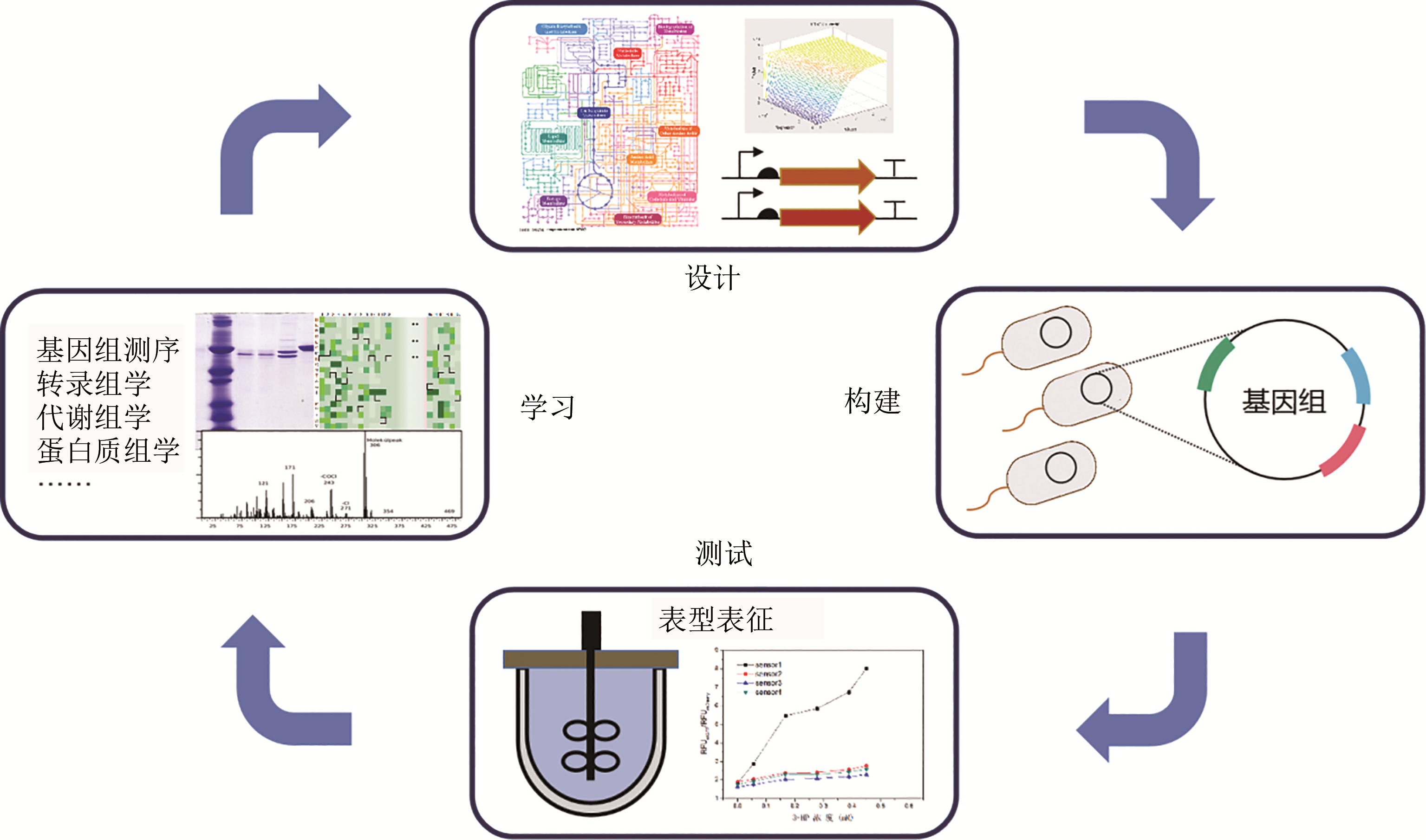
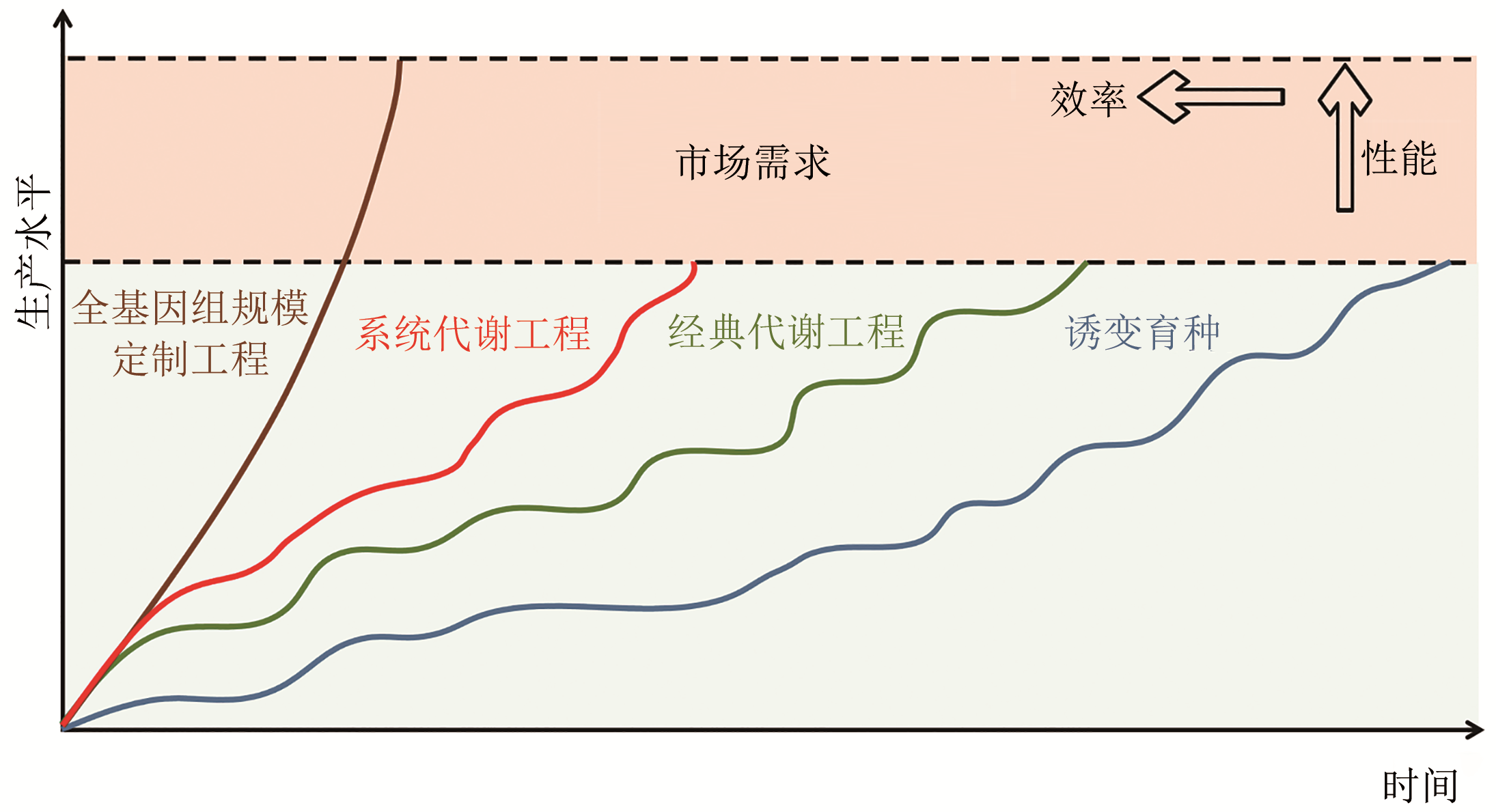
Synthetic Biology Journal ›› 2020, Vol. 1 ›› Issue (6): 656-673.DOI: 10.12211/2096-8208.2020-050
• Invited Review • Previous Articles Next Articles
Progress and prospective of engineering microbial cell factories: from random mutagenesis to customized design in genome scale
YUAN Yaomeng1, XING Xinhui1,2, ZHANG Chong1
- 1.MOE Key Laboratory for Industrial Biocatalysis,Institute of Biochemical Engineering,Department of Chemical Engineering,Center for Synthetic & Systems Biology,Tsinghua University,Beijing 100084,China
2.Institute of Biopharmaceutical and Health Engineering,Tsinghua Shenzhen International Graduate School,Shenzhen 518055,Guangdong,China
-
Received:2020-04-16Revised:2020-09-26Online:2021-01-15Published:2020-12-31 -
Contact:ZHANG Chong
微生物细胞工厂的设计构建:从诱变育种到全基因组定制化创制
袁姚梦1, 邢新会1,2, 张翀1
- 1.工业生物催化教育部重点实验室,清华大学化工系生物化工研究所,清华大学合成与系统生物学研究中心,北京 100084
2.清华大学深圳国际研究生院生物医药与健康工程研究院,广东 深圳 518055
-
通讯作者:张翀 -
作者简介:袁姚梦(1997—),女,博士研究生。主要研究方向为合成生物学、代谢工程。E-mail:2631825401@qq.com
张翀(1979—),男,博士,副教授,博士生导师。主要研究方向为合成生物学、代谢工程、生物化工、系统生物学。E-mail:chongzhang@mail.tsinghua.edu.cn -
基金资助:国家自然科学基金重点项目(2193000018);国家重点研发计划(2019YFA0904802)
CLC Number:
Cite this article
YUAN Yaomeng, XING Xinhui, ZHANG Chong. Progress and prospective of engineering microbial cell factories: from random mutagenesis to customized design in genome scale[J]. Synthetic Biology Journal, 2020, 1(6): 656-673.
袁姚梦, 邢新会, 张翀. 微生物细胞工厂的设计构建:从诱变育种到全基因组定制化创制[J]. 合成生物学, 2020, 1(6): 656-673.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8208.2020-050
| 分类 | 诱变技术/诱变剂 | 描述 | 参考文献 |
|---|---|---|---|
| 物理诱变 | 电离辐射(X射线、γ射线等) | 引起DNA双链或单链断裂,实现DNA的删除或结构改变 | [ |
| 非电离辐射(紫外线) | 使嘧啶形成二聚体,实现GC的删除、移码突变以及GC→AT的转换 | [ | |
| 化学诱变 | 烷化剂(烷基磺酸盐、芥子气等) | 使DNA碱基发生烷化,导致DNA复制时发生配对错误 | [ |
碱基类似物 (嘧啶类似物、嘌呤类似物) | 与DNA碱基结构类似,在DNA复制时掺入并引发配对错误 | [ | |
| 移码诱变剂(原黄素、吖啶橙等) | 与DNA结合导致碱基增添或缺失 | [ | |
| 脱氨剂(亚硝酸) | 引起A、C、G碱基的脱氨,实现GC与AT的相互转换;引起DNA交联作用,引发突变 | [ | |
| 羟化剂(羟胺) | 引起胞嘧啶脱氨,实现GC→AT的转换 | [ | |
| 生物诱变 | 噬菌体、质粒和DNA转座子 | 通过碱基取代和DNA链断裂实现碱基的删除、重复和插入 | [ |
| 原生质体融合 | 将两个亲株的原生质体进行融合,形成杂合二倍体,使两个亲株发生基因组重组 | [ | |
| DNA改组 | 对多个同源序列组成的文库进行随机片段化,再利用PCR使其发生随机的重组,实现多亲本的基因重组 | [ | |
| 基因组重排 | 先利用传统诱变手段获得多个表型改进的菌株,然后模拟DNA改组的反应条件,对原生质体进行多次递推式融合,实现正向突变的富集 | [ | |
| 复合诱变 | 结合多种诱变方式提高诱变效率 | [ | |
| 新型诱变技术 | 离子注入诱变 | 离子注入细胞导致DNA损伤,细胞修复损伤的过程中出现突变 | [ |
| 等离子体诱变 | 等离子体作用于细胞造成DNA损伤,细胞修复损伤的过程中出现突变 | [ |
Tab. 1 Summary of mutagenesis technologies
| 分类 | 诱变技术/诱变剂 | 描述 | 参考文献 |
|---|---|---|---|
| 物理诱变 | 电离辐射(X射线、γ射线等) | 引起DNA双链或单链断裂,实现DNA的删除或结构改变 | [ |
| 非电离辐射(紫外线) | 使嘧啶形成二聚体,实现GC的删除、移码突变以及GC→AT的转换 | [ | |
| 化学诱变 | 烷化剂(烷基磺酸盐、芥子气等) | 使DNA碱基发生烷化,导致DNA复制时发生配对错误 | [ |
碱基类似物 (嘧啶类似物、嘌呤类似物) | 与DNA碱基结构类似,在DNA复制时掺入并引发配对错误 | [ | |
| 移码诱变剂(原黄素、吖啶橙等) | 与DNA结合导致碱基增添或缺失 | [ | |
| 脱氨剂(亚硝酸) | 引起A、C、G碱基的脱氨,实现GC与AT的相互转换;引起DNA交联作用,引发突变 | [ | |
| 羟化剂(羟胺) | 引起胞嘧啶脱氨,实现GC→AT的转换 | [ | |
| 生物诱变 | 噬菌体、质粒和DNA转座子 | 通过碱基取代和DNA链断裂实现碱基的删除、重复和插入 | [ |
| 原生质体融合 | 将两个亲株的原生质体进行融合,形成杂合二倍体,使两个亲株发生基因组重组 | [ | |
| DNA改组 | 对多个同源序列组成的文库进行随机片段化,再利用PCR使其发生随机的重组,实现多亲本的基因重组 | [ | |
| 基因组重排 | 先利用传统诱变手段获得多个表型改进的菌株,然后模拟DNA改组的反应条件,对原生质体进行多次递推式融合,实现正向突变的富集 | [ | |
| 复合诱变 | 结合多种诱变方式提高诱变效率 | [ | |
| 新型诱变技术 | 离子注入诱变 | 离子注入细胞导致DNA损伤,细胞修复损伤的过程中出现突变 | [ |
| 等离子体诱变 | 等离子体作用于细胞造成DNA损伤,细胞修复损伤的过程中出现突变 | [ |
| 类别 | 分析方法 | 描述 | 参考文献 |
|---|---|---|---|
| 动力学分析 | ODE&米氏方程 | 利用常微分方程(ODE)和米氏方程,能够描述胞内代谢物浓度随时间的变化趋势,从而建立代谢网络的动力学模型 | [ |
| 代谢控制分析(MCA) | MCA可用于估算动力学模型中各个通量的控制系数,从而确定代谢途径中需要过表达的目标基因,增加通过途径的通量 | [ | |
| 代谢网络分析 | 代谢通量分析(MFA) | MFA根据胞内代谢物的质量平衡确定线性方程组并求解,能够计算特定培养条件下细胞内的实际代谢通量分布。MFA需要依赖大量实验数据来增加可测量通量的数量,从而计算出不可测量通量向量 | [ |
| 通量平衡分析(FBA) | FBA可用于确定细胞代谢网络中每个反应的最佳通量,该方法基于凸分析,通过对系统施加最大化(最小化)的目标函数来确定代谢通量矢量 | [ | |
| 代谢途径分析(MPA) | MPA可用于识别代谢网络中存在的所有代谢通量向量,该方法仅以化学计量学和反应热力学为约束条件,不需要动力学参数进行计算(例如基元模式分析) | [ | |
| 整合了热力学的代谢网络分析 | 网络嵌入的热力学分析(NET) | 以热力学第二定律为依据,可用于检测代谢组学数据和假定的通量方向是否符合热力学一致性 | [ |
| 基于热力学的代谢通量分析(TMFA) | TMFA同时使用热力学方向性约束和质量守恒约束计算代谢通量分布 | [ | |
| 计算机应变优化算法 | 基于约束的重构与分析(COBRA) | COBRA采用基因组规模的计算机模拟,可用于代谢途径预测和优化,从而改善生产速率和产量 | [ |
| 最小化代谢调节(MOMA) | MOMA用于使野生型菌株和缺失突变体之间代谢通量分布的差别最小化,能够预测基因操作对代谢网络的影响 | [ | |
| 开/关最小化调节(ROOM) | 与MOMA的优化目标相同 | [ | |
| OptKnock | 以产品产率为优化目标的基因敲除分析工具 | [ | |
| 生化网络集成计算浏览器(BNICE) | 使用广义酶反应规则发现新代谢途径的计算工具 | [ |
Tab. 2 Analytical methods for metabolic network design
| 类别 | 分析方法 | 描述 | 参考文献 |
|---|---|---|---|
| 动力学分析 | ODE&米氏方程 | 利用常微分方程(ODE)和米氏方程,能够描述胞内代谢物浓度随时间的变化趋势,从而建立代谢网络的动力学模型 | [ |
| 代谢控制分析(MCA) | MCA可用于估算动力学模型中各个通量的控制系数,从而确定代谢途径中需要过表达的目标基因,增加通过途径的通量 | [ | |
| 代谢网络分析 | 代谢通量分析(MFA) | MFA根据胞内代谢物的质量平衡确定线性方程组并求解,能够计算特定培养条件下细胞内的实际代谢通量分布。MFA需要依赖大量实验数据来增加可测量通量的数量,从而计算出不可测量通量向量 | [ |
| 通量平衡分析(FBA) | FBA可用于确定细胞代谢网络中每个反应的最佳通量,该方法基于凸分析,通过对系统施加最大化(最小化)的目标函数来确定代谢通量矢量 | [ | |
| 代谢途径分析(MPA) | MPA可用于识别代谢网络中存在的所有代谢通量向量,该方法仅以化学计量学和反应热力学为约束条件,不需要动力学参数进行计算(例如基元模式分析) | [ | |
| 整合了热力学的代谢网络分析 | 网络嵌入的热力学分析(NET) | 以热力学第二定律为依据,可用于检测代谢组学数据和假定的通量方向是否符合热力学一致性 | [ |
| 基于热力学的代谢通量分析(TMFA) | TMFA同时使用热力学方向性约束和质量守恒约束计算代谢通量分布 | [ | |
| 计算机应变优化算法 | 基于约束的重构与分析(COBRA) | COBRA采用基因组规模的计算机模拟,可用于代谢途径预测和优化,从而改善生产速率和产量 | [ |
| 最小化代谢调节(MOMA) | MOMA用于使野生型菌株和缺失突变体之间代谢通量分布的差别最小化,能够预测基因操作对代谢网络的影响 | [ | |
| 开/关最小化调节(ROOM) | 与MOMA的优化目标相同 | [ | |
| OptKnock | 以产品产率为优化目标的基因敲除分析工具 | [ | |
| 生化网络集成计算浏览器(BNICE) | 使用广义酶反应规则发现新代谢途径的计算工具 | [ |
| 模型范围 | 模型名称 | 菌株 | 反应/代谢物 | 参考文献 |
|---|---|---|---|---|
| 核心代谢模型 | 未报道 | 大肠杆菌 | 95/94 | [ |
| 未报道 | 谷氨酸棒杆菌 | 未报道 | [ | |
| 未报道 | 酿酒酵母 | 70/83 | [ | |
| 未报道 | 酿酒酵母 | 78/98 | [ | |
| 未报道 | 酿酒酵母 | 37/27 | [ | |
| 全基因组代谢模型(GSMM) | iFF708 | 酿酒酵母 | 1145/825 +708基因 | [ |
| iND750 | 酿酒酵母 | 1149/646 +750基因 | [ | |
| Yeast 4.0 | 酿酒酵母 | 1865/1319 +932基因 | [ | |
| iJO1366 | 大肠杆菌 | 2583/1805 +1366基因 | [ | |
| 未报道 | 谷氨酸棒杆菌 | 495/408 +411基因 | [ | |
| 未报道 | 乳酸菌 | 621/509 +358基因 | [ | |
| 未报道 | 枯草芽孢杆菌 | 754/637 +614基因 | [ | |
全基因组代谢模型 +基因表达水平 | T. maritima | 海栖热袍菌 | 17535/18209 | [ |
| iOL1650-ME | 大肠杆菌 | 76414/56902 | [ | |
全基因组代谢模型 +蛋白质结构特性 | T. maritima | 海栖热袍菌 | 645/503 +478酶结构 | [ |
| E. coli GEM-PRO | 大肠杆菌 | 2583/1805 +1268酶结构 | [ | |
全基因组代谢模型 +转录调控 | iAF1260 PROM | 大肠杆菌 | 2583/1805 +1773转录调控作用 | [ |
| iMH805/837 | 酿酒酵母 | 1489/972 +805基因+837转录调控作用 | [ | |
| 全细胞模型 | ‘Whole-cell’ M. genitalium | 生殖支原体 | 28个细胞过程子模块 +525基因 | [ |
Tab. 3 Summary of microbial metabolism models
| 模型范围 | 模型名称 | 菌株 | 反应/代谢物 | 参考文献 |
|---|---|---|---|---|
| 核心代谢模型 | 未报道 | 大肠杆菌 | 95/94 | [ |
| 未报道 | 谷氨酸棒杆菌 | 未报道 | [ | |
| 未报道 | 酿酒酵母 | 70/83 | [ | |
| 未报道 | 酿酒酵母 | 78/98 | [ | |
| 未报道 | 酿酒酵母 | 37/27 | [ | |
| 全基因组代谢模型(GSMM) | iFF708 | 酿酒酵母 | 1145/825 +708基因 | [ |
| iND750 | 酿酒酵母 | 1149/646 +750基因 | [ | |
| Yeast 4.0 | 酿酒酵母 | 1865/1319 +932基因 | [ | |
| iJO1366 | 大肠杆菌 | 2583/1805 +1366基因 | [ | |
| 未报道 | 谷氨酸棒杆菌 | 495/408 +411基因 | [ | |
| 未报道 | 乳酸菌 | 621/509 +358基因 | [ | |
| 未报道 | 枯草芽孢杆菌 | 754/637 +614基因 | [ | |
全基因组代谢模型 +基因表达水平 | T. maritima | 海栖热袍菌 | 17535/18209 | [ |
| iOL1650-ME | 大肠杆菌 | 76414/56902 | [ | |
全基因组代谢模型 +蛋白质结构特性 | T. maritima | 海栖热袍菌 | 645/503 +478酶结构 | [ |
| E. coli GEM-PRO | 大肠杆菌 | 2583/1805 +1268酶结构 | [ | |
全基因组代谢模型 +转录调控 | iAF1260 PROM | 大肠杆菌 | 2583/1805 +1773转录调控作用 | [ |
| iMH805/837 | 酿酒酵母 | 1489/972 +805基因+837转录调控作用 | [ | |
| 全细胞模型 | ‘Whole-cell’ M. genitalium | 生殖支原体 | 28个细胞过程子模块 +525基因 | [ |
| 产物 | 宿主 | 原料 | 公司 | 参考文献 |
|---|---|---|---|---|
| 琥珀酸 | 大肠杆菌 | 玉米糖 | BioAmber | [ |
大肠杆菌 克鲁斯假丝酵母 | 蔗糖 | Myriant(现名GC Innovation America) | ||
| 酿酒酵母 | 淀粉、糖类 | Reverdia | ||
| 巴斯夫产琥珀酸菌 | 甘油、糖类 | Succinity | ||
| 1,4-丁二醇 | 大肠杆菌 | 糖类 | Genomatica和DuPont Tate & Lyle | [ |
| 1,3-丙二醇 | 大肠杆菌 | 糖类 | DuPont Tate & Lyle | [ |
| 聚羟基链烷酸酯(PHA) | 大肠杆菌 | 糖类 | Metabolix(现名 Yeild10 science) | [ |
| 3-羟基丙酸 | 大肠杆菌 | 未报道 | OPXbio & Dow Chemical | [ |
| 未报道 | 未报道 | Perstorp | ||
| 乙醇 | 酿酒酵母 运动发酵单胞菌 马克斯克鲁维酵母 | 蔗糖、玉米糖、木质纤维素 | [ | |
| 异丁醇 | 酿酒酵母 | 糖类 | Gevo Butalco Butamax | [ |
| 法尼烯 | 酿酒酵母 | 未报道 | Amyris | [ |
| 青蒿素(半合成) | 酿酒酵母 | 未报道 | Amyris | [ |
Tab. 4 Commercial application of classic design strategies guided by metabolic engineering
| 产物 | 宿主 | 原料 | 公司 | 参考文献 |
|---|---|---|---|---|
| 琥珀酸 | 大肠杆菌 | 玉米糖 | BioAmber | [ |
大肠杆菌 克鲁斯假丝酵母 | 蔗糖 | Myriant(现名GC Innovation America) | ||
| 酿酒酵母 | 淀粉、糖类 | Reverdia | ||
| 巴斯夫产琥珀酸菌 | 甘油、糖类 | Succinity | ||
| 1,4-丁二醇 | 大肠杆菌 | 糖类 | Genomatica和DuPont Tate & Lyle | [ |
| 1,3-丙二醇 | 大肠杆菌 | 糖类 | DuPont Tate & Lyle | [ |
| 聚羟基链烷酸酯(PHA) | 大肠杆菌 | 糖类 | Metabolix(现名 Yeild10 science) | [ |
| 3-羟基丙酸 | 大肠杆菌 | 未报道 | OPXbio & Dow Chemical | [ |
| 未报道 | 未报道 | Perstorp | ||
| 乙醇 | 酿酒酵母 运动发酵单胞菌 马克斯克鲁维酵母 | 蔗糖、玉米糖、木质纤维素 | [ | |
| 异丁醇 | 酿酒酵母 | 糖类 | Gevo Butalco Butamax | [ |
| 法尼烯 | 酿酒酵母 | 未报道 | Amyris | [ |
| 青蒿素(半合成) | 酿酒酵母 | 未报道 | Amyris | [ |
| 分类 | 名称 | 描述 | 参考 文献 |
|---|---|---|---|
| 不可追踪技术 | MAGE | 基于重组的基因组编辑技术,可使用多个ssDNA同时对多个目标位点进行修饰。与其他基因编辑工具(如CRISPR/Cas9)联用可进一步提高基因编辑效率。主要用于原核基因组编辑 | [ |
| YOGE | 原理与MAGE类似,主要用于真核基因组编辑 | [ | |
| 可追踪技术 | TRMR | 基于同源重组的基因组编辑技术,能够同时对基因组上千个基因位点进行修饰 | [ |
| CREATE | 该技术基于同源重组和CRISPR/Cas9基因编辑技术,能够在全基因组范围内实现可追踪编辑 | [ | |
| Prime Editor | 使用融合了工程逆转录酶的催化活性受损的Cas9和pegRNA,以更高的效率、更低的脱靶率在全基因组范围内实现所有12种单碱基的自由转换以及多碱基的精准插入和删除 | [ | |
| Target AID | 激活诱导的胞嘧啶脱氨酶(AID)可实现C到T的突变,Target AID技术以核酸酶活性缺失的CRISPR/Cas9系统作为AID的DNA靶向模块,实现定点诱变 | [ | |
| TAM | 该系统将AID-P182X与dCas9蛋白融合,可将G或C突变为另外三个碱基,从而在目标位点处产生大量突变 | [ | |
| CRISPR-X | 使用dCas9募集更高活性的AIDΔ和MS2修饰的sgRNA变体,能以较低的脱靶率同时实现多个靶基因的特异性突变 | [ | |
| EVOLVR | 该系统由一个CRISPR引导的切口酶和一个易错DNA聚合酶组成,可在靶位点处可调窗口长度内实现所有核苷酸的突变 | [ | |
| CRISPRi | 使用dCas9蛋白及其sgRNA阻断靶基因转录,实现基因表达水平的下调 | [ | |
| CRISPRa | 使用与转录激活因子融合的dCas9蛋白实现靶基因表达水平的上调 | [ |
Tab. 5 High-throughput genotype construction technologies
| 分类 | 名称 | 描述 | 参考 文献 |
|---|---|---|---|
| 不可追踪技术 | MAGE | 基于重组的基因组编辑技术,可使用多个ssDNA同时对多个目标位点进行修饰。与其他基因编辑工具(如CRISPR/Cas9)联用可进一步提高基因编辑效率。主要用于原核基因组编辑 | [ |
| YOGE | 原理与MAGE类似,主要用于真核基因组编辑 | [ | |
| 可追踪技术 | TRMR | 基于同源重组的基因组编辑技术,能够同时对基因组上千个基因位点进行修饰 | [ |
| CREATE | 该技术基于同源重组和CRISPR/Cas9基因编辑技术,能够在全基因组范围内实现可追踪编辑 | [ | |
| Prime Editor | 使用融合了工程逆转录酶的催化活性受损的Cas9和pegRNA,以更高的效率、更低的脱靶率在全基因组范围内实现所有12种单碱基的自由转换以及多碱基的精准插入和删除 | [ | |
| Target AID | 激活诱导的胞嘧啶脱氨酶(AID)可实现C到T的突变,Target AID技术以核酸酶活性缺失的CRISPR/Cas9系统作为AID的DNA靶向模块,实现定点诱变 | [ | |
| TAM | 该系统将AID-P182X与dCas9蛋白融合,可将G或C突变为另外三个碱基,从而在目标位点处产生大量突变 | [ | |
| CRISPR-X | 使用dCas9募集更高活性的AIDΔ和MS2修饰的sgRNA变体,能以较低的脱靶率同时实现多个靶基因的特异性突变 | [ | |
| EVOLVR | 该系统由一个CRISPR引导的切口酶和一个易错DNA聚合酶组成,可在靶位点处可调窗口长度内实现所有核苷酸的突变 | [ | |
| CRISPRi | 使用dCas9蛋白及其sgRNA阻断靶基因转录,实现基因表达水平的下调 | [ | |
| CRISPRa | 使用与转录激活因子融合的dCas9蛋白实现靶基因表达水平的上调 | [ |
| 技术 | 通量 | 功能 | 案例 | 参考文献 |
|---|---|---|---|---|
| FACS | 约105细胞/s | 分选 | 利用色氨酸传感器,实现紫色杆菌素高产大肠杆菌筛选 | [ |
| 利用甲羟戊酸细胞传感器,实现甲羟戊酸高产甲基杆菌筛选 | [ | |||
| 利用赖氨酸传感器,实现赖氨酸高产谷氨酸棒杆菌筛选 | [ | |||
| FADS | 约104液滴/s | 培养+分选 | 将胞外重组酶活性与液滴荧光强度耦合,实现酶活测定及胞外重组酶高产菌株筛选 | [ |
| 将传感器菌与MCF菌株共培养,实现对香豆酸高产酵母的筛选 | [ | |||
| 将β-半乳糖苷酶产量与液滴荧光强度耦合,实现β-半乳糖苷酶高产大肠杆菌筛选 | [ | |||
| Droplet-FACS | 培养+分选 | 通过检测核黄素的荧光,实现核黄素高产解脂耶氏酵母筛选 | [ | |
| Gel FACS | 约3000液滴/s | 培养+分选 | 将胞外木聚糖酶的产量与凝胶液滴的荧光强度耦合,实现胞外木聚糖酶高产毕赤酵母筛选 | [ |
| RADS | 260细胞/min | 培养+分选 | 利用虾青素的拉曼光谱,实现虾青素高产雨生红球藻的筛选 | [ |
Tab. 6 High-throughput selection/screening technologies in single-cell level
| 技术 | 通量 | 功能 | 案例 | 参考文献 |
|---|---|---|---|---|
| FACS | 约105细胞/s | 分选 | 利用色氨酸传感器,实现紫色杆菌素高产大肠杆菌筛选 | [ |
| 利用甲羟戊酸细胞传感器,实现甲羟戊酸高产甲基杆菌筛选 | [ | |||
| 利用赖氨酸传感器,实现赖氨酸高产谷氨酸棒杆菌筛选 | [ | |||
| FADS | 约104液滴/s | 培养+分选 | 将胞外重组酶活性与液滴荧光强度耦合,实现酶活测定及胞外重组酶高产菌株筛选 | [ |
| 将传感器菌与MCF菌株共培养,实现对香豆酸高产酵母的筛选 | [ | |||
| 将β-半乳糖苷酶产量与液滴荧光强度耦合,实现β-半乳糖苷酶高产大肠杆菌筛选 | [ | |||
| Droplet-FACS | 培养+分选 | 通过检测核黄素的荧光,实现核黄素高产解脂耶氏酵母筛选 | [ | |
| Gel FACS | 约3000液滴/s | 培养+分选 | 将胞外木聚糖酶的产量与凝胶液滴的荧光强度耦合,实现胞外木聚糖酶高产毕赤酵母筛选 | [ |
| RADS | 260细胞/min | 培养+分选 | 利用虾青素的拉曼光谱,实现虾青素高产雨生红球藻的筛选 | [ |
| 1 | NIELSEN J. Cell factory engineering for improved production of natural products[J]. Natural Product Reports, 2019, 36(9): 1233-1236. |
| 2 | SANO C. History of glutamate production[J]. The American Journal of Clinical Nutrition, 2009, 90(3), 728S-732S. |
| 3 | OTERO J M, NIELSEN J. Industrial systems biology[J]. Biotechnology and Bioengineering, 2010, 105(3), 439-460. |
| 4 | Presidential green chemistry challenge awards[EB/OL]. . |
| 5 | ATSUMI S, HIGASHIDE W, LIAO J C. Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde[J]. Nature Biotechnology, 2009, 27(12), 1177-1180. |
| 6 | GALANIE S, THODEY K, TRENCHARD I J, et al. Complete biosynthesis of opioids in yeast[J]. Science, 2015, 349(6252): 1095-1100. |
| 7 | PARK J H, LEE K H, KIM T Y, et al. Metabolic engineering of Escherichia coli for the production of L-valine based on transcriptome analysis and in silico gene knockout simulation[J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(19): 7797-7802. |
| 8 | XIA X X, QIAN Z G, KI C S, et al. Native-sized recombinant spider silk protein produced in metabolically engineered Escherichia coli results in a strong fiber[J]. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(32):14059-14063. |
| 9 | STEEN E J, KANG Y, BOKINSKY G, et al. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass[J]. Nature, 2010, 463(7280): 559-562. |
| 10 | LEE G, NA J. Future of microbial polyesters[J]. Microbial Cell Factories, 2013, 12(1): 54. |
| 11 | BROWN S W, OLIVER S G. Isolation of ethanol-tolerant mutants of yeast by continuous selection[J]. European Journal of Applied Microbiology and Biotechnology, 1982, 16(2/3): 119-122. |
| 12 | MEADOWS L, HAWKINS K M, TSEGAYE Y, et al. Rewriting yeast central carbon metabolism for industrial isoprenoid production[J]. Nature, 2016, 537(7622): 694-697. |
| 13 | WEIKERT C, SAUER U, BAILEY J. Use of a glycerol-limited, long-term chemostat for isolation of Escherichia coli mutants with improved physiological properties[J]. Microbiology, 1997, 143(5): 1567-1574. |
| 14 | SILMAN N J, CARVER M A, JONES C W. Physiology of amidase production by methylophilus methylotrophus: isolation of hyperactive strains using continuous culture[J]. Microbiology, 1989, 135(11): 3153-3164. |
| 15 | CORNISH A, GREENWOOD J A, JONES C W. Binding-protein-dependent sugar transport by Agrobacterium radiobacter and A. tumefaciens grown in continuous culture[J]. Microbiology, 1989, 135(11): 3001-3013. |
| 16 | BAILEY J. Toward a science of metabolic engineering[J]. Science, 1991, 252(5013): 1668-1675. |
| 17 | STEPHANOPOULOS G, VALLINO J. Network rigidity and metabolic engineering in metabolite overproduction[J]. Science, 1991, 252(5013): 1675-1681. |
| 18 | STEPHANOPOULOS G, SINSKEY A J. Metabolic engineering-methodologies and future prospects[J]. Trends in Biotechnology, 1993, 11(9): 392-396. |
| 19 | FURUSAWA C, HORINOUCHI T, HIRSAWA T, et al. Systems metabolic engineering: the creation of microbial cell factories by rational metabolic design and evolution[J]. Advances in Biochemical Engineering/Biotechnology, 2012, 131(3): 1-23. |
| 20 | NIELSEN J. Production of bio-pharmaceutical proteins by yeast: advances through metabolic engineering[J]. Bioengineered, 2012, 4(4): 207-211. |
| 21 | HONG K K, NIELSEN J. Metabolic engineering of Saccharomyces cerevisiae: a key cell factory platform for future biorefineries[J]. Cellular and Molecular Life Sciences, 2012, 69(16),2671-2690. |
| 22 | CHEN Z, RAPPERT S, SUN J, et al. Integrating molecular dynamics and co-evolutionary analysis for reliable target prediction and deregulation of the allosteric inhibition of aspartokinase for amino acid production[J]. Journal of Biotechnology, 2011, 154(4): 248-254. |
| 23 | CHO B K, FEDEROWICZ S, PARK Y S, et al. Deciphering the transcriptional regulatory logic of amino acid metabolism[J]. Nature Chemical Biology, 2011, 8(1): 65-71. |
| 24 | JIN Y S, STEPHANOPOULOS G. Multi-dimensional gene target search for improving lycopene biosynthesis in Escherichia coli [J]. Metabolic Engineering, 2007, 9(4): 337-347. |
| 25 | ÖZAYDIN B, BURD H, LEE T S, et al. Carotenoid-based phenotypic screen of the yeast deletion collection reveals new genes with roles in isoprenoid production[J]. Metabolic Engineering, 2013, 15: 174-183. |
| 26 | ZHANG Y X, PERRY K, VINCI V A, et al. Genome shuffling leads to rapid phenotypic improvement in bacteria[J]. Nature, 2002, 415(6872), 644-646. |
| 27 | ZHANG X, ZHANG X F, LI H P, et al. Atmospheric and room temperature plasma (ARTP) as a new powerful mutagenesis tool[J]. Applied Microbiology and Biotechnology, 2014, 98(12): 5387-5396. |
| 28 | NICOLAOU S A, GAIDA S M, PAPOUTSAKIS E T. Coexisting/Coexpressing genomic libraries (CoGeL) identify interactions among distantly located genetic loci for developing complex microbial phenotypes[J]. Nucleic Acids Research, 2011, 39(22): e152. |
| 29 | WANG T, CHANG G G, JIA H G, et al. Pooled CRISPR interference screening enables genome-scale functional genomics study in bacteria with superior performance[J]. Nature Communications, 2018, 9(1):2475. |
| 30 | 李翔. 微生物诱变育种技术[J]. 现代商贸工业, 2017(34): 185-187. |
| LI X. Microbial mutation breeding technology[J]. Modern Business Trade Industry, 2017(34): 185-187. | |
| 31 | DEMAIN A L. From natural products discovery to commercialization: a success story[J]. Journal of Industrial Microbiology & Biotechnology, 2006, 33(7), 486-495. |
| 32 | 季宇彬, 朱兴杰, 徐昶儒, 等. 微生物诱变育种方法的研究进展[C]// 中国毒理学会环境与生态毒理学专业委员会成立大会. 2008. |
| JI Y B, ZHU X J, XU C R, et al. Research progress of microbial mutation breeding methods [C] // The Founding Conference of the Professional Committee of Environmental and Ecotoxicology of the Chinese Society of Toxicology. 2008. | |
| 33 | 微生物诱变育种编写组.微生物诱变育种[M].北京:科学出版社, 1973: 25-32. |
| Microbiology Mutation Breeding Compilation Group. Microbial mutation breeding [M]. Beijing: Science Press, 1973: 25-32. | |
| 34 | NESS J E, WELCH M, GIVER L, et al. DNA shuffling of subgenomic sequences of subtilisin[J]. Nature Biotechnology, 1999, 17(9): 893-896. |
| 35 | 施巧琴, 吴松钢. 工业微生物育种[M]. 福州: 福建科学技术出版社, 1991: 54-55. |
| SHI Q Q, WU S G. Breeding of industrial microbes[M]. Fuzhou: Fujian Science and Technology Publishing House, 1991: 54-55. | |
| 36 | CVIJOVIC M, BORDEL S, NIELSEN J. Mathematical models of cell factories: moving towards the core of industrial biotechnology[J]. Microbial Biotechnology, 2010, 4(5): 572-584. |
| 37 | FELL D A. Metabolic control analysis: a survey of its theoretical and experimental development[J]. Biochemical Journal, 1992, 286(2): 313-330. |
| 38 | WIECHERT W. 13C metabolic flux analysis[J]. Metabolic Engineering, 2001, 3(3): 195-206. |
| 39 | STEPHANOPOULOS G. Metabolic fluxes and metabolic engineering[J]. Metabolic Engineering, 1999, 1(1): 1-11. |
| 40 | VARMA A, PALSSON B O. Stoichiometric flux balance models quantitatively predict growth and metabolic by-product secretion in wild-type Escherichia coli W3110[J]. Applied and Environmental Microbiology, 1994, 60 (10): 3724-3731. |
| 41 | PFEIFFER T, SANCHEZ-VALDENEBRO I, NUNO J, et al. METATOOL: for studying metabolic networks[J]. Bioinformatics, 1999, 15(3): 251-257. |
| 42 | TRINH C T, WLASCHIN A, SRIENC F. Elementary mode analysis: a useful metabolic pathway analysis tool for characterizing cellular metabolism[J]. Applied Microbiology and Biotechnology, 2009, 81(5): 813-826. |
| 43 | ZAMBONI N, KÜMMEL A, HEINEMANN M. anNET: a tool for network-embedded thermodynamic analysis of quantitative metabolome data[J]. BMC Bioinformatics, 2008, 9(1): 199. |
| 44 | HENRY C S, BROADBELT L J, HATZIMANIKATIS V. Thermodynamics-based metabolic flux analysis[J]. Biophysical Journal, 2007, 92(5), 1792-1805. |
| 45 | BECKER S A, FEIST A M, MO M L, et al. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox[J]. Nature Protocols, 2007, 2(3): 727-738. |
| 46 | SEGRE D, VITKUP D, CHURCH G M. Analysis of optimality in natural and perturbed metabolic networks[J]. Proceedings of the National Academy of Sciences of the United States of America, 2002, 99(23): 15112-15117. |
| 47 | SHLOMI T, BERKMAN O, RUPPIN E. Regulatory on/off minimization of metabolic flux changes after genetic perturbations[J]. Proceedings of the National Academy of Sciences of the United States of America, 2005, 102(21): 7695-7700. |
| 48 | BURGARD A P, PHARKYA P, MARANAS C D. Optknock: A bilevel programming framework for identifying gene knockout strategies for microbial strain optimization[J]. Biotechnology and Bioengineering, 2003, 84(6): 647-657. |
| 49 | HATZIMANIKATIS V, LI C IONITA J A, et al. Exploring the diversity of complex metabolic networks[J]. Bioinformatics, 2004, 21(8): 1603-1609. |
| 50 | EDWARDS J S, PALSSON B O. The Escherichia coli MG1655 in silico metabolic genotype: Its definition, characteristics, and capabilities[J]. Proceedings of the National Academy of Sciences of the United States of America, 2000, 97(10): 5528-5533. |
| 51 | COVERT M W, SCHILLING C H, FAMILI I, et al. Metabolic modeling of microbial strains in silico[J]. Trends in Biochemical Sciences, 2001, 26(3): 179-186. |
| 52 | ÖSTERLUND T, NOOKAEW I, NIELSEN J. Fifteen years of large scale metabolic modeling of yeast: developments and impacts[J]. Biotechnology Advances, 2012, 30(5): 979-988. |
| 53 | FÖRSTER J, FAMILI I, FU P, et al. Genome-Scale Reconstruction of the Saccharomyces cerevisiae metabolic network[J]. Genome Research, 2003, 13(2): 244-253. |
| 54 | VARMA A, BOESCH B W, PALSSON B O. Biochemical production capabilities of Escherichia coli [J]. Biotechnology and Bioengineering, 1993, 42(1): 59-73. |
| 55 | VALLINO J J, STEPHANOPOULOS G. Carbon flux distributions at the glucose 6-phosphate branch point in Corynebacterium glutamicum during lysine overproduction[J]. Biotechnology Progress, 1994, 10(3): 327-334. |
| 56 | GULIK W M VAN, HEIJNEN J J. A metabolic network stoichiometry analysis of microbial growth and product formation[J]. Biotechnology and Bioengineering, 1995, 48(6): 681-698. |
| 57 | VANROLLEGHEM P A, DE JONG-GUBBELS P, GULIK W M VAN, et al. Validation of a metabolic network for Saccharomyces cerevisiae using mixed substrate studies[J]. Biotechnology Progress, 1996, 12(4): 434-448. |
| 58 | NISSEN T L, SCHULZE U, NIELSEN J, et al. Flux distributions in anaerobic, glucose-limited continuous cultures of Saccharomyces cerevisiae [J]. Microbiology, 1997, 143(1): 203-218. |
| 59 | DUARTE N C. Reconstruction and validation of Saccharomyces cerevisiae iND750, a fully compartmentalized genome-scale metabolic model[J]. Genome Research, 2004, 14(7): 1298-1309. |
| 60 | DOBSON P D, SMALLBONE K, JAMESON D, et al. Further developments towards a genome-scale metabolic model of yeast[J]. BMC Systems Biology, 2010, 4(1): 145. |
| 61 | ORTH J D, CONRAD T M, NA J, et al. A comprehensive genome-scale reconstruction of Escherichia coli metabolism—2011[J]. Molecular Systems Biology, 2011, 7(1): 535. |
| 62 | KJELDSEN K R, NIELSEN J. In silico genome-scale reconstruction and validation of the Corynebacterium glutamicum metabolic network[J]. Biotechnology and Bioengineering, 2009, 102(2): 583-597. |
| 63 | OLIVEIRA A, NIELSEN J, FORSTER J. Modeling Lactococcus lactis using a genome-scale flux model[J]. BMC Microbiology, 2005, 5(1): 39. |
| 64 | PARK S, SCHILLING C, PALSSON B O : Compositions and methods for modeling Bacillus subtilis metabolism: US 20030224363[P]. 2003-12-04. |
| 65 | LERMAN J A, HYDUKE D R, LATIF H, et al. In silico method for modelling metabolism and gene product expression at genome scale[J]. Nature Communications, 2012, 3(1): 929. |
| 66 | LIU J K, O'BRIEN E J, LERMAN J A, et al. Reconstruction and modeling protein translocation and compartmentalization in Escherichia coli at the genome-scale[J]. BMC Systems Biology, 2014, 8:110. |
| 67 | ZHANG Y, THIELE I, WEEKES D, et al. Three-dimensional structural view of the central metabolic network of Thermotoga maritima [J]. Science, 2009, 325(5947): 1544-1549. |
| 68 | CHANG R L, ANDREWS K, KIM D, et al. Structural systems biology evaluation of metabolic thermotolerance in Escherichia coli [J]. Science, 2013, 340(6137): 1220-1223. |
| 69 | CHANG R L, XIE L, BOURNE P E, et al. Antibacterial mechanisms identified through structural systems pharmacology[J]. BMC Systems Biology, 2013, 7(1): 102. |
| 70 | CHANDRASEKARAN S, PRICE N D. Probabilistic integrative modeling of genome-scale metabolic and regulatory networks in Escherichia coli and Mycobacterium tuberculosis [J]. Proceedings of the National Academy Sciences of the United States of America, 2010, 107(41): 17845-17850. |
| 71 | HERRGARD M J. Integrated analysis of regulatory and metabolic networks reveals novel regulatory mechanisms in Saccharomyces cerevisiae [J]. Genome Research, 2006, 16(5): 627-635. |
| 72 | KARR J R, SANGHVI J C, MACKLIN D N, et al. A whole-cell computational model predicts phenotype from genotype[J]. Cell, 2012, 150(2): 389-401. |
| 73 | BECKER J, ZELDER O, HäFNER STEFAN, et al. From zero to hero-design-based systems metabolic engineering of Corynebacterium glutamicum for L-lysine production[J]. Metabolic Engineering, 2011, 13(2): 159-168. |
| 74 | CHOI K R, JANG W D, YANG D, et al. Systems metabolic engineering strategies: integrating systems and synthetic biology with metabolic engineering[J]. Trends in Biotechnology, 2019, 37(8): 817-837. |
| 75 | JANSET M L, GULIK W M VAN. Towards large scale fermentative production of succinic acid[J]. Current Opinion in Biotechnology, 2014,30: 190-197. |
| 76 | BURK M J. Sustainable production of industrial chemicals from sugars[J]. International Sugar Journal, 2010, 112(1333): 30-35. |
| 77 | DATTA J, KASPRZYK P, BŁAŻEK K, et al. Synthesis, structure and properties of poly(ester-urethane)s obtained using bio-based and petrochemical 1,3-propanediol and 1,4-butanediol[J]. Journal of Thermal Analysis and Calorimetry, 2017, 130: 261-276. |
| 78 | NAKAMURA C E, WHITE G M. Metabolic engineering for the microbial production of 1,3-propanediol[J]. Current Opinion in Biotechnology, 2003, 14(5): 454-459. |
| 79 | Yield 10 bioscience agricultural company-Corporate Overview[OL]. . |
| 80 | BECKER J, LANGE A, FABARIUS J. Top value platform chemicals: bio-based production of organic acids[J]. Current Opinion in Biotechnology, 2015, 36: 168-175. |
| 81 | GUSTAVSSON M, LEE S Y. Prospects of microbial cell factories developed through systems metabolic engineering[J]. Microbial Biotechnology, 2016, 9(5): 610-617. |
| 82 | BUIJS N A, SIEWERS V, NIELSEN J. Advanced biofuel production by the yeast Saccharomyces cerevisiae [J]. Current Opinion in Chemical Biology, 2013, 17(3): 480-488. |
| 83 | GEORGE K W, ALONSO-GUTIERREZ J, KEASLING J D, et al. Isoprenoid drugs, biofuels, and chemicals-artemisinin, farnesene, and beyond[J]. Advances in Biochemical Engineering/Biotechnology, 2015, 355-389. |
| 84 | PADDON C J, KEASLING J D.Semi-synthetic artemisinin: a model for the use of synthetic biology inpharmaceutical development[J]. Nature Reviews Microbiology, 2014, 12(5): 355-367. |
| 85 | PADDON C J, WESTFALL P J, PITERA D J, et al. High level semi-synthetic production of the potent antimalarial artemisinin[J]. Nature, 2013, 496(7446): 528-532. |
| 86 | FREED E F, WINKLER J D, WEISS S J, et al. Genome-wide tuning of protein expression levels to rapidly engineer microbial traits[J]. ACS Synthetic Biology, 2015, 4(11): 1244-1253. |
| 87 | BABA T, ARA T, HASEGAWA M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection[J]. Molecular Systems Biology, 2006. DOI: 10.1038/msb4100050 . |
| 88 | KOO B M, KRITIKOS G, FARELLI J D, et al. Construction and analysis of two genome-scale deletion libraries for Bacillus subtilis [J]. Cell systems, 2017, 4(3): 291-305. |
| 89 | WANG H H, ISAACS F J, CARR P A, et al. Programming cells by multiplex genome engineering and accelerated evolution[J]. Nature, 2009, 460(7257): 894-898. |
| 90 | WARNER J R, REEDER P J, KARIMPOUR-FARD A, et al. Rapid profiling of a microbial genome using mixtures of barcoded oligonucleotides[J]. Nature Biotechnology, 2010, 28(8): 856-862. |
| 91 | DOUDNA J A, CHARPENTIER E. The new frontier of genome engineering with CRISPR-Cas9[J]. Science, 2014, 346(6213): 1077. |
| 92 | QI L S, LARSON M H, GILBERT L A, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression[J]. Cell, 2013, 152(5): 1173-1183. |
| 93 | DONG C, FONTANA J, PATEL A, et al. Synthetic CRISPR-Cas gene activators for transcriptional reprogramming in bacteria[J]. Nature Communications, 2018, 9(1): 2489. |
| 94 | MANS R, ROSSUM H M VAN, WIJSMAN M, et al. CRISPR/Cas9: a molecular Swiss army knife for simultaneous introduction of multiple genetic modifications in Saccharomyces cerevisiae [J]. FEMS Yeast Research, 2015,15(2). DOI: 1093/femsyr/fov004 . |
| 95 | HALPERIN S O, TOU C J, WONG E B, et al. CRISPR-guided DNA polymerases enable diversification of all nucleotides in a tunable window[J]. Nature, 2018, 560(7717): 248-252. |
| 96 | JIANG W, BIKARD D, COX D, et al. RNA-guided editing of bacterial genomes using CRISPR-Cas systems[J]. Nature Biotechnology, 2013, 31: 233-239. |
| 97 | OH J H, van PIJKEREN J P. CRISPR-Cas9-assisted recombineering in Lactobacillus reuteri [J]. Nucleic Acids Research 2014, 42: e131. |
| 98 | CHOUDHARY E, THAKUR P, PAREEK M, et al. Gene silencing by CRISPR interference in mycobacteria[J]. Nature Communications, 2015, 6(1): 6267. |
| 99 | YAO L, CENGIC I, ANFELT J, et al. Multiple gene repression in cyanobacteria using CRISPRi[J]. ACS Synthetic Biology 2016, 5: 207-212. |
| 100 | XU T, LI Y, SHI Z, et al. Efficient genome editing in Clostridium cellulolyticum via CRISPR-Cas9 nickase.[J]. Applied and Environmental Microbiology, 2015, 81: 4423-4431. |
| 101 | PETERS J M, COLAVIN A, SHI H, et al. A comprehensive, CRISPR-based functional analysis of essential genes in bacteria[J]. Cell, 2016, 165: 1493-1506. |
| 102 | JIANG Y, QIAN F, YANG J, et al. CRISPR-Cpf1 assisted genome editing of Corynebacterium glutamicum [J]. Nature Communications, 2017, 8(1): 15179. |
| 103 | NAYAK D D, METCALF W W. Cas9-mediated genome editing in the methanogenic archaeon Methanosarcina acetivorans [J]. Proceedings of the National Academy of Sciences of the United States of America, 2017, 114: 2976-2981. |
| 104 | 王天民.基于 CRISPR 干扰技术的微生物高通量功能基因组学方法[D]. 北京: 清华大学, 2018. |
| WANG T M. CRISPR interference based high-throughput functional genomics platform method in bacteria[D]. Beijing: Tsinghua University, 2018. | |
| 105 | DICARLO J E, CONLEY A J, PENTTILA M, et al. Yeast oligo-mediated genome engineering (YOGE)[J]. ACS Synthetic Biology, 2013, 2(12): 741-749. |
| 106 | GARST A D, BASSALO M C, PINES G,et al. Genome-wide mapping of mutations at single-nucleotide resolution for protein, metabolic and genome engineering[J]. Nature Biotechnology, 2016, 35(1): 48-55. |
| 107 | ANZALONE A V, RANDOLPH P B, DAVIS J R, et al. Search-and-replace genome editing without double-strand breaks or donor DNA[J]. Nature, 2019, 576(7785): 149-157. |
| 108 | NISHIDA K, ARAZOE T, YACHIE N, et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems[J]. Science, 2016, 353(6305): aaf8729. |
| 109 | MA Y, ZHANG J, YIN W, et al. Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells[J]. Nature Methods, 2016, 13(12): 1029-1035. |
| 110 | HESS G T, FRESARD L, HAN K, et al. Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells[J]. Nature Methods, 2016, 13(12): 1036-1042. |
| 111 | LI Q, SEO J H, STRANGER B, et al. Integrative eQTL-Based analyses reveal the biology of breast cancer risk loci[J]. Cell, 2013, 152(3): 633-641. |
| 112 | WU J, ZHOU P, ZHANG X, et al. Efficient de novo synthesis of resveratrol by metabolically engineered Escherichia coli [J]. Journal of Industrial Microbiology & Biotechnology, 2017, 44(7): 1083-1095. |
| 113 | GILBERT L A, LARSON M H, MORSUT L, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes[J]. Cell, 2013, 154(2): 442-451. |
| 114 | HALPERIN S O, TOU C J, WANG E B, et al. CRISPR-guided DNA polymerases enable diversification of all nucleotides in a tunable window[J]. Nature, 2018, 560: 248-252. |
| 115 | WONG B G, MANCUSO C P, KIRIAKOV S, et al. Precise, automated control of conditions for high-throughput growth of yeast and bacteria with eVOLVER[J]. Nature Biotechnology, 2018, 36(7): 614-623. |
| 116 | JIAN X, GUO X, WANG J, et al. Microbial microdroplet culture system (MMC): an integrated platform for automated, high-throughput microbial cultivation and adaptive evolution[J]. Biotechnology and Bioengineering, 2019. DOI: 10.1101/2019.12. 19.883561 . |
| 117 | MICHENER J K, THODEY K, LIANG J C, et al. Applications of genetically-encoded biosensors for the construction and control of biosynthetic pathways[J]. Metabolic Engineering, 2012, 14(3): 212-222. |
| 118 | LIU D, EVANS T, ZHANG F Z, et al. Applications and advances of metabolite biosensors for metabolic engineering[J]. Metabolic Engineering, 2015, 31: 15-22. |
| 119 | FANG M, WANG T, ZHANG C, et al. Intermediate-sensor assisted push-pull strategy and its application in heterologous deoxyviolacein production in Escherichia coli [J]. Metabolic Engineering, 2016, 33: 41-51. |
| 120 | LIANG W, CUI L, CUI J, et al. Biosensor-assisted transcriptional regulator engineering for Methylobacterium extorquens AM1 to improve mevalonate synthesis by increasing the acetyl-CoA supply[J]. Metabolic Engineering, 2017, 39: 159-168. |
| 121 | BINDER S, SCHENDZIELORZ G, STABLER N, et al. A high-throughput approach to identify genomic variants of bacterial metabolite producers at the single-cell level[J]. Genome Biology, 2012, 13: R40. |
| 122 | BENEYTON T, THOMAS S, GRIFFITHS A D, et al. Droplet-based microfluidic high-throughput screening of heterologous enzymes secreted by the yeast Yarrowia lipolytica [J]. Microbial Cell Factories, 2017, 16(1): 18. |
| 123 | SIEDLER S, KHATRI N K, ZSOHAR A, et al. Development of a bacterial biosensor for rapid screening of yeast p-coumaric acid production[J]. ACS Synthetic Biology, 2017, 6(10): 1860-1869. |
| 124 | BARET J C, MILLER O J, TALY V, et al. Fluorescence-activated droplet sorting (FADS): efficient microfluidic cell sorting based on enzymatic activity[J]. Lab on a Chip, 2009, 9(13): 1850. |
| 125 | WAGNER J M, LIU L, YUAN S et. al. A comparative analysis of single cell and droplet-based FACS for improving production phenotypes : riboflavin overproduction in Yarrowia lipolytica [J]. Metabolic Engineering, 2018, 47: 346-356. |
| 126 | MA C, TAN Z L, LIN Y, et al. Gel microdroplet-based high-throughput screening for directed evolution of xylanase-producing Pichia pastoris [J]. Journal of Bioscience and Bioengineering, 2019, 128(6): 662-668. |
| 127 | WANG X, REN L, SU Y, et al. Raman-activated droplet sorting (RADS) for label-free high-throughput screening of microalgal single-cells[J]. Analytical Chemistry, 2017, 89(22): 12569-12577. |
| 128 | DIXIT A, PARNAS O, LI B, et al. Perturb-seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens[J]. Cell, 2016, 167: 1853-1866. |
| 129 | TROPINI C, EARLE K A, HUANG K C, et al. The gut microbiome: connecting spatial organization to function[J]. Cell Host Microbe, 2017, 21: 433-442. |
| 130 | SHI H, COLAVIN A, LEE T K, et al. Strain library imaging protocol for high-throughput, automated single-cell microscopy of large bacterial collections arrayed on multiwell plates[J]. Nature Protocols Erecipes for Researchers, 2017, 12: 429-438. |
| 131 | LAWSON M J, CAMSUND D, LARSSON J, et al. In situ genotyping of a pooled strain library after characterizing complex phenotypes[J]. Molecular Systems Biology, 2017, 13: 947. |
| 132 | ZONG Y, ZHANG H M, LYU C, et al. Insulated transcriptional elements enable precise design of genetic circuits[J]. Nature Communications, 2017, 8: 52. |
| 133 | BASSALO M C, GARST A D, CHOUDHURY A, et al. Deep scanning lysine metabolism in Escherichia coli [J]. Molecular Systems Biology, 2018, 14(11): e8371. |
| 134 | LIANG L, LIU R, GARST A D, et al. CRISPR EnAbled trackable genome engineering for isopropanol production in Escherichia coli [J]. Metabolic Engineering, 2017, 41: 1-10. |
| 135 | LIU R, LIANG L, GARST A D, et al. Directed combinatorial mutagenesis of Escherichia coli for complex phenotype engineering[J]. Metabolic Engineering, 2018, 47: 10-20. |
| 136 | LIANG L, LIU R, FOSTER K E, et al. Genome engineering of E. coli for improved styrene production[J]. Metabolic Engineering, 2020, 57: 74-84. |
| 137 | ANGERMUELLER C, PÄRNAMAA T, PARTS L, et al. Deep learning for computational biology[J]. Molecular Systems Biology, 2016, 12(7): 878. |
| 138 | COSTELLO Z, MARTIN H G. A machine learning approach to predict metabolic pathway dynamics from time-series multiomics data[J]. NPJ Systems Biology and Applications, 2018, 4(1): 1-14. |
| 139 | CUPERUS J T, GROVES B, KUCHINA A, et al. Deep learning of the regulatory grammar of yeast 5' untranslated regions from 500,000 random sequences[J]. Genome Research, 2017, 27(12): 2015-2024. |
| 140 | ZHOU Y, LI G, DONG J, et al. MiYA, an efficient machine-learning workflow in conjunction with the YeastFab assembly strategy for combinatorial optimization of heterologous metabolic pathways in Saccharomyces cerevisiae [J]. Metabolic Engineering, 2018, 47: 294-302. |
| 141 | DENBY C M, LI R A, VU V T, et al. Industrial brewing yeast engineered for the production of primary flavor determinants in hopped beer[J]. Nature Communications, 2018, 9(1): 965. |
| 142 | ZHANG J, PSTERSEN S D, RADIVOJEVIC T, et al. Combining mechanistic and machine learning models for predictive engineering and optimization of tryptophan metabolism[J]. Nature Communications, 2020, 11(1): 4880. |
| 143 | GUO J, WANG T, GUAN C, et al. Improved sgRNA design in bacteria via genome-wide activity profiling[J]. Nucleic Acids Research, 2018, 46(14): 7052-7069. |
| [1] | GUO Shuyuan, ZHANG Qiannan, Gulikezi· MAIMAITIREXIATI, YANG Yiqun, YU Tao. Advances in microbial production of liquid biofuels [J]. Synthetic Biology Journal, 2025, 6(1): 18-44. |
| [2] | SHAO Mingwei, SUN Simian, YANG Shimao, CHEN Guoqiang. Bioproduction based on extremophiles [J]. Synthetic Biology Journal, 2024, 5(6): 1419-1436. |
| [3] | ZHAO Liang, LI Zhenshuai, FU Liping, LYU Ming, WANG Shi’an, ZHANG Quan, LIU Licheng, LI Fuli, LIU Ziyong. Progress in biomanufacturing of lipids and single cell protein from one-carbon compounds [J]. Synthetic Biology Journal, 2024, 5(6): 1300-1318. |
| [4] | ZHU Fanghuan, CEN Xuecong, CHEN Zhen. Research progress of diols production by microbes [J]. Synthetic Biology Journal, 2024, 5(6): 1367-1385. |
| [5] | YU Wei, GAO Jiaoqi, ZHOU Yongjin. Bioconversion of one carbon feedstocks for producing organic acids [J]. Synthetic Biology Journal, 2024, 5(5): 1169-1188. |
| [6] | CHEN Xiwei, ZHANG Huaran, ZOU Yi. Biosynthesis and metabolic engineering of fungal non-ribosomal peptides [J]. Synthetic Biology Journal, 2024, 5(3): 571-592. |
| [7] | XIE Huang, ZHENG Yilei, SU Yiting, RUAN Jingyi, LI Yongquan. An overview on reconstructing the biosynthetic system of actinomycetes for polyketides production [J]. Synthetic Biology Journal, 2024, 5(3): 612-630. |
| [8] | ZHAO Jingyu, ZHANG Jian, QI Qingsheng, WANG Qian. Research progress in biosensors based on bacterial two-component systems [J]. Synthetic Biology Journal, 2024, 5(1): 38-52. |
| [9] | SUN Huili, CUI Jinyu, LUAN Guodong, LYU Xuefeng. Progress of cyanobacterial synthetic biotechnology for efficient light-driven carbon fixation and ethanol production [J]. Synthetic Biology Journal, 2023, 4(6): 1161-1177. |
| [10] | YAN Xiongying, WANG Zhen, LOU Jiyun, ZHANG Haoyu, HUANG Xingyu, WANG Xia, YANG Shihui. Progress in the construction of microbial cell factories for efficient biofuel production [J]. Synthetic Biology Journal, 2023, 4(6): 1082-1121. |
| [11] | CHENG Zhenzhen, ZHANG Jian, GAO Cong, LIU Liming, CHEN Xiulai. Progress in metabolic engineering of microorganisms for the utilization of formate [J]. Synthetic Biology Journal, 2023, 4(4): 756-778. |
| [12] | LIU Jiayu, YANG Zhihan, YANG Lei, ZHU Liying, ZHU Zhengming, JIANG Ling. Advances in the development of Clostridium tyrobutyricum cell factories driven by synthetic biotechnology [J]. Synthetic Biology Journal, 2022, 3(6): 1174-1200. |
| [13] | WANG Qian, QI Qingsheng. Low-carbon biomanufacturing of polyhydroxyalkanoates: analysis and application based on carbon conversion rate [J]. Synthetic Biology Journal, 2022, 3(4): 748-762. |
| [14] | GUO Shuyuan, WU Lianghuan, LIU Xiangjian, WANG Bo, YU Tao. Developing C1-based metabolic network in methylotrophy for biotransformation [J]. Synthetic Biology Journal, 2022, 3(1): 116-137. |
| [15] | CHEN Jiuzhou, WANG Yu, PU Wei, ZHENG Ping, SUN Jibin. Advances and perspective on bioproduction of 5-aminolevulinic acid [J]. Synthetic Biology Journal, 2021, 2(6): 1000-1016. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||