeast Ogataea polymorpha for lactate production from methanol [J]. Frontiers in Bioengineering and Biotechnology, 2023, 11: 1223726. |
| [109] | LIU H Y, CHEN Y, LI J, et al. Scavenging intracellular reactive oxygen species to boost methanol assimilation [J]. Chemical Engineering Journal, 2025, 516: 164002. |
| [110] | ZHU C, CHEN Y, SUN W J, et al. Repair of DNA and protein damages caused by formaldehyde improves methanol assimilation [J]. Fundamental Research, 2025. |
| [111] | GUO F, LIU K, QIAO Y Y, et al. Evolutionary engineering of Saccharomyces cerevisiae: Crafting a synthetic methylotroph via self-reprogramming [J]. Science Advances, 2024, 10: 1-15. |
| [112] | QIAN J, FAN L, YANG J, et al. Directed evolution of a neutrophilic and mesophilic methanol dehydrogenase based on high-throughput and accurate measurement of formaldehyde [J]. Synthetic and Systems Biotechnology, 2023, 8(3): 386-395. |
| [113] | PHAM D N, NGUYEN A D, LEE E Y. Outlook on engineering methylotrophs for one-carbon-based industrial biotechnology [J]. Chemical Engineering Journal, 2022, 449: 137769. |
| [114] | LI J J, GAO J Q, YE M, et al. Engineering yeast for high-level production of β-farnesene from sole methanol [J]. Metabolic Engineering, 2024, 85: 194-200. |
| [115] | SRISAWAT P, HIGUCHI-TAKEUCHI M, NUMATA K. Microbial autotrophic biorefineries: Perspectives for biopolymer production [J]. polymer journal, 2022, 54(10): 1139-1151. |
| [116] | SHIN J, BAE J, LEE H, et al. Genome-wide CRISPRi screen identifies enhanced autolithotrophic phenotypes in acetogenic bacterium Eubacterium limosum [J]. Proceedings of the National Academy of Sciences, 2023, 120 (6): e2216244120. |
| [117] | LI J, ZHANF L Y, XU Q, et al. CRISPR-cas9 toolkit for genome editing in an autotrophic CO2-fixing Methanogenic Archaeon [J]. Microbiology Spectrum, 2022, 10(4): e01165-22. |
| [118] | LEE J, YU H E, LEE S Y. Metabolic engineering of microorganisms for carbon dioxide utilization [J]. Current Opinion in Biotechnology, 2025, 91: 103244. |
| [119] | MICHAEL BAUMSCHABL, Ata Özge, BERND M. MITIC,et al. Conversion of CO2 into organic acids by engineered autotrophic yeast [J]. Proceedings of the National Academy of Sciences, 2022, 119 (47): e2211827119. |
| [120] | YU M J, LI M L, ZHANG X Z, et al. Coupling Photocatalytic Reduction and Biosynthesis Towards Sustainable CO2 Upcycling [J]. Angewandte Chemie International Edition, 2025, 64(20): e202423995. |
| [121] | GEWEDA A E, ZAYED M E, KHAN M Y, et al. Mitigating CO2 emissions: A review on emerging technologies/strategies for CO2 capture [J]. Journal of the Energy Institute, 2025, 118: 101911. |
| [122] | TEDEEVA M A, KUSTOV A L, BATKIN A M, et al. Catalytic systems for hydrogenation of CO2 to methanol [J]. Molecular Catalysis, 2024, 566: 114403. |
| [123] | WEI C J, DING H L, ZHANG Z Y, et al. Research progress of bimetallic catalysts for CO2 hydrogenation to methane [J]. Int J Hydrogen Energ, 2024, 58: 872-891. |
| [124] | GAN Y M, MENG X, GAO C, et al. Metabolic engineering strategies for microbial utilization of methanol [J]. Engineering Microbiology, 2023, 3: 100081. |
| [125] | PACESA M, PELEA O, JINEK M. Past, present, and future of CRISPR genome editing technologies [J]. Cell, 2024, 187(5): 1076-1100. |
| [126] | DIXIT S, KUMAR A, SRINIVASAN K, et al. Advancing genome editing with artificial intelligence: opportunities, challenges, and future directions [J]. Frontiers in Bioengineering and Biotechnology, 2024, 11: 1335901. |
| [127] | PETZOLD C J, MUKHOPADHYAY A. From bench to biofactory: high-throughput technologies and automated workflows to accelerate biomanufacturing [J]. Current Opinion in Biotechnology, 2025, 94: 103320. |
| [128] | LANDWEHR G M, BOGART J W, MAGALHAES C, et al. Accelerated enzyme engineering by machine-learning guided cell-free expression [J]. Nature Communications, 2025, 16: 865. |
| [129] | WATSON J L, JUERGENS D, BENNETT N R, et al. De novo design of protein structure and function with RFdiffusion [J]. Nature, 2023, 620(7976): 1089-1100. |
| [1] | FANG Xinyi, SUN Lichao, HUO Yixin, WANG Ying, YUE Haitao. Trends and challenges in microbial synthesis of higher alcohols [J]. Synthetic Biology Journal, 2025, 6(4): 873-898. |
| [2] | WU Xiaoyan, SONG Qi, XU Rui, DING Chenjun, CHEN Fang, GUO Qing, ZHANG Bo. A comparative analysis of global research and development competition in synthetic biology [J]. Synthetic Biology Journal, 2025, 6(4): 940-955. |
| [3] | ZHANG Jiankang, WANG Wenjun, GUO Hongju, BAI Beichen, ZHANG Yafei, YUAN Zheng, LI Yanhui, LI Hang. Development and application of a high-throughput microbial clone picking workstation based on machine vision [J]. Synthetic Biology Journal, 2025, 6(4): 956-971. |
| [4] | MA Muqing, WU Yan, QU Maohua, LU Xiafeng, CAO Min, DU Feng, JI Rongtao, DONG Leichi, LUO Zhibo. Extracellular multi-enzyme assembly and biocatalytic cascade: advances and prospects [J]. Synthetic Biology Journal, 2025, 6(4): 920-939. |
| [5] | LI Quanfei, CHEN Qian, LIU Hao, HE Kundong, PAN Liang, LEI Peng, GU Yi’an, SUN Liang, LI Sha, QIU Yibin, WANG Rui, XU Hong. Synthetic biology and applications of high-adhesion protein materials [J]. Synthetic Biology Journal, 2025, 6(4): 806-828. |
| [6] | WU Ke, LUO Jiahao, LI Feiran. Applications of machine learning in the reconstruction and curation of genome-scale metabolic models [J]. Synthetic Biology Journal, 2025, 6(3): 566-584. |
| [7] | TIAN Xiao-jun, ZHANG Rixin. “Economics Paradox” with cells in synthetic gene circuits [J]. Synthetic Biology Journal, 2025, 6(3): 532-546. |
| [8] | LI Yongzhu, CHEN Yu. Advances and prospects in genome-scale models of yeast [J]. Synthetic Biology Journal, 2025, 6(3): 585-602. |
| [9] | ZHANG Yiqing, LIU Gaowen. Exploration of gene functions and library construction for engineering strains from a synthetic biology perspective [J]. Synthetic Biology Journal, 2025, 6(3): 685-700. |
| [10] | YANG Ying, LI Xia, LIU Lizhong. Applications of synthetic biology to stem-cell-derived modeling of early embryonic development [J]. Synthetic Biology Journal, 2025, 6(3): 669-684. |
| [11] | HUANG Yi, SI Tong, LU Anjing. Standardization for biomanufacturing: global landscape, critical challenges, and pathways forward [J]. Synthetic Biology Journal, 2025, 6(3): 701-714. |
| [12] | SONG Chengzhi, LIN Yihan. AI-enabled directed evolution for protein engineering and optimization [J]. Synthetic Biology Journal, 2025, 6(3): 617-635. |
| [13] | GAO Qi, XIAO Wenhai. Advances in the biosynthesis of monoterpenes by yeast [J]. Synthetic Biology Journal, 2025, 6(2): 357-372. |
| [14] | ZHANG Mengyao, CAI Peng, ZHOU Yongjin. Synthetic biology drives the sustainable production of terpenoid fragrances and flavors [J]. Synthetic Biology Journal, 2025, 6(2): 334-356. |
| [15] | ZHANG Lu’ou, XU Li, HU Xiaoxu, YANG Ying. Synthetic biology ushers cosmetic industry into the “bio-cosmetics” era [J]. Synthetic Biology Journal, 2025, 6(2): 479-491. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||