合成生物学 ›› 2024, Vol. 5 ›› Issue (3): 658-671.DOI: 10.12211/2096-8280.2023-110
CRISPR/Cas9编辑系统在微生物天然产物研究中的应用
惠真1,2, 唐啸宇2
- 1.香港科技大学理学院化学系,清水湾校区,香港 999077
2.深圳湾实验室,化学生物学研究所,广东 深圳 518132
-
收稿日期:2023-12-26修回日期:2024-03-17出版日期:2024-06-30发布日期:2024-07-12 -
通讯作者:唐啸宇 -
作者简介:惠真 (1989—), 男, 博士研究生。研究方向为微生物天然产物基因挖掘和生物合成。E-mail:zhuiaa@connect.ust.hk唐啸宇 (1984—),男,博士,研究员,博士生导师。研究方向为微生物天然产物化学生物学。E-mail:xtang@szbl.ac.cn -
基金资助:国家自然科学基金面上项目(82173719)
Applications of the CRISPR/Cas9 editing system in the study of microbial natural products
HUI Zhen1,2, TANG Xiaoyu2
- 1.Department of Chemistry,School of Science,The Hong Kong University of Science and Technology,Clearwater Bay Campus,Hong Kong 999077,China
2.Institute of Chemical Biology,Shenzhen Bay Laboratory,Shenzhen 518132,Guangdong,China
-
Received:2023-12-26Revised:2024-03-17Online:2024-06-30Published:2024-07-12 -
Contact:TANG Xiaoyu
摘要:
微生物作为天然产物的巨大宝库,一直以来都是研究人员挖掘和开发新的活性化合物的重要来源。目前,利用基因编辑工具发现、生物合成和代谢调控天然产物的研究方法受到该领域研究者的广泛关注。CRISPR/Cas9遗传编辑系统以其独特的灵活靶向优势克服了其他遗传编辑方法常见的对序列同源或位点限制,简化了实验步骤,提高了实验效率,促进了天然产物研究领域的发展。本文主要介绍CRISPR/Cas9系统在微生物天然产物发现、生物合成和工程改造方面的应用,分别从CRISPR/Cas9系统的发展、天然产物生物合成基因簇的克隆和遗传编辑、天然产物结构衍生化和代谢调节、沉默天然产物基因簇的激活这几个方面阐述CRISPR/Cas9系统在微生物天然产物研究领域的优势。最后,针对CRISPR/Cas9系统无法克服的重组效率和宿主适应性问题提供了可行的解决思路。相信随着合成生物学和信息技术的发展,越来越多的与CRISPR/Cas9系统相关的遗传操作工具和方法会被开发,将不断推动天然产物领域的发展进步。
中图分类号:
引用本文
惠真, 唐啸宇. CRISPR/Cas9编辑系统在微生物天然产物研究中的应用[J]. 合成生物学, 2024, 5(3): 658-671.
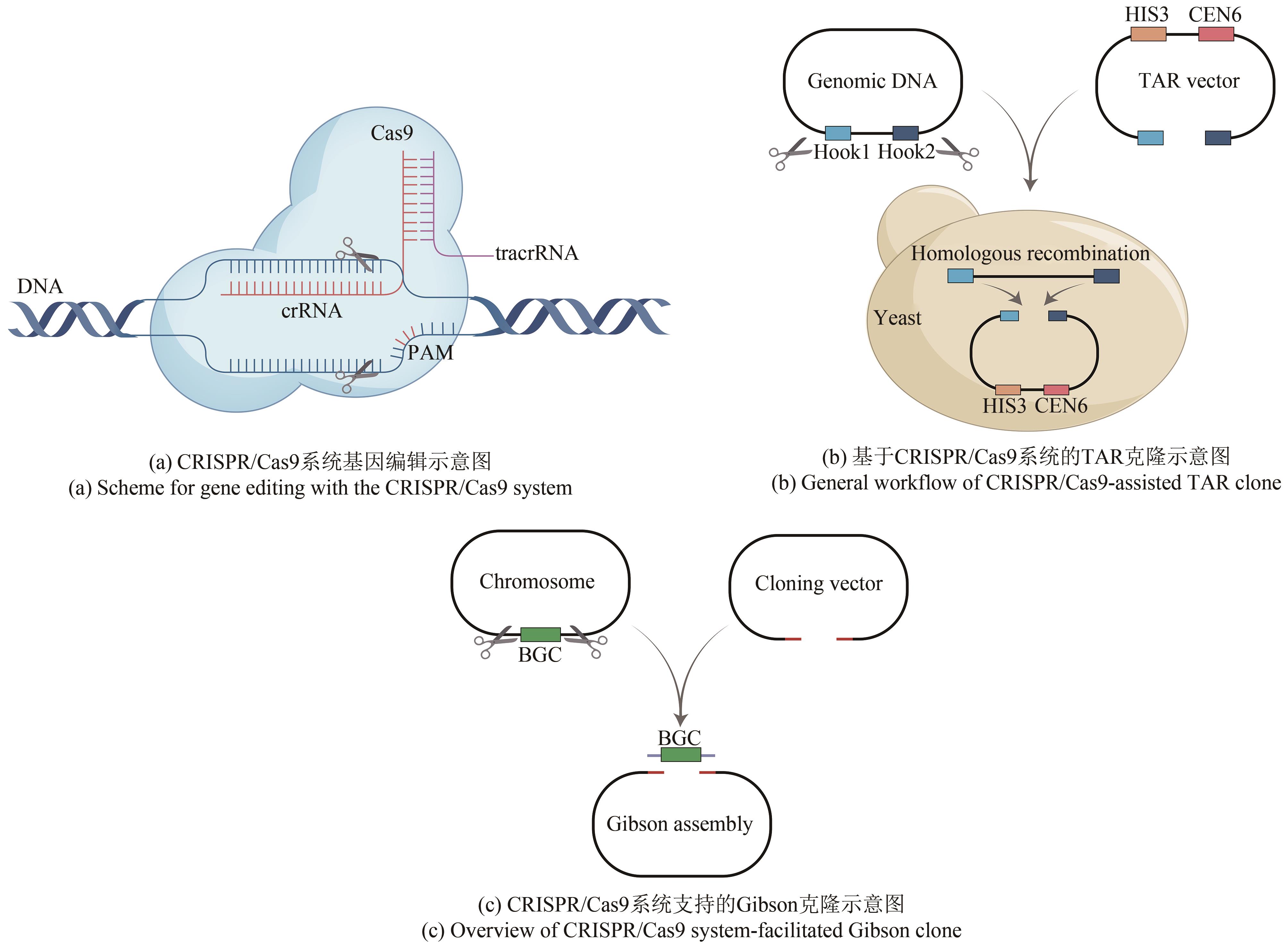
HUI Zhen, TANG Xiaoyu. Applications of the CRISPR/Cas9 editing system in the study of microbial natural products[J]. Synthetic Biology Journal, 2024, 5(3): 658-671.
| 策略 | 生物合成基因簇 | 功能 | 生物合成基因簇来源/宿主 | 参考 文献 | |
|---|---|---|---|---|---|
| 基因簇 克隆 | CATCH | Bacillaene | 基因簇线性化 | Bacillus subtilis str. 168 | [ |
| Jadomycin | Streptomyces venezuelae ISP52030 | ||||
| Chlortetracycline | S. aureofaciens ATCC 10762 | ||||
| Pentaminomycins A-H | S. cacaoi CA-170360 | [ | |||
| BH-18257 A-C | |||||
| ICE & λ packaging system | Tu3010 | S. thiolactonus NRRL 15439 | [ | ||
| Sisomicin | Micromonospora inyonensis DSM 46123 | ||||
| CAPTURE | 43 uncharacterized BGCs | Streptomyces,Bacillus | [ | ||
| CAT-FISHING | Marinolactam A | Micromonospora sp. 181 | [ | ||
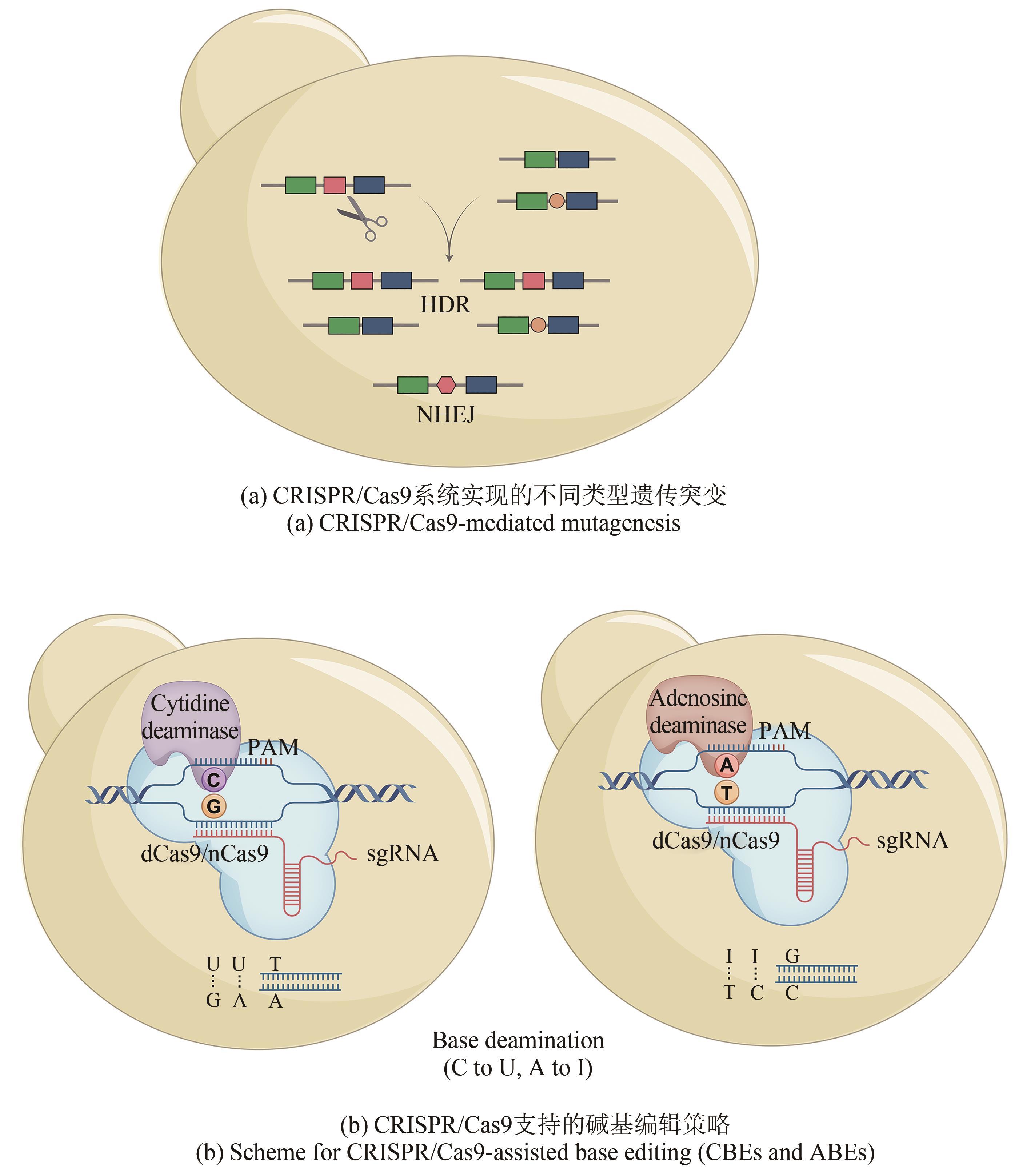
| 基因簇 遗传编辑 | ICE | Tetronate RK-682 | 遗传突变 | Streptomyces sp. Strain 88-682 | [ |
| Holomycin | S. clavuligerus TK24 | ||||
| pCRISPomyces | Undecylprodigiosin, Actinorhodin | S. lividans 66 | [ | ||
| Phosphinothricin tripeptide | S. viridochromogenes DSM 40736 | ||||
| Macrolactam, Lanthipeptide | S. albus J1074 | ||||
| CRISPR/Cas9 | Actinorhodin, Undecylprodigiosin | S. coelicolor M14 | [ | ||
| CRISPR/Cas9-LigD | Actinorhodin | S. coelicolor A3(2) | [ | ||
| CRISPRi | |||||
| CRISPR/ Cas9-CodA(sm) | Actinorhodin | S. coelicolor M145 | [ | ||
| CRISPR/Cas9 | Violacein | E. coli HME68 | [ | ||
| Thalassospiramides | Pseudomonas putida EM383 | ||||
| CBE/ABE | Undecylprodigiosin, Actinorhodin | S. coelicolor M145 | [ | ||
| Avermectin | S. avermitilis MA4680 | ||||
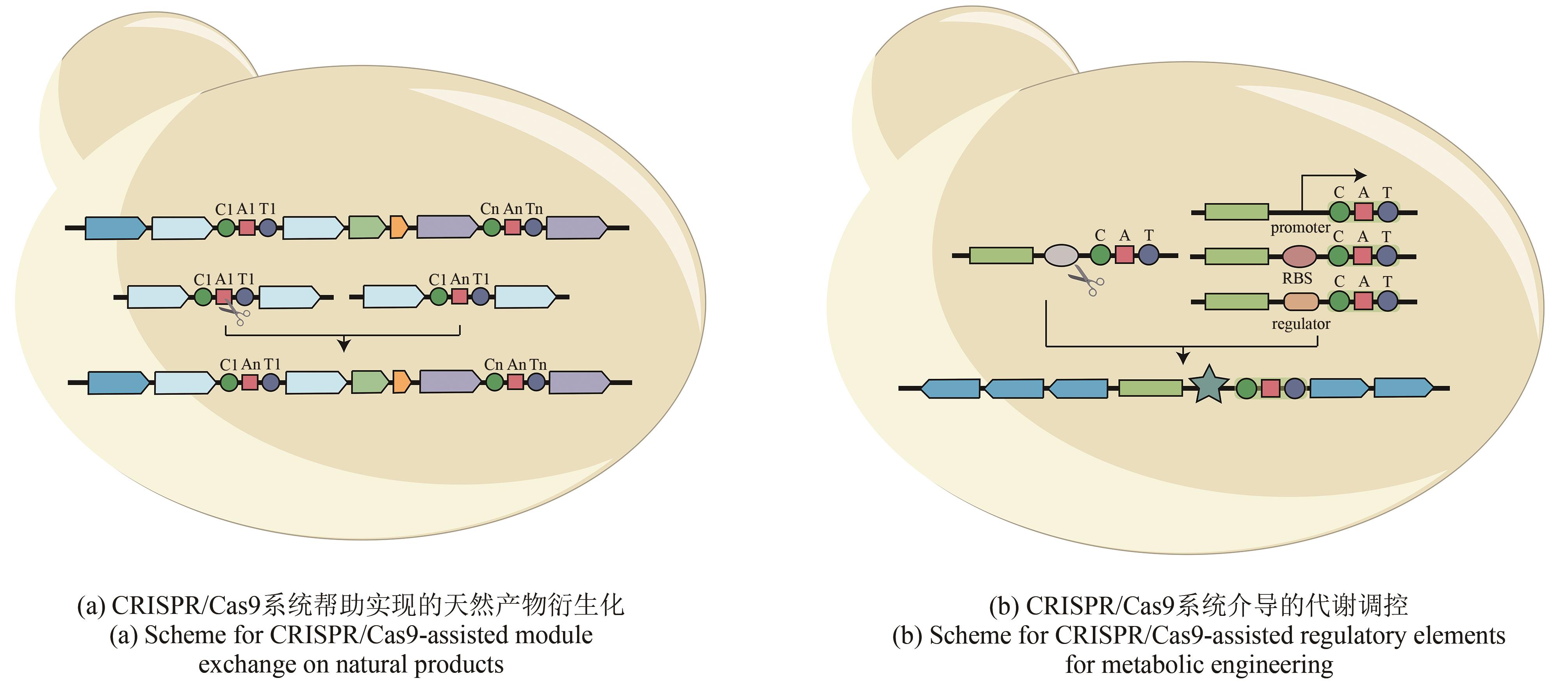
| 产物 衍生化 | CRISPR/Cas9 & Gibson Assembly | Rapamycin | 组装模块编辑 | S. avermitilis SUKA | [ |
| CRISPR/Cas9 | Enduracidin | Streptomyces fungicidicus ATCC 21013 | [ | ||
| 产物代谢调节 | CRISPR/Cas9 & TAR | Actinorhodin | 启动子工程 | S. albus J1074 | [ |
| MSGE | Pristinamyicn Ⅱ, Chloramphenicol, YM-216391 | 多拷贝 | S. pristinaespiralis HCCB10218 S. coelicolor M145 | [ | |
| CRISPR/Cas9 | Amorphadiene | 启动子工程,遗传突变, 多拷贝 | Bacillus subtilis | [ | |
| CCTL | Actinorhodin | 启动子工程 | Streptomyces sp. 4F | [ | |
| 基因簇 激活 | CRISPR/Cas9 | Alteramide A, Macrolactam 2, FR-900098 | 启动子工程 | S. roseosporus NRRL15998 | [ |
| mCRISTAR | Tetarimycin, Lazarimide, AB1210 | S. albus | [ | ||
| mpCRISTAR | Acinorhodin | S. cerevisiae BY4727 | [ | ||
| miCASTAR | Atolypene | Amycolatopsis tolypomycina NRRL B-24205 | [ | ||
| CRISPR/Cas9-LigD | Amicetin | 遗传突变 | Streptomyces WAC6237 | [ | |
| Thiolactomycin, 5-chloro-3-formylindole | Streptomyces WAC5374 | ||||
| Phenanthroviridin aglycone | Streptomyces WAC8241 | ||||
| CRISPRi/CRISPRa | Jadomycinb | 转录调控 | Streptomyces venezuelae | [ |
表1 CRISPR/Cas相关的生物合成基因簇编辑策略
Table 1 CRISPR/Cas-assisted biosynthetic gene cluster editing strategies
| 策略 | 生物合成基因簇 | 功能 | 生物合成基因簇来源/宿主 | 参考 文献 | |
|---|---|---|---|---|---|
| 基因簇 克隆 | CATCH | Bacillaene | 基因簇线性化 | Bacillus subtilis str. 168 | [ |
| Jadomycin | Streptomyces venezuelae ISP52030 | ||||
| Chlortetracycline | S. aureofaciens ATCC 10762 | ||||
| Pentaminomycins A-H | S. cacaoi CA-170360 | [ | |||
| BH-18257 A-C | |||||
| ICE & λ packaging system | Tu3010 | S. thiolactonus NRRL 15439 | [ | ||
| Sisomicin | Micromonospora inyonensis DSM 46123 | ||||
| CAPTURE | 43 uncharacterized BGCs | Streptomyces,Bacillus | [ | ||
| CAT-FISHING | Marinolactam A | Micromonospora sp. 181 | [ | ||
| 基因簇 遗传编辑 | ICE | Tetronate RK-682 | 遗传突变 | Streptomyces sp. Strain 88-682 | [ |
| Holomycin | S. clavuligerus TK24 | ||||
| pCRISPomyces | Undecylprodigiosin, Actinorhodin | S. lividans 66 | [ | ||
| Phosphinothricin tripeptide | S. viridochromogenes DSM 40736 | ||||
| Macrolactam, Lanthipeptide | S. albus J1074 | ||||
| CRISPR/Cas9 | Actinorhodin, Undecylprodigiosin | S. coelicolor M14 | [ | ||
| CRISPR/Cas9-LigD | Actinorhodin | S. coelicolor A3(2) | [ | ||
| CRISPRi | |||||
| CRISPR/ Cas9-CodA(sm) | Actinorhodin | S. coelicolor M145 | [ | ||
| CRISPR/Cas9 | Violacein | E. coli HME68 | [ | ||
| Thalassospiramides | Pseudomonas putida EM383 | ||||
| CBE/ABE | Undecylprodigiosin, Actinorhodin | S. coelicolor M145 | [ | ||
| Avermectin | S. avermitilis MA4680 | ||||
| 产物 衍生化 | CRISPR/Cas9 & Gibson Assembly | Rapamycin | 组装模块编辑 | S. avermitilis SUKA | [ |
| CRISPR/Cas9 | Enduracidin | Streptomyces fungicidicus ATCC 21013 | [ | ||
| 产物代谢调节 | CRISPR/Cas9 & TAR | Actinorhodin | 启动子工程 | S. albus J1074 | [ |
| MSGE | Pristinamyicn Ⅱ, Chloramphenicol, YM-216391 | 多拷贝 | S. pristinaespiralis HCCB10218 S. coelicolor M145 | [ | |
| CRISPR/Cas9 | Amorphadiene | 启动子工程,遗传突变, 多拷贝 | Bacillus subtilis | [ | |
| CCTL | Actinorhodin | 启动子工程 | Streptomyces sp. 4F | [ | |
| 基因簇 激活 | CRISPR/Cas9 | Alteramide A, Macrolactam 2, FR-900098 | 启动子工程 | S. roseosporus NRRL15998 | [ |
| mCRISTAR | Tetarimycin, Lazarimide, AB1210 | S. albus | [ | ||
| mpCRISTAR | Acinorhodin | S. cerevisiae BY4727 | [ | ||
| miCASTAR | Atolypene | Amycolatopsis tolypomycina NRRL B-24205 | [ | ||
| CRISPR/Cas9-LigD | Amicetin | 遗传突变 | Streptomyces WAC6237 | [ | |
| Thiolactomycin, 5-chloro-3-formylindole | Streptomyces WAC5374 | ||||
| Phenanthroviridin aglycone | Streptomyces WAC8241 | ||||
| CRISPRi/CRISPRa | Jadomycinb | 转录调控 | Streptomyces venezuelae | [ |
| 1 | BAKER D D, CHU M, OZA U, et al. The value of natural products to future pharmaceutical discovery[J]. Natural Product Reports, 2007, 24(6): 1225-1244. |
| 2 | NEWMAN D J, CRAGG G M. Natural products as sources of new drugs from 1981 to 2014[J]. Journal of Natural Products, 2016, 79(3): 629-661. |
| 3 | MOLONEY M G. Natural products as a source for novel antibiotics[J]. Trends in Pharmacological Sciences, 2016, 37(8): 689-701. |
| 4 | KIM H U, BLIN K, LEE S Y, et al. Recent development of computational resources for new antibiotics discovery[J]. Current Opinion in Microbiology, 2017, 39: 113-120. |
| 5 | TRACANNA V, JONG A D, MEDEMA M H, et al. Mining prokaryotes for antimicrobial compounds: from diversity to function[J]. FEMS Microbiology Reviews, 2017, 41(3): 417-429. |
| 6 | JIANG F G, DOUDNA J A. CRISPR-Cas9 structures and mechanisms[J]. Annual Review of Biophysics, 2017, 46: 505-529. |
| 7 | BARRANGOU R, FREMAUX C, DEVEAU H, et al. CRISPR provides acquired resistance against viruses in prokaryotes[J]. Science, 2007, 315(5819): 1709-1712. |
| 8 | MOJICA F J M, DÍEZ-VILLASEÑOR C, GARCÍA-MARTÍNEZ J, et al. Short motif sequences determine the targets of the prokaryotic CRISPR defence system[J]. Microbiology, 2009, 155(Pt 3): 733-740. |
| 9 | SAPRANAUSKAS R, GASIUNAS G, FREMAUX C, et al. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli [J]. Nucleic Acids Research, 2011, 39(21): 9275-9282. |
| 10 | DELTCHEVA E, CHYLINSKI K, SHARMA C M, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase Ⅲ[J]. Nature, 2011, 471(7340): 602-607. |
| 11 | STERNBERG S H, REDDING S, JINEK M, et al. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9[J]. Nature, 2014, 507(7490): 62-67. |
| 12 | JINEK M, CHYLINSKI K, FONFARA I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity[J]. Science, 2012, 337(6096): 816-821. |
| 13 | GASIUNAS G, BARRANGOU R, HORVATH P, et al. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(39): E2579-E2586. |
| 14 | JIANG W Y, BIKARD D, COX D, et al. RNA-guided editing of bacterial genomes using CRISPR-Cas systems[J]. Nature Biotechnology, 2013, 31(3): 233-239. |
| 15 | THOMASON L C, COSTANTINO N, LI X T, et al. Recombineering: genetic engineering in Escherichia coli using homologous recombination[J]. Current Protocols, 2023, 3(2): e656. |
| 16 | QI L S, LARSON M H, GILBERT L A, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression[J]. Cell, 2013(5): 1173-1183. |
| 17 | BIKARD D, JIANG W Y, SAMAI P, et al. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system[J]. Nucleic Acids Research, 2013, 41(15): 7429-7437. |
| 18 | DOVE S L, HOCHSCHILD A. Conversion of the omega subunit of Escherichia coli RNA polymerase into a transcriptional activator or an activation target[J]. Genes & Development, 1998, 12(5): 745-754. |
| 19 | HILTON I B, GERSBACH C A. Enabling functional genomics with genome engineering[J]. Genome Research, 2015, 25(10): 1442-1455. |
| 20 | KOMOR A C, KIM Y B, PACKER M S, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage[J]. Nature, 2016, 533(7603): 420-424. |
| 21 | GAUDELLI N M, KOMOR A C, REES H A, et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage[J]. Nature, 2017, 551(7681): 464-471. |
| 22 | LIU H B, JIANG H, HALTLI B, et al. Rapid cloning and heterologous expression of the meridamycin biosynthetic gene cluster using a versatile Escherichia coli-Streptomyces artificial chromosome vector, pSBAC[J]. Journal of Natural Products, 2009, 72(3): 389-395. |
| 23 | NAH H J, WOO M W, CHOI S S, et al. Precise cloning and tandem integration of large polyketide biosynthetic gene cluster using Streptomyces artificial chromosome system[J]. Microbial Cell Factories, 2015, 14: 140. |
| 24 | KIM J H, FENG Z Y, BAUER J D, et al. Cloning large natural product gene clusters from the environment: piecing environmental DNA gene clusters back together with TAR[J]. Biopolymers, 2010, 93(9): 833-844. |
| 25 | KOUPRINA N, LARIONOV V. Transformation-associated recombination (TAR) cloning for genomics studies and synthetic biology[J]. Chromosoma, 2016, 125(4): 621-632. |
| 26 | BIAN X Y, HUANG F, STEWART F A, et al. Direct cloning, genetic engineering, and heterologous expression of the syringolin biosynthetic gene cluster in E. coli through Red/ET recombineering[J]. Chembiochem, 2012, 13(13): 1946-1952. |
| 27 | FU J, BIAN X Y, HU S B, et al. Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting[J]. Nature Biotechnology, 2012, 30(5): 440-446. |
| 28 | MURPHY K C. Phage recombinases and their applications[J]. Advances in Virus Research, 2012, 83: 367-414. |
| 29 | JIANG W J, ZHAO X J, GABRIELI T, et al. Cas9-Assisted targeting of chromosome segments CATCH enables one-step targeted cloning of large gene clusters[J]. Nature Communications, 2015, 6: 8101. |
| 30 | ROMÁN-HURTADO F, SÁNCHEZ-HIDALGO M, MARTÍN J, et al. One pathway, two cyclic non-ribosomal pentapeptides: heterologous expression of BE-18257 antibiotics and pentaminomycins from Streptomyces cacaoi CA-170360[J]. Microorganisms, 2021, 9(1): 135. |
| 31 | TAO W X, CHEN L, ZHAO C H, et al. In vitro packaging mediated one-step targeted cloning of natural product pathway[J]. ACS Synthetic Biology, 2019, 8(9): 1991-1997. |
| 32 | ENGHIAD B, HUANG C S, GUO F, et al. Cas12a-assisted precise targeted cloning using in vivo Cre-lox recombination[J]. Nature Communications, 2021, 12(1): 1171. |
| 33 | LIANG M D, LIU L S, XU F, et al. Activating cryptic biosynthetic gene cluster through a CRISPR-Cas12a-mediated direct cloning approach[J]. Nucleic Acids Research, 2022, 50(6): 3581-3592. |
| 34 | LIU Y K, TAO W X, WEN S S, et al. In vitro CRISPR/Cas9 system for efficient targeted DNA editing[J]. mBio, 2015, 6(6): e01714-15. |
| 35 | COBB R E, WANG Y J, ZHAO H M. High-efficiency multiplex genome editing of Streptomyces species using an engineered CRISPR/Cas system[J]. ACS Synthetic Biology, 2015, 4(6): 723-728. |
| 36 | HUANG H, ZHENG G S, JIANG W H, et al. One-step high-efficiency CRISPR/Cas9-mediated genome editing in Streptomyces [J]. Acta Biochimica et Biophysica Sinica, 2015, 47(4): 231-243. |
| 37 | TONG Y J, CHARUSANTI P, ZHANG L X, et al. CRISPR-Cas9 based engineering of actinomycetal genomes[J]. ACS Synthetic Biology, 2015, 4(9): 1020-1029. |
| 38 | ZENG H, WEN S S, XU W, et al. Highly efficient editing of the actinorhodin polyketide chain length factor gene in Streptomyces coelicolor M145 using CRISPR/Cas9-CodA(sm) combined system[J]. Applied Microbiology and Biotechnology, 2015, 99(24): 10575-10585. |
| 39 | ZHANG J J, MOORE B S. Site-directed mutagenesis of large biosynthetic gene clusters via oligonucleotide recombineering and CRISPR/Cas9 targeting[J]. ACS Synthetic Biology, 2020, 9(7): 1917-1922. |
| 40 | ZHONG Z Y, GUO J H, DENG L, et al. Base editing in Streptomyces with Cas9-deaminase fusions[EB/OL]. bioRxiv. (2019-05-07)[2023-11-01]. . |
| 41 | KUDO K, HASHIMOTO T, HASHIMOTO J, et al. In vitro Cas9-assisted editing of modular polyketide synthase genes to produce desired natural product derivatives[J]. Nature Communications, 2020, 11(1): 4022. |
| 42 | THONG W L, ZHANG Y X, ZHUO Y, et al. Gene editing enables rapid engineering of complex antibiotic assembly lines[J]. Nature Communications, 2021, 12(1): 6872. |
| 43 | JI C H, KIM H, KANG H S. Synthetic inducible regulatory systems optimized for the modulation of secondary metabolite production in Streptomyces [J]. ACS Synthetic Biology, 2019, 8(3): 577-586. |
| 44 | LI L, ZHENG G S, CHEN J, et al. Multiplexed site-specific genome engineering for overproducing bioactive secondary metabolites in actinomycetes[J]. Metabolic Engineering, 2017, 40: 80-92. |
| 45 | SONG Y F, HE S Q, ABDALLAH I I, et al. Engineering of multiple modules to improve amorphadiene production in Bacillus subtilis using CRISPR-Cas9[J]. Journal of Agricultural and Food Chemistry, 2021, 69(16): 4785-4794. |
| 46 | LEI C, LI S Y, LIU J K, et al. The CCTL (Cpf1-assisted cutting and Taq DNA ligase-assisted ligation) method for efficient editing of large DNA constructs in vitro [J]. Nucleic Acids Research, 2017, 45(9): e74. |
| 47 | ZHANG M M, WONG F T, WANG Y J, et al. CRISPR-Cas9 strategy for activation of silent Streptomyces biosynthetic gene clusters[J]. Nature Chemical Biology, 2017, 13: 607-609. |
| 48 | KANG H S, CHARLOP-POWERS Z, BRADY S F. Multiplexed CRISPR/Cas9- and TAR-mediated promoter engineering of natural product biosynthetic gene clusters in yeast[J]. ACS Synthetic Biology, 2016, 5(9): 1002-1010. |
| 49 | KIM H Y, JI C H, JE H W, et al. mpCRISTAR: multiple plasmid approach for CRISPR/Cas9 and TAR-mediated multiplexed refactoring of natural product biosynthetic gene clusters[J]. ACS Synthetic Biology, 2020, 9(1): 175-180. |
| 50 | KIM S H, LU W L, AHMADI M K, et al. Atolypenes, tricyclic bacterial sesterterpenes discovered using a multiplexed in vitro Cas9-TAR gene cluster refactoring approach[J]. ACS Synthetic Biology, 2019, 8(1): 109-118. |
| 51 | CULP E J, YIM G, WAGLECHNER N, et al. Hidden antibiotics in actinomycetes can be identified by inactivation of gene clusters for common antibiotics[J]. Nature Biotechnology, 2019, 37(10): 1149-1154. |
| 52 | AMERUOSO A, VILLEGAS KCAM M C, COHEN K P, et al. Activating natural product synthesis using CRISPR interference and activation systems in Streptomyces [J]. Nucleic Acids Research, 2022, 50(13): 7751-7760. |
| 53 | WANG J W, WANG A, LI K Y, et al. CRISPR/Cas9 nuclease cleavage combined with Gibson assembly for seamless cloning[J]. BioTechniques, 2015, 58(4): 161-170. |
| 54 | LEE N C O, LARIONOV V, KOUPRINA N. Highly efficient CRISPR/Cas9-mediated TAR cloning of genes and chromosomal loci from complex genomes in yeast[J]. Nucleic Acids Research, 2015, 43(8): e55. |
| 55 | ZHOU J T, WU R H, XUE X L, et al. CasHRA (Cas9-facilitated homologous recombination assembly) method of constructing megabase-sized DNA[J]. Nucleic Acids Research, 2016, 44(14): e124. |
| 56 | HOHN B, WURTZ M, KLEIN B, et al. Phage lambda DNA packaging, in vitro [J]. Journal of Supramolecular Structure, 1974, 2(2-4): 302-317. |
| 57 | DATSENKO K A, WANNER B L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products[J]. Proceedings of the National Academy of Sciences of the United States of America, 2000, 97(12): 6640-6645. |
| 58 | TISCHER B K, VON EINEM J, KAUFER B, et al. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli [J]. BioTechniques, 2006, 40(2): 191-197. |
| 59 | SIEGL T, PETZKE L, WELLE E, et al. I-SceI endonuclease: a new tool for DNA repair studies and genetic manipulations in Streptomyces [J]. Applied Microbiology & Biotechnology, 2010, 87(4): 1525-1532. |
| 60 | HERRMANN S, SIEGL T, LUZHETSKA M, et al. Site-specific recombination strategies for engineering actinomycete genomes[J]. Applied & Environmental Microbiology, 2012, 78(6): 1804-1812. |
| 61 | WOLF T, GREN T, THIEME E, et al. Targeted genome editing in the rare actinomycete Actinoplanes sp. SE50/110 by using the CRISPR/Cas9 System[J]. Journal of Biotechnology, 2016, 231: 122-128. |
| 62 | ZHANG Y, YUN K Y, HUANG H M, et al. Antisense RNA interference-enhanced CRISPR/Cas9 base editing method for improving base editing efficiency in Streptomyces lividans 66[J]. ACS Synthetic Biology, 2021, 10(5): 1053-1063. |
| 63 | SHAFEE T, LOWE R. Eukaryotic and prokaryotic gene structure[J]. WikiJournal of Medicine, 2017, 4(1): 2. |
| 64 | JIN L Q, JIN W R, MA Z C, et al. Promoter engineering strategies for the overproduction of valuable metabolites in microbes[J]. Applied Microbiology and Biotechnology, 2019, 103(21-22): 8725-8736. |
| 65 | WANG Y, CHENG H J, LIU Y, et al. In-situ generation of large numbers of genetic combinations for metabolic reprogramming via CRISPR-guided base editing[J]. Nature Communications, 2021, 12(1): 678. |
| 66 | RUTLEDGE P J, CHALLIS G L. Discovery of microbial natural products by activation of silent biosynthetic gene clusters[J]. Nature Reviews Microbiology, 2015, 13(8): 509-523. |
| 67 | ZETSCHE B, GOOTENBERG J S, ABUDAYYEH O O, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system[J]. Cell, 2015, 163(3): 759-771. |
| 68 | FONFARA I, RICHTER H, BRATOVIČ M, et al. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA[J]. Nature, 2016, 532(7600): 517-521. |
| 69 | ZETSCHE B, HEIDENREICH M, MOHANRAJU P, et al. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array[J]. Nature Biotechnology, 2017, 35: 31-34. |
| 70 | LI L, WEI K K, ZHENG G S, et al. CRISPR-Cpf1-assisted multiplex genome editing and transcriptional repression in Streptomyces [J]. Applied and Environmental Microbiology, 2018, 84(18): e00827-18. |
| 71 | SCHILLING C, KOFFAS M A G, SIEBER V, et al. Novel prokaryotic CRISPR-Cas12a-based tool for programmable transcriptional activation and repression[J]. ACS Synthetic Biology, 2020, 9(12): 3353-3363. |
| 72 | LI S Y, ZHAO G P, WANG J. C-brick: a new standard for assembly of biological parts using Cpf1[J]. ACS Synthetic Biology, 2016, 5(12): 1383-1388. |
| 73 | BAUMAN K D, BUTLER K S, MOORE B S, et al. Genome mining methods to discover bioactive natural products[J]. Natural Product Reports, 2021, 38(11): 2100-2129. |
| [1] | 郭姝媛, 张倩楠, 姑丽克孜·买买提热夏提, 杨一群, 于涛. 液体生物燃料合成与炼制的研究进展[J]. 合成生物学, 2025, 6(1): 18-44. |
| [2] | 高歌, 边旗, 王宝俊. 合成基因线路的工程化设计研究进展与展望[J]. 合成生物学, 2025, 6(1): 45-64. |
| [3] | 李冀渊, 吴国盛. 合成生物学视域下有机体的两种隐喻[J]. 合成生物学, 2025, 6(1): 190-202. |
| [4] | 焦洪涛, 齐蒙, 邵滨, 蒋劲松. DNA数据存储技术的法律治理议题[J]. 合成生物学, 2025, 6(1): 177-189. |
| [5] | 唐兴华, 陆钱能, 胡翌霖. 人类世中对合成生物学的哲学反思[J]. 合成生物学, 2025, 6(1): 203-212. |
| [6] | 徐怀胜, 石晓龙, 刘晓光, 徐苗苗. DNA存储的关键技术:编码、纠错、随机访问与安全性[J]. 合成生物学, 2025, 6(1): 157-176. |
| [7] | 石婷, 宋展, 宋世怡, 张以恒. 体外生物转化(ivBT):生物制造的新前沿[J]. 合成生物学, 2024, 5(6): 1437-1460. |
| [8] | 柴猛, 王风清, 魏东芝. 综合利用木质纤维素生物转化合成有机酸[J]. 合成生物学, 2024, 5(6): 1242-1263. |
| [9] | 邵明威, 孙思勉, 杨时茂, 陈国强. 基于极端微生物的生物制造[J]. 合成生物学, 2024, 5(6): 1419-1436. |
| [10] | 赵亮, 李振帅, 付丽平, 吕明, 王士安, 张全, 刘立成, 李福利, 刘自勇. 生物转化一碳化合物原料产油脂与单细胞蛋白研究进展[J]. 合成生物学, 2024, 5(6): 1300-1318. |
| [11] | 竺方欢, 岑雪聪, 陈振. 微生物合成二元醇研究进展[J]. 合成生物学, 2024, 5(6): 1367-1385. |
| [12] | 陈雨, 张康, 邱以婧, 程彩云, 殷晶晶, 宋天顺, 谢婧婧. 微生物电合成技术转化二氧化碳研究进展[J]. 合成生物学, 2024, 5(5): 1142-1168. |
| [13] | 郑皓天, 李朝风, 刘良叙, 王嘉伟, 李恒润, 倪俊. 负碳人工光合群落的设计、优化与应用[J]. 合成生物学, 2024, 5(5): 1189-1210. |
| [14] | 夏孔晨, 徐维华, 吴起. 光酶催化混乱性反应的研究进展[J]. 合成生物学, 2024, 5(5): 997-1020. |
| [15] | 程中玉, 李付琸. 基于P450选择性氧化的天然产物化学-酶法合成进展[J]. 合成生物学, 2024, 5(5): 960-980. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||