Synthetic Biology Journal ›› 2025, Vol. 6 ›› Issue (5): 1184-1202.DOI: 10.12211/2096-8280.2025-032
• Invited Review • Previous Articles Next Articles
Construction of microbial cell factories for aspartate-family feed amino acids
ZHAO Xinyu, SHENG Qi, LIU Kaifang, LIU Jia, LIU Liming
- Key Laboratory of Industrial Biotechnology,Ministry of Education,School of Biotechnology,Jiangnan University,Wuxi 214122,Jiangsu,China
-
Received:2025-04-02Revised:2025-06-18Online:2025-11-05Published:2025-10-31 -
Contact:LIU Jia, LIU Liming
天冬氨酸族饲用氨基酸微生物细胞工厂的创制
赵欣雨, 盛琦, 刘开放, 刘佳, 刘立明
- 江南大学生物工程学院,工业生物技术教育部重点实验室,江苏 无锡 214122
-
通讯作者:刘佳,刘立明 -
作者简介:赵欣雨 (2001—),女,硕士研究生。研究方向为微生物细胞工厂。E-mail:zxy_201710@163.com刘佳 (1988—),女,高级实验师。研究方向为酶催化工程技术。E-mail:liujia@jiangnan.edu.cn刘立明 (1976—),男,教授,博士生导师,教育部高层次人才。研究方向为合成生物学技术和微生物细胞工厂。E-mail:mingll@jiangnan.edu.cn -
基金资助:江苏省农业科技自主创新资金项目(CX(23)2005)
CLC Number:
Cite this article
ZHAO Xinyu, SHENG Qi, LIU Kaifang, LIU Jia, LIU Liming. Construction of microbial cell factories for aspartate-family feed amino acids[J]. Synthetic Biology Journal, 2025, 6(5): 1184-1202.
赵欣雨, 盛琦, 刘开放, 刘佳, 刘立明. 天冬氨酸族饲用氨基酸微生物细胞工厂的创制[J]. 合成生物学, 2025, 6(5): 1184-1202.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2025-032
| 产品 | 菌种 | |
|---|---|---|
| 液体L-赖氨酸碱 | 大肠杆菌KCCM 80190 | 大肠杆菌NITE BP-02917 |
| 谷氨酸棒状杆菌KCCM 80216 | 谷氨酸棒状杆菌KCTC 12307BP | |
| 谷氨酸棒状杆菌NRRL-B-67439 | 谷氨酸棒状杆菌NRRL-B-67535 | |
| 液体L-赖氨酸单盐酸盐 | 大肠杆菌NITE BP-02917 | |
| 技术纯L-赖氨酸单盐酸盐 | 大肠杆菌NITE BP-02917 | 谷氨酸棒状杆菌NRRL-B-67439 |
| 谷氨酸棒状杆菌DSM 32932 | 谷氨酸棒状杆菌CGMCC 7.266 | |
| 谷氨酸棒状杆菌KCCM 80183 | 谷氨酸棒状杆菌CGMCC 17927 | |
| 谷氨酸棒状杆菌NRRL B-67535 | 谷氨酸棒状杆菌CCTCC M 2015595 | |
| L-赖氨酸硫酸盐 | 大肠杆菌CGMCC 7.398 | 谷氨酸棒状杆菌CGMCC 7.266 |
| 谷氨酸棒状杆菌KFCC 11043 | 谷氨酸棒状杆菌CGMCC 17927 | |
| 谷氨酸棒状杆菌KCCM 80227 | 谷氨酸棒状杆菌CCTCC M 2015595 | |
| L-苏氨酸 | 大肠杆菌DSM 25085 | 大肠杆菌FERM BP-11383 |
| 大肠杆菌DSM 25086 | 大肠杆菌FERM BP-10942 | |
| 大肠杆菌CGMCC 3703 | 大肠杆菌NRRL B-30843 | |
| 大肠杆菌CGMCC 7.58 | 大肠杆菌KCCM 11133P | |
| 大肠杆菌CGMCC 7.232 | 谷氨酸棒状杆菌KCCM 80117 | |
| 大肠杆菌CGMCC 13325 | 谷氨酸棒状杆菌KCCM 80118 | |
| L-甲硫氨酸 | 大肠杆菌KCCM 80245 | 谷氨酸棒状杆菌KCCM 80246 |
| L-异亮氨酸 | 大肠杆菌FERM ABP-1064 | 谷氨酸棒杆菌KCCM 80189 |
| 谷氨酸棒杆菌KCCM 80185 | 谷氨酸棒杆菌CGMCC 20437 | |
Table 1 EU-authorized microbial strains for production of aspartate-family amino acids approved as feed additives
| 产品 | 菌种 | |
|---|---|---|
| 液体L-赖氨酸碱 | 大肠杆菌KCCM 80190 | 大肠杆菌NITE BP-02917 |
| 谷氨酸棒状杆菌KCCM 80216 | 谷氨酸棒状杆菌KCTC 12307BP | |
| 谷氨酸棒状杆菌NRRL-B-67439 | 谷氨酸棒状杆菌NRRL-B-67535 | |
| 液体L-赖氨酸单盐酸盐 | 大肠杆菌NITE BP-02917 | |
| 技术纯L-赖氨酸单盐酸盐 | 大肠杆菌NITE BP-02917 | 谷氨酸棒状杆菌NRRL-B-67439 |
| 谷氨酸棒状杆菌DSM 32932 | 谷氨酸棒状杆菌CGMCC 7.266 | |
| 谷氨酸棒状杆菌KCCM 80183 | 谷氨酸棒状杆菌CGMCC 17927 | |
| 谷氨酸棒状杆菌NRRL B-67535 | 谷氨酸棒状杆菌CCTCC M 2015595 | |
| L-赖氨酸硫酸盐 | 大肠杆菌CGMCC 7.398 | 谷氨酸棒状杆菌CGMCC 7.266 |
| 谷氨酸棒状杆菌KFCC 11043 | 谷氨酸棒状杆菌CGMCC 17927 | |
| 谷氨酸棒状杆菌KCCM 80227 | 谷氨酸棒状杆菌CCTCC M 2015595 | |
| L-苏氨酸 | 大肠杆菌DSM 25085 | 大肠杆菌FERM BP-11383 |
| 大肠杆菌DSM 25086 | 大肠杆菌FERM BP-10942 | |
| 大肠杆菌CGMCC 3703 | 大肠杆菌NRRL B-30843 | |
| 大肠杆菌CGMCC 7.58 | 大肠杆菌KCCM 11133P | |
| 大肠杆菌CGMCC 7.232 | 谷氨酸棒状杆菌KCCM 80117 | |
| 大肠杆菌CGMCC 13325 | 谷氨酸棒状杆菌KCCM 80118 | |
| L-甲硫氨酸 | 大肠杆菌KCCM 80245 | 谷氨酸棒状杆菌KCCM 80246 |
| L-异亮氨酸 | 大肠杆菌FERM ABP-1064 | 谷氨酸棒杆菌KCCM 80189 |
| 谷氨酸棒杆菌KCCM 80185 | 谷氨酸棒杆菌CGMCC 20437 | |
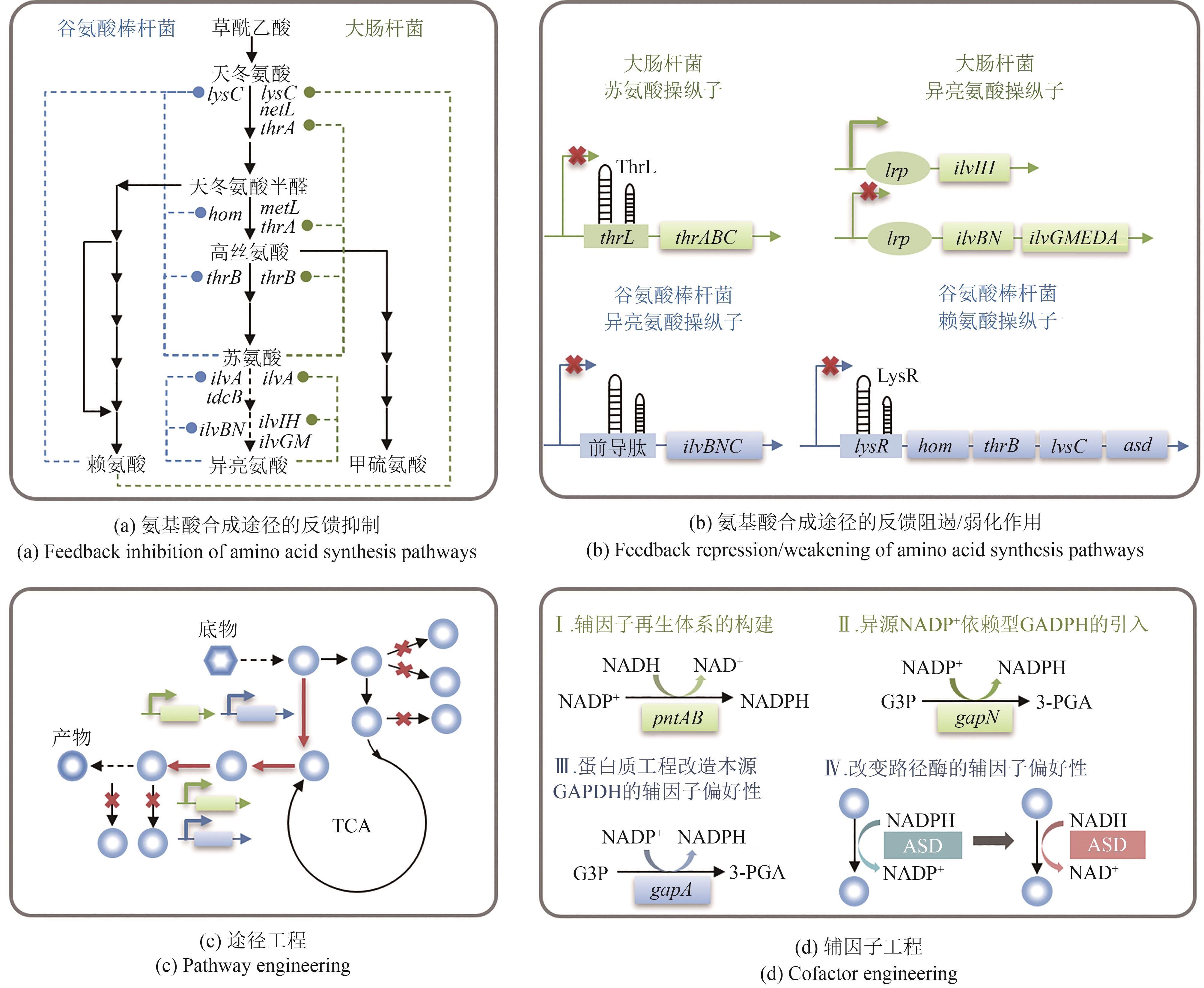
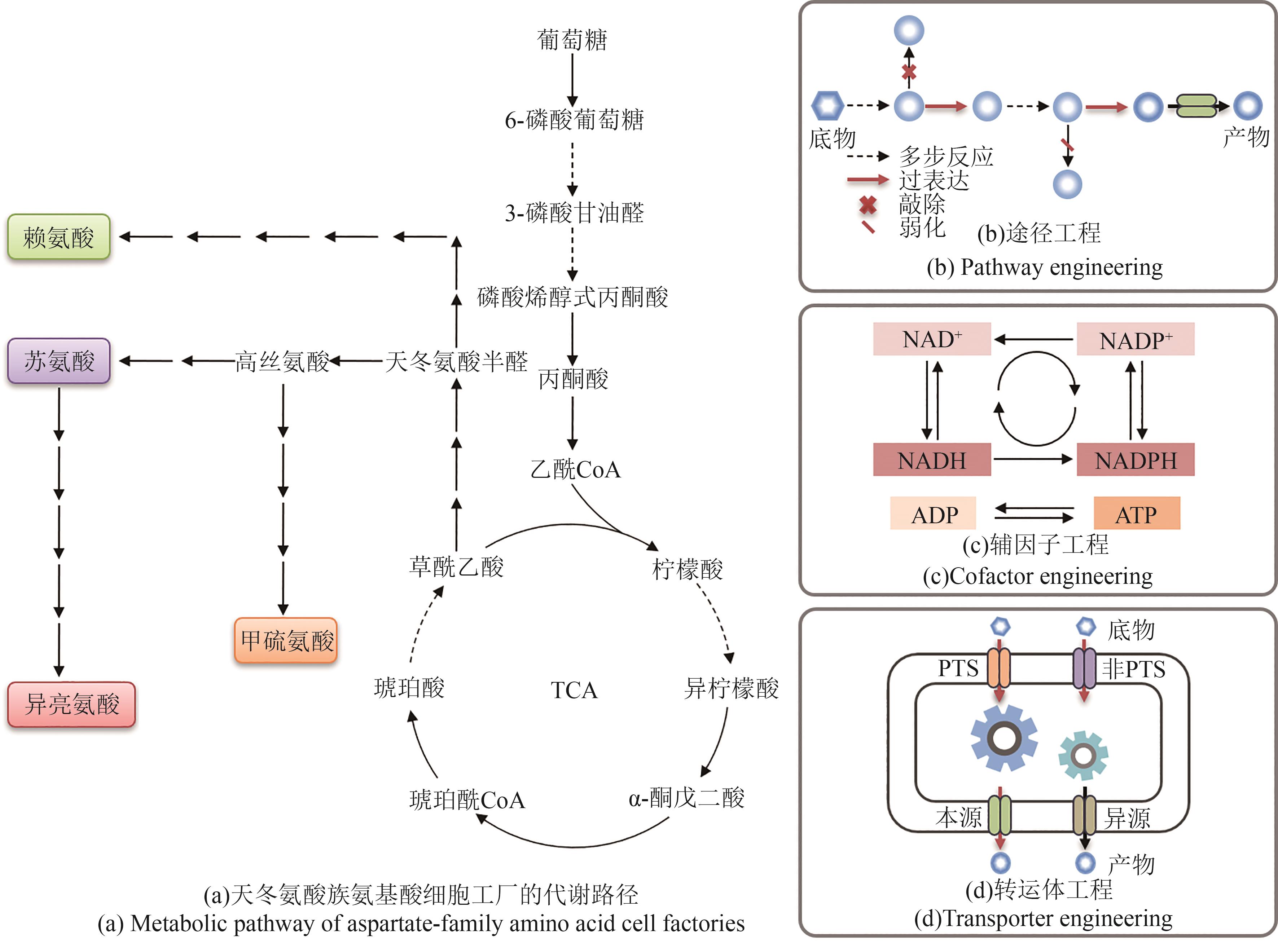
Fig. 3 Feedback regulation mechanism and metabolic pathway optimization strategy of aspartate-family amino acids(thrA—Encoding aspartate kinase Ⅰ; metL—Encoding aspartate kinase Ⅱ; lysC—Encoding aspartate kinase Ⅲ; hom—Encoding homoserine dehydrogenase; thrB—Encoding homoserine kinase; thrB—Encoding homoserine kinase; ilvA—Encoding threonine dehydrase Ⅰ; tdcB—Encoding threonine dehydrase Ⅱ; ilvBN—Encoding acetolactate synthase Ⅰ; ilvGM—Encoding acetolactate synthase Ⅱ; ilvIH—Encoding acetolactate synthase Ⅲ; asd—Encoding aspartate semialdehyde dehydrogenase; pntAB—Encoding subunits of NAD(P) transhydrogenase; gapN/gapA—Encoding glyceraldehyde-3-phosphate dehydrogenase; lrp—Encoding transcriptional regulatory factor Lrp; G3P—Glyceraldehyde 3-phosphate; 3-PGA—3-Phosphoglyceric acid; ASD—Aspartate semialdehyde dehydrogenase)
| 酶/前导肽/阻遏蛋白 | 基因 | 菌株 | 策略 | 效果 | 参考文献 |
|---|---|---|---|---|---|
| AK | lysC | E. coli | T344M | L-赖氨酸产量6.3 g/L | [ |
| T352I | L-苏氨酸产量14.4 g/L,提升30.9% | [ | |||
| C. glutamicum | T311I | L-苏氨酸产量0.27 g/L | [ | ||
| S301F | L-苏氨酸产量14.17 g/L,提升4.2% | [ | |||
| thrA | E. coli | S345F | L-苏氨酸产量82.4 mmol/L | [ | |
| G433R | L-苏氨酸产量36.61 mg/L | [ | |||
| AHAS | ilvH | E. coli | G14D, S17F | L-异亮氨酸产量0.322 g/L | [ |
| ilvN | E. coli | H47L | L-异亮氨酸产量5.1 g/L,提升59.4% | [ | |
| ilvB | E. coli | K30Q, N156D, V233I | L-异亮氨酸产量5.1 g/L,提升59.4% | [ | |
| C. glutamicum | P176S, D426E, L575W | L-异亮氨酸产量4.17 g/L,提升61.0% | [ | ||
| I47Y | L-异亮氨酸产量22.7 g/L,提升10.2% | [ | |||
| TD | ilvA | E. coli | S97F | L-异亮氨酸产量0.322 g/L | [ |
| C. glutamicum | G96D | L-异亮氨酸产量12 g/L | [ | ||
| ThrL | thrL | E. coli | 敲除thrL基因 | L-苏氨酸产量1.63 g/L | [ |
| MetJ | metJ | E. coli | 敲除metJ基因 | L-甲硫氨酸产量提升11% | [ |
Table 2 Strategies for relieving feedback regulation mechanisms
| 酶/前导肽/阻遏蛋白 | 基因 | 菌株 | 策略 | 效果 | 参考文献 |
|---|---|---|---|---|---|
| AK | lysC | E. coli | T344M | L-赖氨酸产量6.3 g/L | [ |
| T352I | L-苏氨酸产量14.4 g/L,提升30.9% | [ | |||
| C. glutamicum | T311I | L-苏氨酸产量0.27 g/L | [ | ||
| S301F | L-苏氨酸产量14.17 g/L,提升4.2% | [ | |||
| thrA | E. coli | S345F | L-苏氨酸产量82.4 mmol/L | [ | |
| G433R | L-苏氨酸产量36.61 mg/L | [ | |||
| AHAS | ilvH | E. coli | G14D, S17F | L-异亮氨酸产量0.322 g/L | [ |
| ilvN | E. coli | H47L | L-异亮氨酸产量5.1 g/L,提升59.4% | [ | |
| ilvB | E. coli | K30Q, N156D, V233I | L-异亮氨酸产量5.1 g/L,提升59.4% | [ | |
| C. glutamicum | P176S, D426E, L575W | L-异亮氨酸产量4.17 g/L,提升61.0% | [ | ||
| I47Y | L-异亮氨酸产量22.7 g/L,提升10.2% | [ | |||
| TD | ilvA | E. coli | S97F | L-异亮氨酸产量0.322 g/L | [ |
| C. glutamicum | G96D | L-异亮氨酸产量12 g/L | [ | ||
| ThrL | thrL | E. coli | 敲除thrL基因 | L-苏氨酸产量1.63 g/L | [ |
| MetJ | metJ | E. coli | 敲除metJ基因 | L-甲硫氨酸产量提升11% | [ |
| 产品 | 竞争/降解路径 | 菌株 | 策略 | 参考文献 |
|---|---|---|---|---|
| L-苏氨酸 | L-赖氨酸、L-甲硫氨酸 | E. coli | 敲除lysA和metA基因 | [ |
| L-赖氨酸、L-甲硫氨酸、L-异亮氨酸、L-甘氨酸 | E. coli | 敲除lysA、metA和tdh基因,引入ilvAS97F基因 | [ | |
| L-赖氨酸、L-异亮氨酸 | C. glutamicum | 异源表达Streptococcus pneumoniae来源的dapA基因和E. coli来源的ilvA基因 | [ | |
| L-赖氨酸、L-异亮氨酸、L-丙氨酸 | C. glutamicum | 引入ilvAG96D和dapAK68H基因,敲除alaT和avtA基因 | [ |
Table 3 Strategies for blocking competing pathways and attenuating degradation pathways
| 产品 | 竞争/降解路径 | 菌株 | 策略 | 参考文献 |
|---|---|---|---|---|
| L-苏氨酸 | L-赖氨酸、L-甲硫氨酸 | E. coli | 敲除lysA和metA基因 | [ |
| L-赖氨酸、L-甲硫氨酸、L-异亮氨酸、L-甘氨酸 | E. coli | 敲除lysA、metA和tdh基因,引入ilvAS97F基因 | [ | |
| L-赖氨酸、L-异亮氨酸 | C. glutamicum | 异源表达Streptococcus pneumoniae来源的dapA基因和E. coli来源的ilvA基因 | [ | |
| L-赖氨酸、L-异亮氨酸、L-丙氨酸 | C. glutamicum | 引入ilvAG96D和dapAK68H基因,敲除alaT和avtA基因 | [ |
| 产品 | 菌株 | 策略 | 参考文献 |
|---|---|---|---|
| L-苏氨酸 | E. coli | 过表达thrB、thrC和编码抗反馈抑制突变体的thrAS345F基因 | [ |
| P cysJ 启动子动态调控asd和thrABC基因的表达量 | [ | ||
| C. glutamicum | 过表达asd、thrB以及编码抗反馈抑制突变体的lysCT311I和homG378E基因 | [ | |
| L-甲硫氨酸 | E. coli | 过表达metC和编码抗反馈抑制突变体的metAR27C,I296S,P298L基因 | [ |
| C. glutamicum | 过表达lysC、asd、hom、metH、aecD和metYX基因 | [ | |
| L-赖氨酸 | E. coli | 过表达lysC和lysA基因 | [ |
| C. glutamicum | 过表达lysC、asd、dapA、dapB、ddh和lysA基因 | [ | |
| L-异亮氨酸 | E. coli | 过表达编码抗反馈抑制突变体的ilvAP363L、ilvNK30Q,N156D,V233I、metAG189C和metBQ5R, L29H, G69D, F87I, E136G, N148Y, K273G, A346T基因 | [ |
| C. glutamicum | 过表达编码抗反馈抑制突变体的ilvAF383V和ilvBP176S,D426EE,L575W基因 | [ |
Table 4 Strategies for reconstructing primary anabolic pathways
| 产品 | 菌株 | 策略 | 参考文献 |
|---|---|---|---|
| L-苏氨酸 | E. coli | 过表达thrB、thrC和编码抗反馈抑制突变体的thrAS345F基因 | [ |
| P cysJ 启动子动态调控asd和thrABC基因的表达量 | [ | ||
| C. glutamicum | 过表达asd、thrB以及编码抗反馈抑制突变体的lysCT311I和homG378E基因 | [ | |
| L-甲硫氨酸 | E. coli | 过表达metC和编码抗反馈抑制突变体的metAR27C,I296S,P298L基因 | [ |
| C. glutamicum | 过表达lysC、asd、hom、metH、aecD和metYX基因 | [ | |
| L-赖氨酸 | E. coli | 过表达lysC和lysA基因 | [ |
| C. glutamicum | 过表达lysC、asd、dapA、dapB、ddh和lysA基因 | [ | |
| L-异亮氨酸 | E. coli | 过表达编码抗反馈抑制突变体的ilvAP363L、ilvNK30Q,N156D,V233I、metAG189C和metBQ5R, L29H, G69D, F87I, E136G, N148Y, K273G, A346T基因 | [ |
| C. glutamicum | 过表达编码抗反馈抑制突变体的ilvAF383V和ilvBP176S,D426EE,L575W基因 | [ |
| 前体 | 产品 | 菌株 | 策略 | 参考文献 |
|---|---|---|---|---|
| 草酰乙酸 | L-赖氨酸 | C. glutamicum | 过表达ppc和pyc基因,敲除pck基因 | [ |
| L-苏氨酸 | E. coli | 过表达ppc基因,异源表达Rhizobium etli来源的pyc基因,利用P flic 启动子动态调控gltA基因 | [ | |
| 过表达ppc和acs基因,敲除iclR基因 | [ | |||
| C. glutamicum | 过表达ppc、pyc和aspB基因,异源表达E. coli来源的aspA基因 | [ | ||
| L-甲硫氨酸 | E. coli | 敲除pykA和pykF基因 | [ | |
| 过表达sucA基因,敲除sucD基因 | [ | |||
| C. glutamicum | 过表达pycP458S基因,敲除pyk2基因 | [ | ||
| L-异亮氨酸 | E. coli | 敲除ptsG、pykF和iclR基因,过表达ppc基因,异源表达Methanococcus jannaschii来源的cimAI47V, E114V, H126Q, T204A, L238S, V373STOP基因 | [ | |
| 敲除aceA和sucCD基因,过表达metA和metB基因 | [ | |||
| C. glutamicum | 敲除alaT和alr基因 | [ |
Table 5 Strategies for enhancing precursor supply at key metabolic nodes
| 前体 | 产品 | 菌株 | 策略 | 参考文献 |
|---|---|---|---|---|
| 草酰乙酸 | L-赖氨酸 | C. glutamicum | 过表达ppc和pyc基因,敲除pck基因 | [ |
| L-苏氨酸 | E. coli | 过表达ppc基因,异源表达Rhizobium etli来源的pyc基因,利用P flic 启动子动态调控gltA基因 | [ | |
| 过表达ppc和acs基因,敲除iclR基因 | [ | |||
| C. glutamicum | 过表达ppc、pyc和aspB基因,异源表达E. coli来源的aspA基因 | [ | ||
| L-甲硫氨酸 | E. coli | 敲除pykA和pykF基因 | [ | |
| 过表达sucA基因,敲除sucD基因 | [ | |||
| C. glutamicum | 过表达pycP458S基因,敲除pyk2基因 | [ | ||
| L-异亮氨酸 | E. coli | 敲除ptsG、pykF和iclR基因,过表达ppc基因,异源表达Methanococcus jannaschii来源的cimAI47V, E114V, H126Q, T204A, L238S, V373STOP基因 | [ | |
| 敲除aceA和sucCD基因,过表达metA和metB基因 | [ | |||
| C. glutamicum | 敲除alaT和alr基因 | [ |
| 辅因子 | 菌株 | 产品 | 策略 | 参考文献 |
|---|---|---|---|---|
| NADPH | E. coli | L-苏氨酸 | 过表达pntAB基因,异源表达Tistrella mobilis来源的asd基因 | [ |
| L-异亮氨酸 | 过表达aspA、bcd和pntAB基因 | [ | ||
| C. glutamicum | L-赖氨酸 | 过表达pntAB和zwf基因,P lysE 启动子动态调控gapN基因 | [ | |
| L-甲硫氨酸 | 过表达zwf fbr和gnd fbr基因,异源表达Clostridium acetobutylicum来源的gapC基因 | [ | ||
| ATP | E. coli | L-苏氨酸 | 异源表达Tistrella mobilis来源的asd基因和Pseudomonas aeruginosa来源的adh基因,敲除amn基因,异源表达Mannheimia succiniciproducens来源的pckA基因和Vitreoscilla来源的血红蛋白突变体编码基因vgbH36R, Q66R | [ |
| C. glutamicum | L-赖氨酸 | 过表达pgk和pyk基因,敲除amn基因 | [ | |
| 异源表达M. maripaludis来源的glk/pfk基因,过表达ndh基因,敲除sigH基因 | [ |
Table 6 Cofactor engineering
| 辅因子 | 菌株 | 产品 | 策略 | 参考文献 |
|---|---|---|---|---|
| NADPH | E. coli | L-苏氨酸 | 过表达pntAB基因,异源表达Tistrella mobilis来源的asd基因 | [ |
| L-异亮氨酸 | 过表达aspA、bcd和pntAB基因 | [ | ||
| C. glutamicum | L-赖氨酸 | 过表达pntAB和zwf基因,P lysE 启动子动态调控gapN基因 | [ | |
| L-甲硫氨酸 | 过表达zwf fbr和gnd fbr基因,异源表达Clostridium acetobutylicum来源的gapC基因 | [ | ||
| ATP | E. coli | L-苏氨酸 | 异源表达Tistrella mobilis来源的asd基因和Pseudomonas aeruginosa来源的adh基因,敲除amn基因,异源表达Mannheimia succiniciproducens来源的pckA基因和Vitreoscilla来源的血红蛋白突变体编码基因vgbH36R, Q66R | [ |
| C. glutamicum | L-赖氨酸 | 过表达pgk和pyk基因,敲除amn基因 | [ | |
| 异源表达M. maripaludis来源的glk/pfk基因,过表达ndh基因,敲除sigH基因 | [ |
| 产品 | 菌株 | 策略 | 参考文献 |
|---|---|---|---|
| L-赖氨酸 | C. glutamicum | 敲除pgi基因,过表达ptsG基因 | [ |
| 过表达lysE基因,敲除lysP基因,异源表达E. coli来源的ybjE基因 | [ | ||
| L-苏氨酸 | E. coli | 敲除crr、sstT和tdcC基因,过表达glk、rhtC和rhtA基因,异源表达Zymomonas mobilis来源的glf基因 | [ |
| C. glutamicum | 过表达thrE基因,异源表达E. coli来源的rhtC基因 | [ | |
| L-甲硫氨酸 | E. coli | 过表达yjeH基因,敲除metD和rhtA基因 | [ |
| C. glutamicum | 过表达brnFE基因,敲除metD基因 | [ | |
| L-异亮氨酸 | E. coli | 过表达brnFE基因,敲除brnQ基因 | [ |
| 过表达glk、ygaZH基因,敲除ptsG和rhtC基因 | [ | ||
| 过表达ygaZH基因,敲除rhtC和livJ基因 | [ |
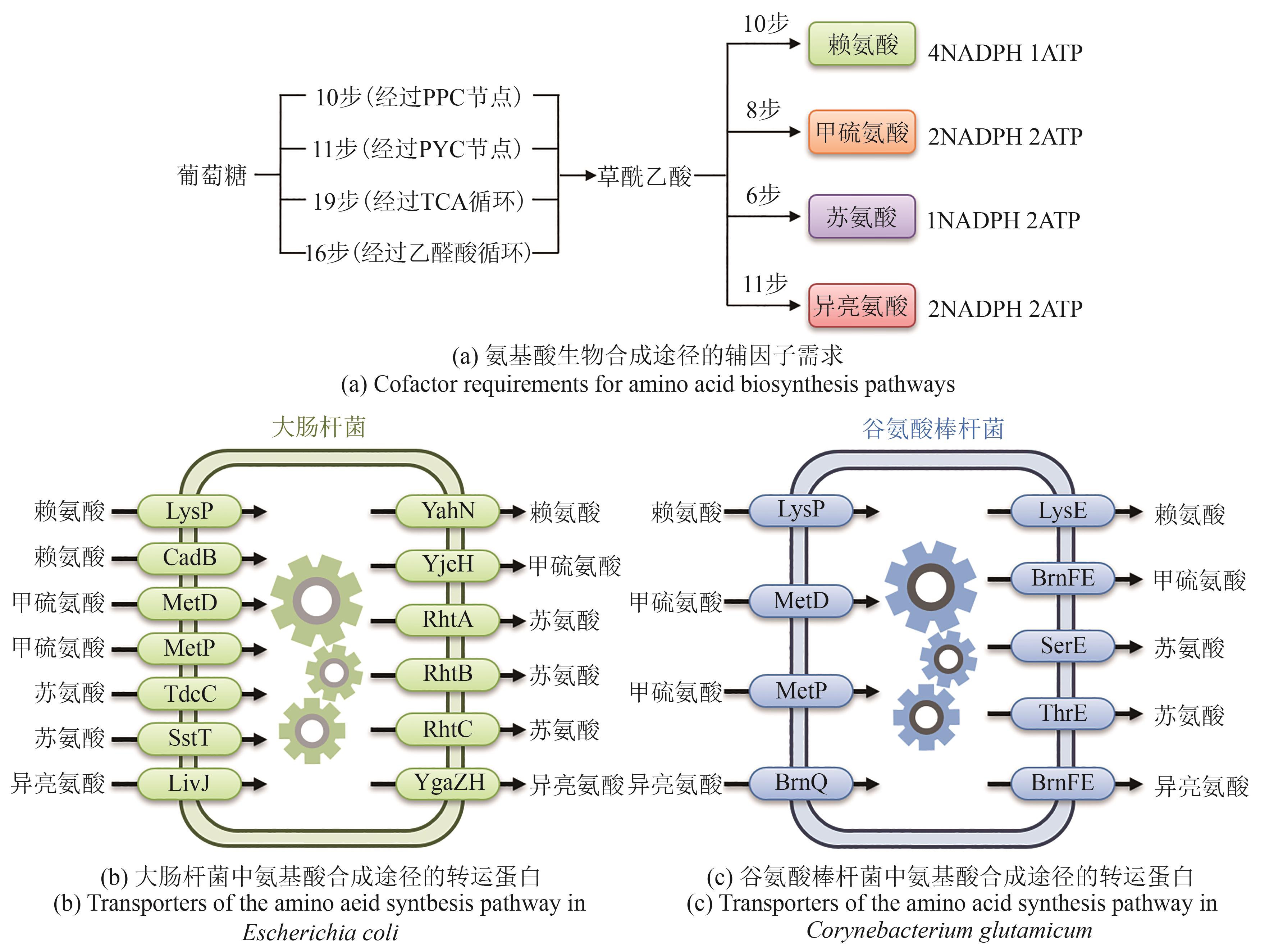
Table 7 Transporter engineering
| 产品 | 菌株 | 策略 | 参考文献 |
|---|---|---|---|
| L-赖氨酸 | C. glutamicum | 敲除pgi基因,过表达ptsG基因 | [ |
| 过表达lysE基因,敲除lysP基因,异源表达E. coli来源的ybjE基因 | [ | ||
| L-苏氨酸 | E. coli | 敲除crr、sstT和tdcC基因,过表达glk、rhtC和rhtA基因,异源表达Zymomonas mobilis来源的glf基因 | [ |
| C. glutamicum | 过表达thrE基因,异源表达E. coli来源的rhtC基因 | [ | |
| L-甲硫氨酸 | E. coli | 过表达yjeH基因,敲除metD和rhtA基因 | [ |
| C. glutamicum | 过表达brnFE基因,敲除metD基因 | [ | |
| L-异亮氨酸 | E. coli | 过表达brnFE基因,敲除brnQ基因 | [ |
| 过表达glk、ygaZH基因,敲除ptsG和rhtC基因 | [ | ||
| 过表达ygaZH基因,敲除rhtC和livJ基因 | [ |
| [31] | ZHANG Q Q, WANG Y H, WANG X L, et al. Metabolic engineering of Escherichia coli for efficient L-isoleucine production based on the citramalate pathway[J]. Journal of Agricultural and Food Chemistry, 2025, 73(19): 11900-11911. |
| [32] | GUILLOUET S, RODAL A, AN G H, et al. Metabolic redirection of carbon flow toward isoleucine by expressing a catabolic threonine dehydratase in a threonine-overproducing Corynebacterium glutamicum [J]. Applied Microbiology and Biotechnology, 2001, 57(5): 667-673. |
| [33] | YU S Z, ZHENG B, CHEN Z Y, et al. Metabolic engineering of Corynebacterium glutamicum for producing branched chain amino acids[J]. Microbial Cell Factories, 2021, 20(1): 230. |
| [34] | GUO Y F, HAN M, XU J Z, et al. Analysis of acetohydroxyacid synthase variants from branched-chain amino acids-producing strains and their effects on the synthesis of branched-chain amino acids in Corynebacterium glutamicum [J]. Protein Expression and Purification, 2015, 109: 106-112. |
| [35] | YIN L H, HU X Q, XU D Q, et al. Co-expression of feedback-resistant threonine dehydratase and acetohydroxy acid synthase increase L-isoleucine production in Corynebacterium glutamicum [J]. Metabolic Engineering, 2012, 14(5): 542-550. |
| [36] | SHI J, FENG Z Z, SONG Q, et al. Structural and functional insights into transcription activation of the essential LysR-type transcriptional regulators[J]. Protein Science, 2024, 33(6): e5012. |
| [37] | DE GRAAF A A, EGGELING L, SAHM H. Metabolic engineering for L-lysine production by Corynebacterium glutamicum [J]. Advances in Biochemical Engineering/Biotechnology, 2001, 73: 9-29. |
| [38] | GRUZDEV N, HACHAM Y, HAVIV H, et al. Conversion of methionine biosynthesis in Escherichia coli from trans- to direct-sulfurylation enhances extracellular methionine levels[J]. Microbial Cell Factories, 2023, 22(1): 151. |
| [39] | ZHOU Z, ZHANG X Y, WU J, et al. Targeting cofactors regeneration in methylation and hydroxylation for high level production of Ferulic acid[J]. Metabolic Engineering, 2022, 73: 247-255. |
| [40] | RHEE K Y, PAREKH B S, HATFIELD G W. Leucine-responsive regulatory protein-DNA interactions in the leader region of the ilvGMEDA operon of Escherichia coli [J]. Journal of Biological Chemistry, 1996, 271(43): 26499-26507. |
| [41] | JAFRI S, CHEN S L, CALVO J M. ilvIH operon expression in Escherichia coli requires Lrp binding to two distinct regions of DNA[J]. Journal of Bacteriology, 2002, 184(19): 5293-5300. |
| [42] | MORBACH S, JUNGER C, SAHM H, et al. Attenuation control of ilvBNC in Corynebacterium glutamicum: evidence of leader peptide formation without the presence of a ribosome binding site[J]. Journal of Bioscience and Bioengineering, 2000, 90(5): 501-507. |
| [1] | FANG Q C, ZHANG X Y, DAI G C, et al. Low-opportunity-cost feed can reduce land-use-related environmental impacts by about one-third in China[J]. Nature Food, 2023, 4(8): 677-685. |
| [2] | MA Q, ZHANG Q W, XU Q Y, et al. Systems metabolic engineering strategies for the production of amino acids[J]. Synthetic and Systems Biotechnology, 2017, 2(2): 87-96. |
| [3] | 刘佳, 盛琦, 刘开放, 等. 微生物制造饲用氨基酸助力豆粕减量替代[J]. 中国科学院院刊, 2025, 40(1): 25-35. |
| LIU J, SHENG Q, LIU K F, et al. Microbial production of feed amino acids promotes reduction and replacement of soybean meal[J]. Bulletin of Chinese Academy of Sciences, 2025, 40(1): 25-35. | |
| [4] | KIM S Y. 9 What would be the next feed amino acid based on a microbial point of view?[J]. Journal of Animal Science, 2021, 99(S1): 12-13. |
| [5] | LIAO J L, ZHANG P G, YIN J D, et al. New insights into the effects of dietary amino acid composition on meat quality in pigs: a review[J]. Meat Science, 2025, 221: 109721. |
| [6] | OLUWABIYI C T, SONG Z G. Branched-chain amino acids supplementation in low-protein broiler diets: a review[J]. Animal Feed Science and Technology, 2024, 318: 116114. |
| [7] | LI M H, LI H, ZHANG X, et al. Metabolic engineering of Corynebacterium glutamicum: unlocking its potential as a key cell factory platform for organic acid production[J]. Biotechnology Advances, 2024, 77: 108475. |
| [8] | LIU J, XU J Z, RAO Z M, et al. Industrial production of L-lysine in Corynebacterium glutamicum: progress and prospects[J]. Microbiological Research, 2022, 262: 127101. |
| [9] | LI C Y, ZHANG R H, WANG J, et al. Protein engineering for improving and diversifying natural product biosynthesis[J]. Trends in Biotechnology, 2020, 38(7): 729-744. |
| [10] | DONG X Y, ZHAO Y, HU J Y, et al. Attenuating L-lysine production by deletion of ddh and lysE and their effect on L-threonine and L-isoleucine production in Corynebacterium glutamicum [J]. Enzyme and Microbial Technology, 2016, 93-94: 70-78. |
| [11] | BOMMAREDDY R R, CHEN Z, RAPPERT S, et al. A de novo NADPH generation pathway for improving lysine production of Corynebacterium glutamicum by rational design of the coenzyme specificity of glyceraldehyde 3-phosphate dehydrogenase[J]. Metabolic Engineering, 2014, 25: 30-37. |
| [43] | BOOB A G, CHEN J Y, ZHAO H M. Enabling pathway design by multiplex experimentation and machine learning[J]. Metabolic Engineering, 2024, 81: 70-87. |
| [44] | DING D Q, ZHU Y R, BAI D Y, et al. Monitoring and dynamically controlling glucose uptake rate and central metabolism[J]. Nature Chemical Engineering, 2025, 2(1): 50-62. |
| [45] | SHENG Q, YI L X, ZHONG B, et al. Shikimic acid biosynthesis in microorganisms: current status and future direction[J]. Biotechnology Advances, 2023, 62: 108073. |
| [46] | XU J Z, HAN M, REN X D, et al. Modification of aspartokinase Ⅲ and dihydrodipicolinate synthetase increases the production of L-lysine in Escherichia coli [J]. Biochemical Engineering Journal, 2016, 114: 79-86. |
| [47] | LIU J H, LIU J, LI J J, et al. Reconstruction the feedback regulation of amino acid metabolism to develop a non-auxotrophic L-threonine producing Corynebacterium glutamicum [J]. Bioresources and Bioprocessing, 2024, 11(1): 43. |
| [48] | LEE J H, LEE D E, LEE B U, et al. Global analyses of transcriptomes and proteomes of a parent strain and an L-threonine-overproducing mutant strain[J]. Journal of Bacteriology, 2003, 185(18): 5442-5451. |
| [49] | ZHAO Z Q, YOU J J, SHI X P, et al. Engineering Escherichia coli for L-threonine hyperproduction based on multidimensional optimization strategies[J]. Journal of Agricultural and Food Chemistry, 2024, 72(41): 22682-22691. |
| [50] | PARK J H, OH J E, LEE K H, et al. Rational design of Escherichia coli for L-isoleucine production[J]. ACS Synthetic Biology, 2012, 1(11): 532-540. |
| [51] | ZHANG Y C, LIU Y D, ZHANG S Y, et al. Metabolic engineering of Corynebacterium glutamicum WM001 to improve L-isoleucine production[J]. Biotechnology and Applied Biochemistry, 2021, 68(3): 568-584. |
| [52] | LIU Y D, LI Y Y, WANG X Y. Acetohydroxyacid synthases: evolution, structure, and function[J]. Applied Microbiology and Biotechnology, 2016, 100(20): 8633-8649. |
| [53] | DUAN M, CHEN S, LIU X L, et al. The application of Corynebacterium glutamicum in L-threonine biosynthesis[J]. Fermentation, 2023, 9(9): 822. |
| [54] | JIN X, WANG S M, GAO Y P, et al. Combinatorial metabolic engineering of Escherichia coli to efficiently produce L-threonine from untreated cane molasses[J]. Bioresource Technology, 2025, 419: 132058. |
| [12] | CHEN Y Y, HUANG L G, YU T, et al. Balancing the AspC and AspA pathways of Escherichia coli by systematic metabolic engineering strategy for high-efficient L-homoserine production[J]. ACS Synthetic Biology, 2024, 13(8): 2457-2469. |
| [13] | BASSALO M C, GARST A D, CHOUDHURY A, et al. Deep scanning lysine metabolism in Escherichia coli [J]. Molecular Systems Biology, 2018, 14(11): e8371. |
| [14] | OSIRE T, YANG T W, XU M J, et al. Integrated gene engineering synergistically improved substrate-product transport, cofactor generation and gene translation for cadaverine biosynthesis in E. coli [J]. International Journal of Biological Macromolecules, 2021, 169: 8-17. |
| [15] | XIAO J, WANG D T, WANG L, et al. Increasing L-lysine production in Corynebacterium glutamicum by engineering amino acid transporters[J]. Amino Acids, 2020, 52(10): 1363-1374. |
| [16] | LIU J, OU Y, XU J Z, et al. L-lysine production by systems metabolic engineering of an NADPH auto-regulated Corynebacterium glutamicum [J]. Bioresource Technology, 2023, 387: 129701. |
| [17] | MALLA S, VAN DER HELM E, DARBANI B, et al. A novel efficient L-lysine exporter identified by functional metagenomics[J]. Frontiers in Microbiology, 2022, 13: 855736. |
| [18] | LI Z C, LIU Q, SUN J H, et al. Multivariate modular metabolic engineering for enhanced L-methionine biosynthesis in Escherichia coli [J]. Biotechnology for Biofuels and Bioproducts, 2023, 16(1): 101. |
| [19] | LI Y, CONG H, LIU B N, et al. Metabolic engineering of Corynebacterium glutamicum for methionine production by removing feedback inhibition and increasing NADPH level[J]. Antonie Van Leeuwenhoek, 2016, 109(9): 1185-1197. |
| [20] | ZHOU B Z, ZHAO G H, YU J, et al. Multi-step metabolic engineering Corynebacterium glutamicum ATCC13032 to produce L-methionine[J]. Systems Microbiology and Biomanufacturing, 2025, 5(2): 593-610. |
| [21] | ZHAO Z Q, YOU J J, SHI X P, et al. Multi-module engineering to guide the development of an efficient L-threonine-producing cell factory[J]. Bioresource Technology, 2025, 416: 131802. |
| [22] | LEE K H, PARK J H, KIM T Y, et al. Systems metabolic engineering of Escherichia coli for L-threonine production[J]. Molecular Systems Biology, 2007, 3: 149. |
| [23] | VO T M, PARK J Y, KIM D, et al. Use of acetate as substrate for sustainable production of homoserine and threonine by Escherichia coli W3110: a modular metabolic engineering approach[J]. Metabolic Engineering, 2024, 84: 13-22. |
| [55] | YE C, LUO Q L, GUO L, et al. Improving lysine production through construction of an Escherichia coli enzyme-constrained model[J]. Biotechnology and Bioengineering, 2020, 117(11): 3533-3544. |
| [56] | LV Q L, HU M K, TIAN L Z, et al. Enhancing L-glutamine production in Corynebacterium glutamicum by rational metabolic engineering combined with a two-stage pH control strategy[J]. Bioresource Technology, 2021, 341: 125799. |
| [57] | WANG L J, GUO Y Y, LI M Y, et al. Antibiotic-free high-level L-methionine production in engineered Escherichia coli [J]. Journal of Agricultural and Food Chemistry, 2024, 72(46): 25791-25800. |
| [58] | JIANG S, WANG R R, WANG D H, et al. Metabolic reprogramming and biosensor-assisted mutagenesis screening for high-level production of L-arginine in Escherichia coli [J]. Metabolic Engineering, 2023, 76: 146-157. |
| [59] | YE J W, HU D K, CHE X M, et al. Engineering of Halomonas bluephagenesis for low cost production of poly (3-hydroxybutyrate-co-4-hydroxybutyrate) from glucose[J]. Metabolic Engineering, 2018, 47: 143-152. |
| [60] | YE J W, HU D K, YIN J, et al. Stimulus response-based fine-tuning of polyhydroxyalkanoate pathway in Halomonas [J]. Metabolic Engineering, 2020, 57: 85-95. |
| [61] | LI T, YE J W, SHEN R, et al. Semirational approach for ultrahigh poly(3-hydroxybutyrate) accumulation in Escherichia coli by combining one-step library construction and high-throughput screening[J]. ACS Synthetic Biology, 2016, 5(11): 1308-1317. |
| [62] | SHEN R, YIN J, YE J W, et al. Promoter engineering for enhanced P(3HB-co-4HB) production by Halomonas bluephagenesis [J]. ACS Synthetic Biology, 2018, 7(8): 1897-1906. |
| [63] | DU F, LI Z J, LI X, et al. Optimizing multicopy chromosomal integration for stable high-performing strains[J]. Nature Chemical Biology, 2024, 20(12): 1670-1679. |
| [64] | BAEK J M, MAZUMDAR S, LEE S W, et al. Butyrate production in engineered Escherichia coli with synthetic scaffolds[J]. Biotechnology and Bioengineering, 2013, 110(10): 2790-2794. |
| [65] | DUEBER J E, WU G C, MALMIRCHEGINI G R, et al. Synthetic protein scaffolds provide modular control over metabolic flux[J]. Nature Biotechnology, 2009, 27(8): 753-759. |
| [66] | ZHOU H, VONK B, ROUBOS J A, et al. Algorithmic co-optimization of genetic constructs and growth conditions: application to 6-ACA, a potential nylon-6 precursor[J]. Nucleic Acids Research, 2015, 43(21): 10560-10570. |
| [67] | VENAYAK N, ANESIADIS N, CLUETT W R, et al. Engineering metabolism through dynamic control[J]. Current Opinion in Biotechnology, 2015, 34: 142-152. |
| [68] | CRESS B F, TRANTAS E A, VERVERIDIS F, et al. Sensitive cells: enabling tools for static and dynamic control of microbial metabolic pathways[J]. Current Opinion in Biotechnology, 2015, 36: 205-214. |
| [69] | IMMETHUN C M, DELORENZO D M, FOCHT C M, et al. Physical, chemical, and metabolic state sensors expand the synthetic biology toolbox for Synechocystis sp. PCC 6803[J]. Biotechnology and Bioengineering, 2017, 114(7): 1561-1569. |
| [70] | SHEN X L, WANG J, LI C Y, et al. Dynamic gene expression engineering as a tool in pathway engineering[J]. Current Opinion in Biotechnology, 2019, 59: 122-129. |
| [71] | LALWANI M A, ZHAO E M, AVALOS J L. Current and future modalities of dynamic control in metabolic engineering[J]. Current Opinion in Biotechnology, 2018, 52: 56-65. |
| [72] | PINTO D, VECCHIONE S, WU H, et al. Engineering orthogonal synthetic timer circuits based on extracytoplasmic function σ factors[J]. Nucleic Acids Research, 2018, 46(14): 7450-7464. |
| [73] | MEYER A J, SEGALL-SHAPIRO T H, GLASSEY E, et al. Escherichia coli “Marionette” strains with 12 highly optimized small-molecule sensors[J]. Nature Chemical Biology, 2019, 15(2): 196-204. |
| [74] | 叶健文, 陈江楠, 张旭, 等. 动态调控: 一种高效的细胞工厂工程化代谢改造策略[J]. 生物技术通报, 2020, 36(6): 1-12. |
| YE J W, CHEN J N, ZHANG X, et al. Dynamic control: an efficient strategy for metabolically engineering microbial cell factories[J]. Biotechnology Bulletin, 2020, 36(6): 1-12. | |
| [75] | BORKOWSKI O, BRICIO C, MURGIANO M, et al. Cell-free prediction of protein expression costs for growing cells[J]. Nature Communications, 2018, 9: 1457. |
| [76] | GUPTA A, REIZMAN I M B, REISCH C R, et al. Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit[J]. Nature Biotechnology, 2017, 35(3): 273-279. |
| [77] | DING X H, YANG W J, DU X B, et al. High-level and-yield production of L-leucine in engineered Escherichia coli by multistep metabolic engineering[J]. Metabolic Engineering, 2023, 78: 128-136. |
| [78] | YANG L Y, MU X Q, NIE Y, et al. Improving the production of NAD+ via multi-strategy metabolic engineering in Escherichia coli [J]. Metabolic Engineering, 2021, 64: 122-133. |
| [79] | BOUZON M, DÖRING V, DUBOIS I, et al. Change in cofactor specificity of oxidoreductases by adaptive evolution of an Escherichia coli NADPH-auxotrophic strain[J]. mBio, 2021, 12(4): e0032921. |
| [80] | XU J Z, RUAN H Z, YU H B, et al. Metabolic engineering of carbohydrate metabolism systems in Corynebacterium glutamicum for improving the efficiency of L-lysine production from mixed sugar[J]. Microbial Cell Factories, 2020, 19(1): 39. |
| [81] | WANG K, SONG X T, CUI B Y, et al. Metabolic engineering of Escherichia coli for efficient production of ectoine[J]. Journal of Agricultural and Food Chemistry, 2025, 73(1): 646-654. |
| [82] | LIU B N, SUN X Y, LIU Y, et al. Increased NADPH supply enhances glycolysis metabolic flux and L-methionine production in Corynebacterium glutamicum [J]. Foods, 2022, 11(7): 1031. |
| [83] | YU F, ZHAO X R, ZHOU J W, et al. Biosynthesis of high-active hemoproteins by the efficient heme-supply Pichia pastoris chassis[J]. Advanced Science, 2023, 10(30): 2302826. |
| [84] | WANG Y S, BAI Y L, ZENG Q, et al. Recent advances in the metabolic engineering and physiological opportunities for microbial synthesis of L-aspartic acid family amino acids: a review[J]. International Journal of Biological Macromolecules, 2023, 253: 126916. |
| [85] | 赵阔, 程金宇, 郭亮, 等. 谷氨酸棒杆菌代谢工程高效生产L-缬氨酸[J]. 生物工程学报, 2023, 39(8): 3253-3272. |
| ZHAO K, CHENG J Y, GUO L, et al. Highly efficient production of L-valine by multiplex metabolic engineering of Corynebacterium glutamicum [J]. Chinese Journal of Biotechnology, 2023, 39(8): 3253-3272. | |
| [86] | XU S Y, ZHOU L, XU Y, et al. Recent advances in structure-based enzyme engineering for functional reconstruction[J]. Biotechnology and Bioengineering, 2023, 120(12): 3427-3445. |
| [87] | 贾男, 臧国伟, 李春, 等. 辅因子在微生物细胞工厂中的代谢调控与应用[J]. 中国生物工程杂志, 2022, 42(7): 79-89. |
| JIA N, ZANG G W, LI C, et al. Metabolic regulations and applications of cofactors in microbial cell factories[J]. China Biotechnology, 2022, 42(7): 79-89. | |
| [88] | 陈修来, 刘佳, 罗秋玲, 等. 微生物辅因子平衡的代谢调控[J]. 生物工程学报, 2017, 33(1): 16-26. |
| CHEN X L, LIU J, LUO Q L, et al. Manipulation of cofactor balance in microorganisms[J]. Chinese Journal of Biotechnology, 2017, 33(1): 16-26. | |
| [89] | 陈雅维. ATP调控策略及其在微生物代谢产物合成中的应用[J]. 生物工程学报, 2020, 36(8): 1515-1527. |
| CHEN Y W. ATP regulation strategy and its application in the synthesis of microbial metabolites[J]. Chinese Journal of Biotechnology, 2020, 36(8): 1515-1527. | |
| [90] | WEUSTHUIS R A, FOLCH P L, POZO-RODRÍGUEZ A, et al. Applying non-canonical redox cofactors in fermentation processes[J]. iScience, 2020, 23(9): 101471. |
| [91] | 张鸿伟, 王鹏超. 微生物辅因子工程研究进展[J]. 中国生物工程杂志, 2023, 43(4): 112-122. |
| ZHANG H W, WANG P C. Research progress of microbial cofactor engineering[J]. China Biotechnology, 2023, 43(4): 112-122. | |
| [92] | GAO C, SONG W, YE C, et al. Bifunctional optogenetic switch powered NADPH availability for improving L-valine production in Escherichia coli [J]. ACS Sustainable Chemistry & Engineering, 2024, 12(41): 15103-15113. |
| [93] | 刘开放. 新型微生物辅因子系统的开发与应用[D]. 无锡: 江南大学, 2024. |
| LIU K F. Development and application of novel cofactor systems in microorganisms[D]. Wuxi: Jiangnan University, 2024. | |
| [94] | BLACK W B, ZHANG L Y, MAK W S, et al. Engineering a nicotinamide mononucleotide redox cofactor system for biocatalysis[J]. Nature Chemical Biology, 2020, 16(1): 87-94. |
| [95] | 于袁欢, 周阳, 王欣怡, 等. 光遗传学照进生物医学研究进展[J]. 合成生物学, 2023, 4(1): 102-140. |
| YU Y H, ZHOU Y, WANG X Y, et al. Advances in optogenetics for biomedical research[J]. Synthetic Biology Journal, 2023, 4(1): 102-140. | |
| [96] | CHEN R B, YANG S, ZHANG L, et al. Advanced strategies for production of natural products in yeast[J]. iScience, 2020, 23(3): 100879. |
| [97] | CHEN R B, GAO J Q, YU W, et al. Engineering cofactor supply and recycling to drive phenolic acid biosynthesis in yeast[J]. Nature Chemical Biology, 2022, 18(5): 520-529. |
| [98] | ZHANG Y F, ZHANG G Y, ZHANG H F, et al. Efficient fermentative production of β-alanine from glucose through multidimensional engineering of Escherichia coli [J]. Journal of Agricultural and Food Chemistry, 2024, 72(25): 14274-14283. |
| [99] | LINDNER S N, PETROV D P, HAGMANN C T, et al. Phosphotransferase system-mediated glucose uptake is repressed in phosphoglucoisomerase-deficient Corynebacterium glutamicum strains[J]. Applied and Environmental Microbiology, 2013, 79(8): 2588-2595. |
| [100] | MOLINA-VÁZQUEZ E R, CASPETA L, GOSSET G, et al. Tailoring Escherichia coli BL21 (DE3) for preferential xylose utilization via metabolic and regulatory engineering[J]. Applied Microbiology and Biotechnology, 2025, 109(1): 54. |
| [101] | ZHU X N, FAN F Y, QIU H N, et al. New xylose transporters support the simultaneous consumption of glucose and xylose in Escherichia coli [J]. mLife, 2022, 1(2): 156-170. |
| [102] | LI J, QIU Z T, ZHAO G R. Modular engineering of E. coli coculture for efficient production of resveratrol from glucose and arabinose mixture[J]. Synthetic and Systems Biotechnology, 2022, 7(2): 718-729. |
| [103] | HALLE L, HÖPPNER D, DOSER M, et al. From molasses to purified α-ketoglutarate with engineered Corynebacterium glutamicum [J]. Bioresource Technology, 2025, 416: 131803. |
| [104] | MOON M W, PARK S Y, CHOI S K, et al. The phosphotransferase system of Corynebacterium glutamicum: features of sugar transport and carbon regulation[J]. Journal of Molecular Microbiology and Biotechnology, 2007, 12(1-2): 43-50. |
| [105] | 刘冬冬. 强化葡萄糖代谢途径提高L-赖氨酸发酵水平的研究[D]. 无锡: 江南大学, 2017. |
| LIU D D. Increasing the fermentation level of L-lysine via enhancing glucose metabolism pathways[D]. Wuxi: Jiangnan University, 2017. | |
| [106] | PARCHE S, BURKOVSKI A, SPRENGER G A, et al. Corynebacterium glutamicum: a dissection of the PTS[J]. Journal of Molecular Microbiology and Biotechnology, 2001, 3(3): 423-428. |
| [107] | WANG L, LI N, YU S Q, et al. Enhancing caffeic acid production in Escherichia coli by engineering the biosynthesis pathway and transporter[J]. Bioresource Technology, 2023, 368: 128320. |
| [108] | 郭亮, 高聪, 柳亚迪, 等. 大肠杆菌生产饲用氨基酸的研究进展[J]. 合成生物学, 2021, 2(6): 964-981. |
| GUO L, GAO C, LIU Y D, et al. Advances in bioproduction of feed amino acid by Escherichia coli [J]. Synthetic Biology Journal, 2021, 2(6): 964-981. | |
| [109] | 于勇, 朱欣娜, 毕昌昊, 等. 大肠杆菌细胞工厂的创建技术[J]. 生物工程学报, 2021, 37(5): 1564-1577. |
| YU Y, ZHU X N, BI C H, et al. Construction of Escherichia coli cell factories[J]. Chinese Journal of Biotechnology, 2021, 37(5): 1564-1577. | |
| [110] | 赵静. 几种主要饲用氨基酸的营养研究进展[J]. 中国饲料添加剂, 2016(1): 10-13. |
| ZHAO J. Research progress on nutrition of several main feed amino acids[J]. China Feed Additive, 2016(1): 10-13. | |
| [111] | ISOGAI S, TAKAGI H. Enhancement of lysine biosynthesis confers high-temperature stress tolerance to Escherichia coli cells[J]. Applied Microbiology and Biotechnology, 2021, 105(18): 6899-6908. |
| [112] | LIU J, ZHAO X J, CHENG H J, et al. Comprehensive screening of industrially relevant components at genome scale using a high-quality gene overexpression collection of Corynebacterium glutamicum [J]. Trends in Biotechnology, 2025, 43(1): 220-247. |
| [113] | ZHANG Y W, YANG J J, QIAN F H, et al. Engineering a xylose fermenting yeast for lignocellulosic ethanol production[J]. Nature Chemical Biology, 2025, 21(3): 443-450. |
| [114] | GAO J Q, LI Y X, YU W, et al. Rescuing yeast from cell death enables overproduction of fatty acids from sole methanol[J]. Nature Metabolism, 2022, 4(7): 932-943. |
| [115] | GE C, YU Z, SHENG H K, et al. Redesigning regulatory components of quorum-sensing system for diverse metabolic control[J]. Nature Communications, 2022, 13: 2182. |
| [24] | ZHAO G H, ZHANG D Z, ZHOU B Z, et al. Fine-regulating the carbon flux of L-isoleucine producing Corynebacterium glutamicum WM001 for efficient L-threonine production[J]. ACS Synthetic Biology, 2024, 13(10): 3446-3460. |
| [25] | PU W, FENG J H, CHEN J Z, et al. Engineering of L-threonine and L-proline biosensors by directed evolution of transcriptional regulator SerR and application for high-throughput screening[J]. Bioresources and Bioprocessing, 2025, 12(1): 4. |
| [26] | SONG J, ZHUANG M M, DU C Y, et al. Metabolic engineering of Escherichia coli for self-induced production of L-isoleucine[J]. ACS Synthetic Biology, 2025, 14(1): 179-192. |
| [27] | SHI C R, HUO X J, YOU R, et al. High yield production of L-isoleucine through readjusting the ratio of two direct precursors in Escherichia coli [J]. Bioresource Technology, 2025, 418: 131889. |
| [28] | LU N, WEI M H, YANG X J, et al. Growth-coupled production of L-isoleucine in Escherichia coli via metabolic engineering[J]. Metabolic Engineering, 2024, 86: 181-193. |
| [29] | LI Y J, WEI H B, WANG T, et al. Current status on metabolic engineering for the production of L-aspartate family amino acids and derivatives[J]. Bioresource Technology, 2017, 245(Pt B): 1588-1602. |
| [30] | LIU Z Y, LIU J, ZHANG F, et al. Modifying Corynebacterium glutamicum by metabolic engineering for efficient synthesis of L-lysine[J]. Systems Microbiology and Biomanufacturing, 2025, 5(1): 288-299. |
| [1] | SONG Kainan, ZHANG Liwen, WANG Chao, TIAN Pingfang, LI Guangyue, PAN Guohui, XU Yuquan. Advances in small-molecule biopesticides and their biosynthesis [J]. Synthetic Biology Journal, 2025, 6(5): 1203-1223. |
| [2] | YU Wenwen, LV Xueqin, LI Zhaofeng, LIU Long. Plant synthetic biology and bioproduction of human milk oligosaccharides [J]. Synthetic Biology Journal, 2025, 6(5): 992-997. |
| [3] | YAN Zhaotao, ZHOU Pengfei, WANG Yangzhong, ZHANG Xin, XIE Wenyan, TIAN Chenfei, WANG Yong. Plant synthetic biology: new opportunities for large-scale culture of plant cells [J]. Synthetic Biology Journal, 2025, 6(5): 1107-1125. |
| [4] | SUN Yang, CHEN Lichao, SHI Yanyun, WANG Ke, LV Dandan, XU Xiumei, ZHANG Lixin. Strategies and prospects of synthetic biology in crop photosynthesis [J]. Synthetic Biology Journal, 2025, 6(5): 1025-1040. |
| [5] | HE Yangyu, YANG Kai, WANG Weilin, HUANG Qian, QIU Ziying, SONG Tao, HE Liushang, YAO Jinxin, GAN Lu, HE Yuchi. Design and practice of plant synthetic biology theme in the International Genetically Engineered Machine Competition [J]. Synthetic Biology Journal, 2025, 6(5): 1243-1254. |
| [6] | ZHANG Xuebo, ZHU Chengshu, CHEN Ruiyun, JIN Qingzi, LIU Xiao, XIONG Yan, CHEN Daming. Policy planning and industrial development of agricultural synthetic biology [J]. Synthetic Biology Journal, 2025, 6(5): 1224-1242. |
| [7] | LIU Jie, GAO Yu, MA Yongshuo, SHANG Yi. Progress and challenges of synthetic biology in agriculture [J]. Synthetic Biology Journal, 2025, 6(5): 998-1024. |
| [8] | ZHENG Lei, ZHENG Qiteng, ZHANG Tianjiao, DUAN Kun, ZHANG Ruifu. Engineering rhizosphere synthetic microbial communities to enhance crop nutrient use efficiency [J]. Synthetic Biology Journal, 2025, 6(5): 1058-1071. |
| [9] | LI Chao, ZHANG Huan, YANG Jun, WANG Ertao. Research advances in nitrogen fixation synthetic biology [J]. Synthetic Biology Journal, 2025, 6(5): 1041-1057. |
| [10] | WEI Jiaxiu, JI Peiyun, JIE Qingyu, HUANG Qiuyan, YE Hao, DAI Junbiao. Construction and application of plant artificial chromosomes [J]. Synthetic Biology Journal, 2025, 6(5): 1093-1106. |
| [11] | FANG Xinyi, SUN Lichao, HUO Yixin, WANG Ying, YUE Haitao. Trends and challenges in microbial synthesis of higher alcohols [J]. Synthetic Biology Journal, 2025, 6(4): 873-898. |
| [12] | WU Xiaoyan, SONG Qi, XU Rui, DING Chenjun, CHEN Fang, GUO Qing, ZHANG Bo. A comparative analysis of global research and development competition in synthetic biology [J]. Synthetic Biology Journal, 2025, 6(4): 940-955. |
| [13] | WANG Hong, LU Kongyong, ZHENG Yangyang, CHEN Tao, WANG Zhiwen. Construction and advances in the applications of transcription factor-based biosensors [J]. Synthetic Biology Journal, 2025, 6(4): 829-845. |
| [14] | ZHANG Jiankang, WANG Wenjun, GUO Hongju, BAI Beichen, ZHANG Yafei, YUAN Zheng, LI Yanhui, LI Hang. Development and application of a high-throughput microbial clone picking workstation based on machine vision [J]. Synthetic Biology Journal, 2025, 6(4): 956-971. |
| [15] | LI Quanfei, CHEN Qian, LIU Hao, HE Kundong, PAN Liang, LEI Peng, GU Yi’an, SUN Liang, LI Sha, QIU Yibin, WANG Rui, XU Hong. Synthetic biology and applications of high-adhesion protein materials [J]. Synthetic Biology Journal, 2025, 6(4): 806-828. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||