Synthetic Biology Journal ›› 2022, Vol. 3 ›› Issue (6): 1174-1200.DOI: 10.12211/2096-8280.2022-022
• Invited Review • Previous Articles Next Articles
Advances in the development of Clostridium tyrobutyricum cell factories driven by synthetic biotechnology
LIU Jiayu1, YANG Zhihan2, YANG Lei2, ZHU Liying3, ZHU Zhengming1, JIANG Ling1,4
- 1.School of Food and Light Industry,Nanjing Tech University,Nanjing 211816,Jiangsu,China
2.School of Biological and Pharmaceutical Engineering,Nanjing Tech University,Nanjing 211816,Jiangsu,China
3.School of Chemical and Molecular Engineering,Nanjing Tech University,Nanjing 211816,Jiangsu,China
4.State Key Laboratory of Chemical Engineering of Materials,Nanjing Tech University,Nanjing 211816,Jiangsu,China
-
Received:2022-04-14Revised:2022-05-25Online:2023-01-17Published:2022-12-31 -
Contact:ZHU Zhengming, JIANG Ling
合成生物技术驱动酪丁酸梭菌细胞工厂开发的研究进展
刘家宇1, 杨智晗2, 杨蕾2, 朱丽英3, 朱政明1, 江凌1,4
- 1.南京工业大学食品与轻工学院,江苏 南京 211816
2.南京工业大学生物与制药工程学院,江苏 南京 211816
3.南京工业大学化学与分子工程学院,江苏 南京 211816
4.南京工业大学材料化学工程国家重点实验室,江苏 南京 211816
-
通讯作者:朱政明,江凌 -
作者简介:刘家宇 (1998—),男,硕士研究生。研究方向为微生物系统与合成生物学。E-mail:liujoy@njtech.edu.cn朱政明 (1989—),男,博士,副教授。研究方向为微生物系统与合成生物学。E-mail:zhuzm@njtech.edu.cn江凌 (1982—),男,博士,教授。研究方向为发酵工程与酶工程。E-mail:jiangling@njtech.edu.cn -
基金资助:国家自然科学基金优秀青年科学基金项目(31922070);国家自然科学基金青年科学基金项目(22008114);国家自然科学基金山东联合基金重点项目(U2106228);江苏省自然科学基金青年基金项目(BK20200684);江苏省先进生物制造创新中心资助(XTC2205)
CLC Number:
Cite this article
LIU Jiayu, YANG Zhihan, YANG Lei, ZHU Liying, ZHU Zhengming, JIANG Ling. Advances in the development of Clostridium tyrobutyricum cell factories driven by synthetic biotechnology[J]. Synthetic Biology Journal, 2022, 3(6): 1174-1200.
刘家宇, 杨智晗, 杨蕾, 朱丽英, 朱政明, 江凌. 合成生物技术驱动酪丁酸梭菌细胞工厂开发的研究进展[J]. 合成生物学, 2022, 3(6): 1174-1200.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2022-022
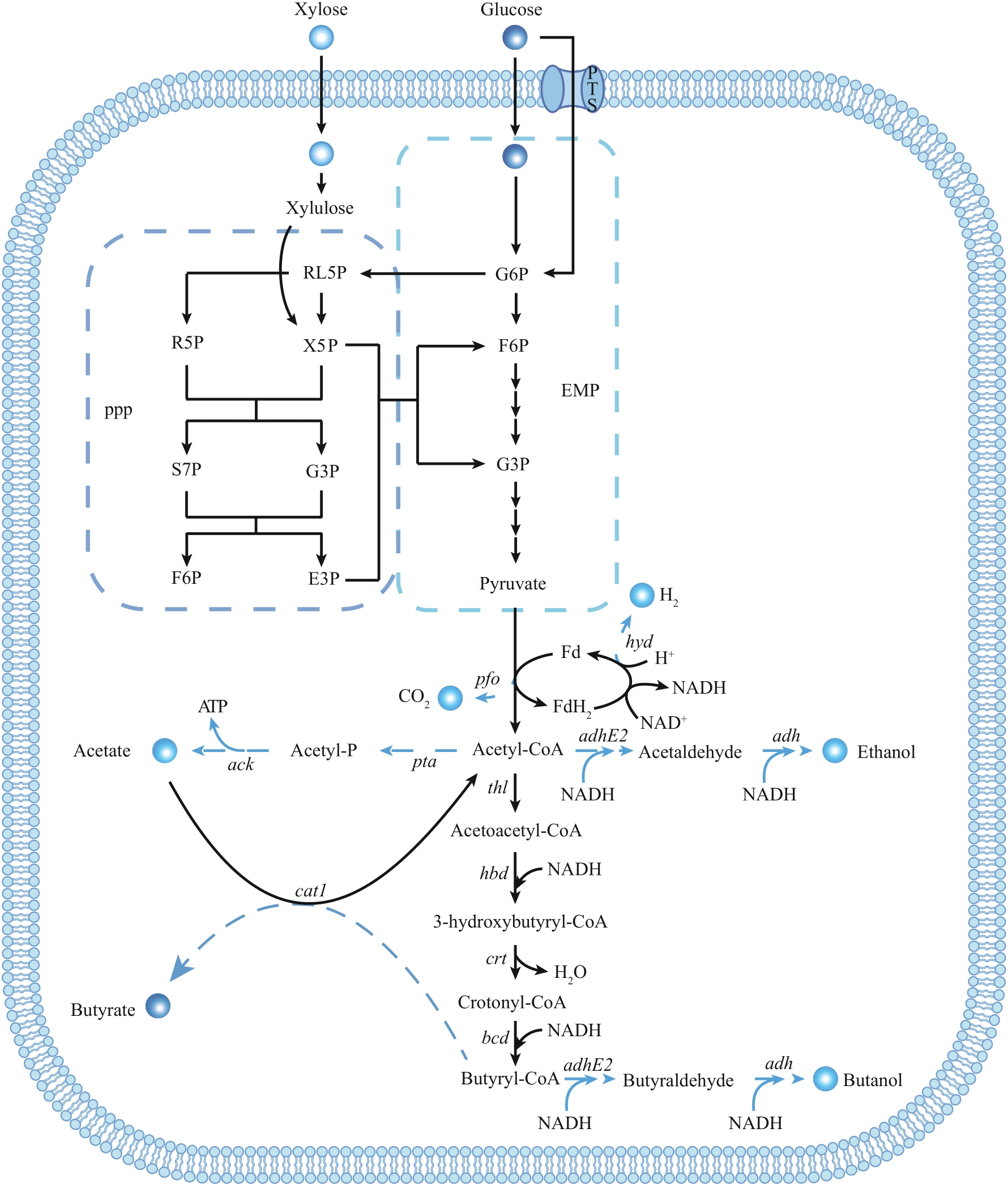
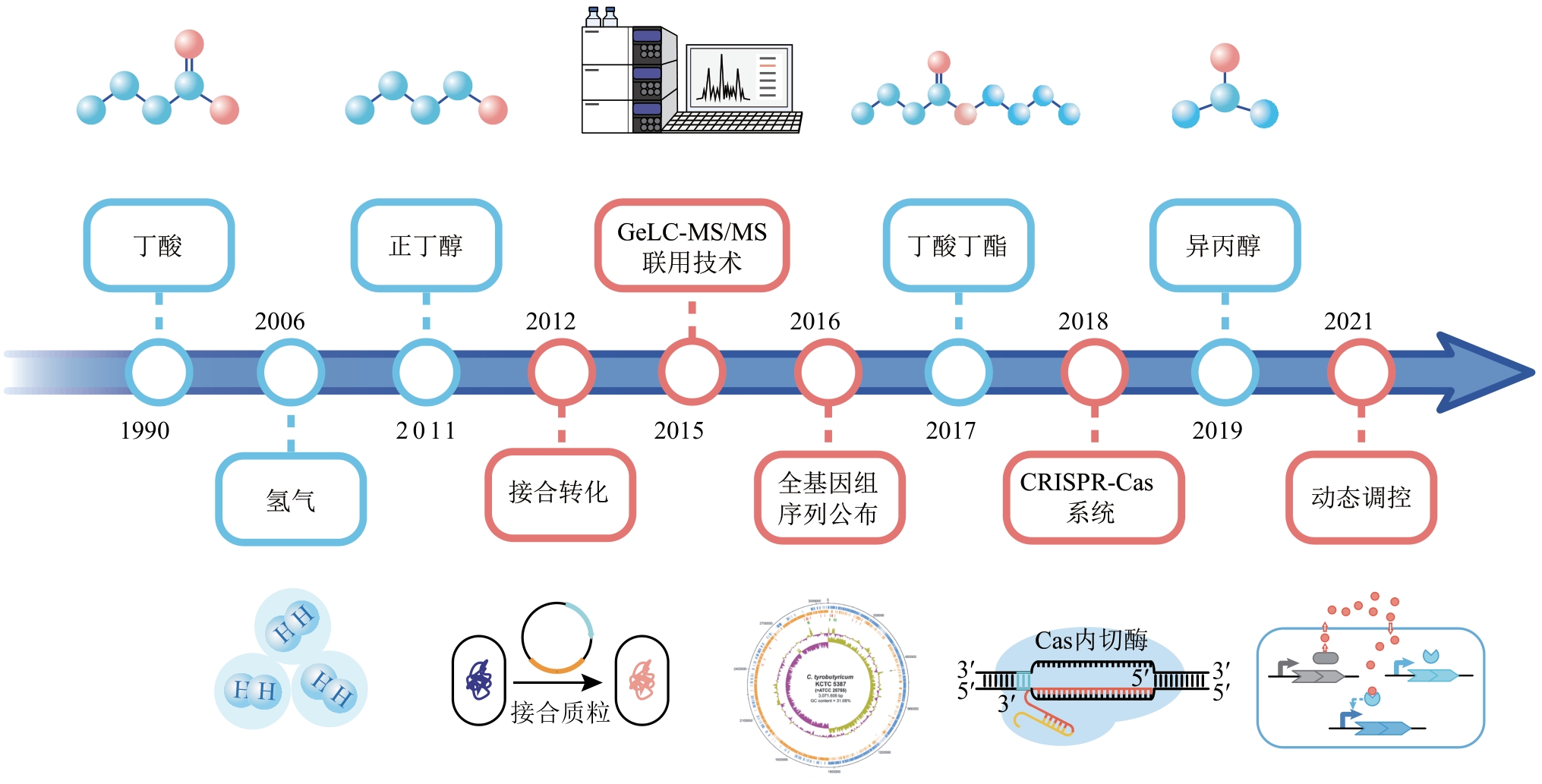
Fig. 3 Metabolic synthesis pathway of butyric acid in C. tyrobutyricum[5]The genes encode key enzymes in pathway: hyd—hydrogenase; pfo—pyruvate:ferredoxin oxidoreductase; ack—acetate kinase; pta—phosphotransacetylase; adhE2—aldehyde-alcohol dehydrogenase; adh—alcohol dehydrogenase; thl—thiolase; hbd—beta-hydroxybutyryl-CoA dehydrogenase; crt—crotonase; bcd—butanoyl-coA dehydrogenase; cat1—butyryl-CoA/acetate CoA transferase
| 底物 | 成分 | 发酵方式 | 产物 | 滴度 /(g/L) | 产率 /[g/(L·h)] | 产量 /(g/g) | 改造方法 | 参考文献 | |
|---|---|---|---|---|---|---|---|---|---|
| 单糖 | 葡萄糖 | — | 补料分批 | 丁酸 | 48.2 | 0.50 | 0.38 | 过表达pfkA、pykA | [ |
| 丁醇 | 约45 | — | 0.30 | 敲除ack,过表达adhE2 | [ | ||||
| FBB反应器 | 丁酸 | 52.2 | 0.41 | 0.37 | 过表达groESL | [ | |||
| 丁醇 | 约54 | — | 0.33 | 敲除ack,过表达adhE2 | [ | ||||
| 分批 | 丁酸 | — | 0.20 | 0.36 | 敲除ptb | [ | |||
| 丁醇 | 约10.5 | 约0.24 | 约0.24 | 敲除ack,过表达adhE2、ctfAB | [ | ||||
| 重复FBB反应器 | 丁酸 | 86.9 | 1.1 | 0.46 | —① | [ | |||
| 木糖 | — | FBB反应器 | 丁酸 | 13.6 | — | 0.44 | 敲除ack | [ | |
| 分批 | 丁酸 | — | 0.22 | 0.35 | 敲除ptb | [ | |||
| 果糖 | — | FBB反应器 | 丁酸 | 49.4 | 2.60 | 0.48 | — | [ | |
| 二糖 | 蔗糖 | 葡萄糖、果糖 | FBB反应器 | 丁酸 | 45.8 | — | 0.43 | — | [ |
| 分批 | 丁醇 | 18.8 | 0.20 | 0.21 | 敲除ack,过表达adhE2、ctfABK | [ | |||
| 麦芽糖 | 葡萄糖 | 分批 | 丁醇 | 17.3 | 0.40 | 0.17 | 敲除ack,过表达adhE2、aglul | [ | |
| 农业原料及农业废弃物 | 咖啡残渣水解物 | 木糖、阿拉伯糖、 半乳糖 | 分批 | 丁酸 | 34.3 | 0.36 | 0.37 | 过表达galKETP | [ |
| 甘蔗糖蜜 | 果糖、葡萄糖 | 补料分批 | 丁酸 | 45.71 | — | 0.39 | 过表达scrBAK | [ | |
| 甘蔗汁 | 果糖、葡萄糖 | 分批 | 丁醇 | 14.8 | 0.15 | 0.21 | 敲除ack,过表达adhE2、scrBAK | [ | |
| 甘蔗渣水解物 | 木糖、阿拉伯糖、 半乳糖 | 补料分批 | 丁酸 | 20.9 | 0.51 | 0.48 | — | [ | |
| 菊粉 | 果糖、葡萄糖 | 分批 | 氢气 | — | 620 ml/(L·h) | 3.7 mol/mol | 过表达外源 菊粉酶 | [ | |
| 补料分批FBB 反应器 | 丁酸 | 60.4 | 1.14 | 0.38 | — | [ | |||
| 玉米秸秆水解物 | 葡萄糖、木糖 | 分批 | 丁酸 | 16.1 | 0.38 | 0.35 | 过表达groESL | [ | |
| 玉米面粉水解物 | 葡萄糖、木糖 | 补料分批FBB 反应器 | 丁酸 | 46.0 | 6.8 | 0.45 | — | [ | |
| 玉米纤维水解物 | 葡萄糖、木糖 | 补料分批FBB 反应器 | 丁酸 | 29.0 | 2.9 | 0.47 | — | [ | |
| 玉米皮水解物 | 葡萄糖、木糖 | 重复FBB反应器 | 丁酸 | 20.8 | 0.42 | 0.39 | — | [ | |
| 小麦秸秆水解物 | 葡萄糖、木糖 | 分批 | 丁酸 | 20.0 | 0.21 | 0.33 | — | [ | |
| 稻草水解物 | 葡萄糖、木糖 | 分批 | 丁酸 | 16.9 | 0.40 | 0.35 | 过表达groESL | [ | |
| 高粱秸秆和甜菜秸秆水解物 | 葡萄糖、木糖 | 分批 | 丁酸 | 58.8 | 1.9 | 0.52 | — | [ | |
Tab. 1 Substrate profile of C. tyrobutyricum[5]
| 底物 | 成分 | 发酵方式 | 产物 | 滴度 /(g/L) | 产率 /[g/(L·h)] | 产量 /(g/g) | 改造方法 | 参考文献 | |
|---|---|---|---|---|---|---|---|---|---|
| 单糖 | 葡萄糖 | — | 补料分批 | 丁酸 | 48.2 | 0.50 | 0.38 | 过表达pfkA、pykA | [ |
| 丁醇 | 约45 | — | 0.30 | 敲除ack,过表达adhE2 | [ | ||||
| FBB反应器 | 丁酸 | 52.2 | 0.41 | 0.37 | 过表达groESL | [ | |||
| 丁醇 | 约54 | — | 0.33 | 敲除ack,过表达adhE2 | [ | ||||
| 分批 | 丁酸 | — | 0.20 | 0.36 | 敲除ptb | [ | |||
| 丁醇 | 约10.5 | 约0.24 | 约0.24 | 敲除ack,过表达adhE2、ctfAB | [ | ||||
| 重复FBB反应器 | 丁酸 | 86.9 | 1.1 | 0.46 | —① | [ | |||
| 木糖 | — | FBB反应器 | 丁酸 | 13.6 | — | 0.44 | 敲除ack | [ | |
| 分批 | 丁酸 | — | 0.22 | 0.35 | 敲除ptb | [ | |||
| 果糖 | — | FBB反应器 | 丁酸 | 49.4 | 2.60 | 0.48 | — | [ | |
| 二糖 | 蔗糖 | 葡萄糖、果糖 | FBB反应器 | 丁酸 | 45.8 | — | 0.43 | — | [ |
| 分批 | 丁醇 | 18.8 | 0.20 | 0.21 | 敲除ack,过表达adhE2、ctfABK | [ | |||
| 麦芽糖 | 葡萄糖 | 分批 | 丁醇 | 17.3 | 0.40 | 0.17 | 敲除ack,过表达adhE2、aglul | [ | |
| 农业原料及农业废弃物 | 咖啡残渣水解物 | 木糖、阿拉伯糖、 半乳糖 | 分批 | 丁酸 | 34.3 | 0.36 | 0.37 | 过表达galKETP | [ |
| 甘蔗糖蜜 | 果糖、葡萄糖 | 补料分批 | 丁酸 | 45.71 | — | 0.39 | 过表达scrBAK | [ | |
| 甘蔗汁 | 果糖、葡萄糖 | 分批 | 丁醇 | 14.8 | 0.15 | 0.21 | 敲除ack,过表达adhE2、scrBAK | [ | |
| 甘蔗渣水解物 | 木糖、阿拉伯糖、 半乳糖 | 补料分批 | 丁酸 | 20.9 | 0.51 | 0.48 | — | [ | |
| 菊粉 | 果糖、葡萄糖 | 分批 | 氢气 | — | 620 ml/(L·h) | 3.7 mol/mol | 过表达外源 菊粉酶 | [ | |
| 补料分批FBB 反应器 | 丁酸 | 60.4 | 1.14 | 0.38 | — | [ | |||
| 玉米秸秆水解物 | 葡萄糖、木糖 | 分批 | 丁酸 | 16.1 | 0.38 | 0.35 | 过表达groESL | [ | |
| 玉米面粉水解物 | 葡萄糖、木糖 | 补料分批FBB 反应器 | 丁酸 | 46.0 | 6.8 | 0.45 | — | [ | |
| 玉米纤维水解物 | 葡萄糖、木糖 | 补料分批FBB 反应器 | 丁酸 | 29.0 | 2.9 | 0.47 | — | [ | |
| 玉米皮水解物 | 葡萄糖、木糖 | 重复FBB反应器 | 丁酸 | 20.8 | 0.42 | 0.39 | — | [ | |
| 小麦秸秆水解物 | 葡萄糖、木糖 | 分批 | 丁酸 | 20.0 | 0.21 | 0.33 | — | [ | |
| 稻草水解物 | 葡萄糖、木糖 | 分批 | 丁酸 | 16.9 | 0.40 | 0.35 | 过表达groESL | [ | |
| 高粱秸秆和甜菜秸秆水解物 | 葡萄糖、木糖 | 分批 | 丁酸 | 58.8 | 1.9 | 0.52 | — | [ | |
| 固碳途径 | 固碳种类 | 酶总数 | 关键酶 | 酶比活力①/[µmol/(min·mg)] | 能量来源 | 产物 | ATP 消耗 |
|---|---|---|---|---|---|---|---|
| WL途径[ | CO2 | 8 | 甲酸脱氢酶 | 439 | 氢气 | 乙酰辅酶A | <1 |
| CO脱氢酶 | 14 000 | ||||||
| 还原性甘氨酸途径[ | CO2 | 5 | 还原性甘氨酸裂解复合物 | — | — | 乙酰辅酶A | 2 |
| 卡尔文循环[ | CO2 | 11 | RuBisCO | 304.3 | 光 | 3-磷酸甘油醛 | 9 |
| 还原性TCA循环[ | CO2 | 8 | 2-酮戊二酸合酶 | 35.2 | 光和硫 | 乙酰辅酶A | 2 |
| ATP-柠檬酸裂合酶 | 26.7 | ||||||
| DC/HB循环[ | CO2/HCO3- | 14 | 4-羟基丁酰辅酶A脱水酶 | — | 氢和硫 | 乙酰辅酶A | 5 |
| HP/HB循环[ | HCO3- | 15 | 4-羟基丁酰辅酶A脱水酶 | — | 氢和氧 | 乙酰辅酶A | 6 |
| 3-HP双循环[ | HCO3- | 18 | 丙二酰辅酶A还原酶 | 80 | 光和硫 | 丙酮酸 | 7 |
| 丙酰辅酶A合酶 | 22 |
Tab. 2 Comparison of CO2 fixation pathways
| 固碳途径 | 固碳种类 | 酶总数 | 关键酶 | 酶比活力①/[µmol/(min·mg)] | 能量来源 | 产物 | ATP 消耗 |
|---|---|---|---|---|---|---|---|
| WL途径[ | CO2 | 8 | 甲酸脱氢酶 | 439 | 氢气 | 乙酰辅酶A | <1 |
| CO脱氢酶 | 14 000 | ||||||
| 还原性甘氨酸途径[ | CO2 | 5 | 还原性甘氨酸裂解复合物 | — | — | 乙酰辅酶A | 2 |
| 卡尔文循环[ | CO2 | 11 | RuBisCO | 304.3 | 光 | 3-磷酸甘油醛 | 9 |
| 还原性TCA循环[ | CO2 | 8 | 2-酮戊二酸合酶 | 35.2 | 光和硫 | 乙酰辅酶A | 2 |
| ATP-柠檬酸裂合酶 | 26.7 | ||||||
| DC/HB循环[ | CO2/HCO3- | 14 | 4-羟基丁酰辅酶A脱水酶 | — | 氢和硫 | 乙酰辅酶A | 5 |
| HP/HB循环[ | HCO3- | 15 | 4-羟基丁酰辅酶A脱水酶 | — | 氢和氧 | 乙酰辅酶A | 6 |
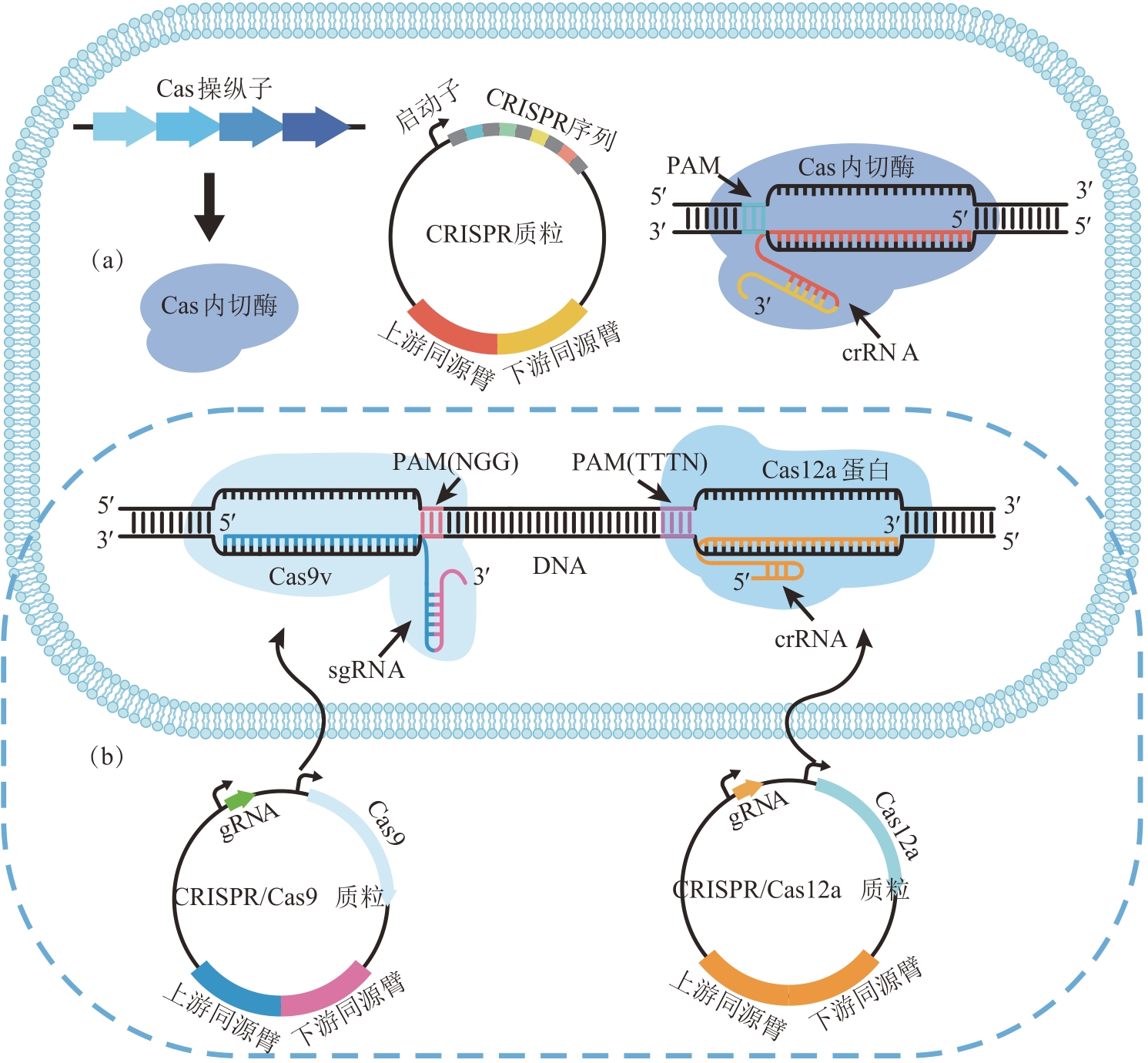
| 3-HP双循环[ | HCO3- | 18 | 丙二酰辅酶A还原酶 | 80 | 光和硫 | 丙酮酸 | 7 |
| 丙酰辅酶A合酶 | 22 |
| 菌株名称 | 测序程度 | 基因组大小/Mb | G+C/% | 蛋白数量 | rRNA | tRNA | 其他RNA |
|---|---|---|---|---|---|---|---|
| KCTC 5387 | Complete | 3.13 | 31.0 | 3015 | 19 | 63 | 5 |
| L319 | Complete | 3.09 | 31.0 | 2922 | 19 | 63 | 5 |
| W428 | Complete | 3.07 | 30.9 | 2945 | 16 | 51 | 5 |
| Cirm BIA 2237 | Chromosome | 3.16 | 30.8 | 3069 | 3 | 52 | 5 |
| FAM22553 | Scaffold | 3.09 | 31.0 | 2992 | 18 | 63 | 5 |
| FAM22552 | Scaffold | 3.05 | 30.9 | 2923 | 18 | 68 | 5 |
| DSM 663 | Scaffold | 3.15 | 30.7 | 3091 | 3 | 35 | 5 |
| 1001713B170207_170306_A1 | Scaffold | 3.02 | 31.0 | 2831 | 4 | 57 | 5 |
| 1001713B170207_170306_A10 | Scaffold | 3.05 | 30.5 | 2881 | 4 | 60 | 5 |
| 1001283B150210_160208_G3 | Scaffold | 3.07 | 30.5 | 2869 | 5 | 52 | 5 |
| 1001283B150210_160208_D6 | Scaffold | 3.00 | 30.5 | 2791 | 4 | 58 | 5 |
| Cl_188 | Scaffold | 3.01 | 30.5 | 2876 | 3 | 40 | 5 |
| Cl_171 | Scaffold | 3.04 | 30.5 | 2938 | 3 | 36 | 5 |
| Cl_239 | Scaffold | 3.33 | 30.5 | 3219 | 4 | 48 | 5 |
| Cl_117 | Scaffold | 3.11 | 31.0 | 3034 | 4 | 46 | 5 |
| Cl_238 | Scaffold | 3.23 | 30.5 | 3115 | 4 | 30 | 5 |
| Cl_84 | Scaffold | 3.05 | 30.5 | 2954 | 3 | 52 | 5 |
| Cl_82 | Scaffold | 3.01 | 30.5 | 2916 | 4 | 45 | 5 |
| Cl_64 | Scaffold | 3.17 | 31.0 | 3087 | 4 | 46 | 5 |
| Cl_52 | Scaffold | 3.09 | 30.5 | 2940 | 2 | 42 | 5 |
| Cl_80 | Scaffold | 3.09 | 30.5 | 3049 | 3 | 34 | 5 |
| Cl_29 | Scaffold | 3.09 | 30.5 | 2988 | 4 | 37 | 5 |
| MGYG-HGUT-00125 | Scaffold | 3.26 | 30.5 | 3101 | 9 | 63 | 5 |
| 24853 | Scaffold | 3.00 | 30.5 | 2664 | 4 | 59 | 5 |
| ATCC 25755 | Contig | 3.01 | 30.5 | 2908 | 4 | 46 | 5 |
| DIVETGP | Contig | 3.02 | 30.5 | 2907 | 4 | 46 | 5 |
| UC7086 | Contig | 3.06 | 30.5 | 2943 | 4 | 48 | 5 |
| IFP923 | Contig | 3.19 | 30.5 | 3090 | 10 | 60 | 5 |
Tab. 3 Genomic information of C. tyrobutyricum
| 菌株名称 | 测序程度 | 基因组大小/Mb | G+C/% | 蛋白数量 | rRNA | tRNA | 其他RNA |
|---|---|---|---|---|---|---|---|
| KCTC 5387 | Complete | 3.13 | 31.0 | 3015 | 19 | 63 | 5 |
| L319 | Complete | 3.09 | 31.0 | 2922 | 19 | 63 | 5 |
| W428 | Complete | 3.07 | 30.9 | 2945 | 16 | 51 | 5 |
| Cirm BIA 2237 | Chromosome | 3.16 | 30.8 | 3069 | 3 | 52 | 5 |
| FAM22553 | Scaffold | 3.09 | 31.0 | 2992 | 18 | 63 | 5 |
| FAM22552 | Scaffold | 3.05 | 30.9 | 2923 | 18 | 68 | 5 |
| DSM 663 | Scaffold | 3.15 | 30.7 | 3091 | 3 | 35 | 5 |
| 1001713B170207_170306_A1 | Scaffold | 3.02 | 31.0 | 2831 | 4 | 57 | 5 |
| 1001713B170207_170306_A10 | Scaffold | 3.05 | 30.5 | 2881 | 4 | 60 | 5 |
| 1001283B150210_160208_G3 | Scaffold | 3.07 | 30.5 | 2869 | 5 | 52 | 5 |
| 1001283B150210_160208_D6 | Scaffold | 3.00 | 30.5 | 2791 | 4 | 58 | 5 |
| Cl_188 | Scaffold | 3.01 | 30.5 | 2876 | 3 | 40 | 5 |
| Cl_171 | Scaffold | 3.04 | 30.5 | 2938 | 3 | 36 | 5 |
| Cl_239 | Scaffold | 3.33 | 30.5 | 3219 | 4 | 48 | 5 |
| Cl_117 | Scaffold | 3.11 | 31.0 | 3034 | 4 | 46 | 5 |
| Cl_238 | Scaffold | 3.23 | 30.5 | 3115 | 4 | 30 | 5 |
| Cl_84 | Scaffold | 3.05 | 30.5 | 2954 | 3 | 52 | 5 |
| Cl_82 | Scaffold | 3.01 | 30.5 | 2916 | 4 | 45 | 5 |
| Cl_64 | Scaffold | 3.17 | 31.0 | 3087 | 4 | 46 | 5 |
| Cl_52 | Scaffold | 3.09 | 30.5 | 2940 | 2 | 42 | 5 |
| Cl_80 | Scaffold | 3.09 | 30.5 | 3049 | 3 | 34 | 5 |
| Cl_29 | Scaffold | 3.09 | 30.5 | 2988 | 4 | 37 | 5 |
| MGYG-HGUT-00125 | Scaffold | 3.26 | 30.5 | 3101 | 9 | 63 | 5 |
| 24853 | Scaffold | 3.00 | 30.5 | 2664 | 4 | 59 | 5 |
| ATCC 25755 | Contig | 3.01 | 30.5 | 2908 | 4 | 46 | 5 |
| DIVETGP | Contig | 3.02 | 30.5 | 2907 | 4 | 46 | 5 |
| UC7086 | Contig | 3.06 | 30.5 | 2943 | 4 | 48 | 5 |
| IFP923 | Contig | 3.19 | 30.5 | 3090 | 10 | 60 | 5 |
| 启动子 | 类型 | 性质 | 参考文献 |
|---|---|---|---|
| P ptb | 组成型 | 控制ptb的表达 | [ |
| P thl | 组成型 | 控制thl的表达 | [ |
| P hydA | 组成型 | 电子代谢关键酶基因hydA的启动子 | [ |
| P ptk | 诱导型 | 阿拉伯糖诱导 | [ |
| P xylA | 诱导型 | 受木糖诱导 | [ |
| P bgaL | 诱导型 | 受乳糖诱导 | [ |
| P celC | 诱导型 | 受纤维素诱导 | [ |
| P Pcm-2tetO1 | 诱导型 | 受四环素诱导 | [ |
Tab. 4 Common promoters of Clostridium expression systems
| 启动子 | 类型 | 性质 | 参考文献 |
|---|---|---|---|
| P ptb | 组成型 | 控制ptb的表达 | [ |
| P thl | 组成型 | 控制thl的表达 | [ |
| P hydA | 组成型 | 电子代谢关键酶基因hydA的启动子 | [ |
| P ptk | 诱导型 | 阿拉伯糖诱导 | [ |
| P xylA | 诱导型 | 受木糖诱导 | [ |
| P bgaL | 诱导型 | 受乳糖诱导 | [ |
| P celC | 诱导型 | 受纤维素诱导 | [ |
| P Pcm-2tetO1 | 诱导型 | 受四环素诱导 | [ |
| 1 | WANG J F, LIN M, XU M M, et al. Anaerobic fermentation for production of carboxylic acids as bulk chemicals from renewable biomass[M]// Hatti-Kaul R, Mamo G, Mattiasson B. Anaerobes in biotechnology. Advances in biochemical engineering/biotechnology, Cham, Swizerland: Springer, 2016 156: 323-361. |
| 2 | CLOMBURG J M, CRUMBLEY A M, GONZALEZ R. Industrial biomanufacturing: the future of chemical production[J]. Science, 2017, 355(6320): aag0804. |
| 3 | YANG X M, TANG S, LU T L, et al. Sulfonic acid resin-catalyzed oxidation of aldehydes to carboxylic acids by hydrogen peroxide[J]. Synthetic Communications, 2013, 43(7): 979-985. |
| 4 | YU T, ZHOU Y J J, HUANG M T, et al. Reprogramming yeast metabolism from alcoholic fermentation to lipogenesis[J]. Cell, 2018, 174(6): 1549-1558.e14. |
| 5 | LINGER J G, FORD L R, RAMNATH K, et al. Development of Clostridium tyrobutyricum as a microbial cell factory for the production of fuel and chemical intermediates from lignocellulosic feedstocks[J]. Frontiers in Energy Research, 2020, 8: 183. |
| 6 | 唐万. 酪丁酸梭菌代谢机理及其应用的探索研究[D]. 杭州: 浙江工业大学, 2018. |
| TANG W. Study on metabolic mechanism of Clostridium tyrobutyricum and its application for butyric acid production[D]. Hangzhou: Zhejiang University of Technology, 2018. | |
| 7 | 江凌. 纤维床固定化酪丁酸梭菌发酵廉价生物质生产丁酸的研究[D]. 广州: 华南理工大学, 2010. |
| JIANG L. Study on butyric acid production from cheap biomass by Clostridium tyrobutyricum immobilized in a fibrous bed bioreactor[D]. Guangzhou: South China University of Technology, 2010. | |
| 8 | ZHU Z M, ZHU L Y, JIANG L. Dynamic regulation of gut Clostridium-derived short-chain fatty acids[J]. Trends in Biotechnology, 2022, 40(3): 266-270. |
| 9 | 索玉凯. 代谢工程改造酪丁酸梭菌(Clostridium tyrobutyricum)强化丁酸生产及木质纤维素利用[D]. 广州: 华南理工大学, 2018. |
| SUO Y K. Metabolic engineering of Clostridium tyrobutyricum for enhanced butyric acid production and lignocellulosic utilization[D]. Guangzhou: South China University of Technology, 2018. | |
| 10 | LEE J, YUN H, FEIST A M, et al. Genome-scale reconstruction and in silico analysis of the Clostridium acetobutylicum ATCC 824 metabolic network[J]. Applied Microbiology and Biotechnology, 2008, 80(5): 849-862. |
| 11 | SENGER R S, PAPOUTSAKIS E T. Genome-scale model for Clostridium acetobutylicum: Part I. Metabolic network resolution and analysis[J]. Biotechnology and Bioengineering, 2008, 101(5): 1036-1052. |
| 12 | XIN B, TAO F, WANG Y, et al. Genome sequence of Clostridium butyricum strain DSM 10702, a promising producer of biofuels and biochemicals[J]. Genome Announcements, 2013, 1(4): e00563-e00513. |
| 13 | LEE J, JANG Y S, HAN M J, et al. Deciphering Clostridium tyrobutyricum metabolism based on the whole-genome sequence and proteome analyses[J]. mBio, 2016, 7(3): e00743-e00716. |
| 14 | SUO Y K, FU H X, REN M M, et al. Butyric acid production from lignocellulosic biomass hydrolysates by engineered Clostridium tyrobutyricum overexpressing Class I heat shock protein GroESL[J]. Bioresource Technology, 2018, 250: 691-698. |
| 15 | HUANG J, CAI J, WANG J, et al. Efficient production of butyric acid from Jerusalem artichoke by immobilized Clostridium tyrobutyricum in a fibrous-bed bioreactor[J]. Bioresource Technology, 2011, 102(4): 3923-3926. |
| 16 | ALAM S, STEVENS D, BAJPAI R. Production of butyric acid by batch fermentation of cheese whey with Clostridium beijerinckii [J]. Journal of Industrial Microbiology, 1988, 2(6): 359-364. |
| 17 | ZHU Y, WU Z T, YANG S T. Butyric acid production from acid hydrolysate of corn fibre by Clostridium tyrobutyricum in a fibrous-bed bioreactor[J]. Process Biochemistry, 2002, 38(5): 657-666. |
| 18 | FAYOLLE F, MARCHAL R, BALLERINI D. Effect of controlled substrate feeding on butyric acid production by Clostridium tyrobutyricum [J]. Journal of Industrial Microbiology, 1990, 6(3): 179-183. |
| 19 | HUANG Y L, WU Z T, ZHANG L K, et al. Production of carboxylic acids from hydrolyzed corn meal by immobilized cell fermentation in a fibrous-bed bioreactor[J]. Bioresource Technology, 2002, 82(1): 51-59. |
| 20 | JIANG L, WANG J F, LIANG S Z, et al. Butyric acid fermentation in a fibrous bed bioreactor with immobilized Clostridium tyrobutyricum from cane molasses[J]. Bioresource Technology, 2009, 100(13): 3403-3409. |
| 21 | HE F F, QIN S W, YANG Z, et al. Butyric acid production from spent coffee grounds by engineered Clostridium tyrobutyricum overexpressing galactose catabolism genes[J]. Bioresource Technology, 2020, 304: 122977. |
| 22 | SUO Y K, FU H X, REN M M, et al. Enhanced butyric acid production in Clostridium tyrobutyricum by overexpression of rate-limiting enzymes in the Embden-Meyerhof-Parnas pathway[J]. Journal of Biotechnology, 2018, 272/273: 14-21. |
| 23 | DU Y M, JIANG W Y, YU M R, et al. Metabolic process engineering of Clostridium tyrobutyricum Δack-adhE2 for enhanced n-butanol production from glucose: effects of methyl viologen on NADH availability, flux distribution, and fermentation kinetics[J]. Biotechnology and Bioengineering, 2015, 112(4): 705-715. |
| 24 | SUO Y K, LUO S, ZHANG Y N, et al. Enhanced butyric acid tolerance and production by Class I heat shock protein-overproducing Clostridium tyrobutyricum ATCC 25755[J]. Journal of Industrial Microbiology and Biotechnology, 2017, 44(8): 1145-1156. |
| 25 | ZHANG Y L, YU M R, YANG S T. Effects of ptb knockout on butyric acid fermentation by Clostridium tyrobutyricum [J]. Biotechnology Progress, 2012, 28(1): 52-59. |
| 26 | YU L, ZHAO J B, XU M M, et al. Metabolic engineering of Clostridium tyrobutyricum for n-butanol production: effects of CoA transferase[J]. Applied Microbiology and Biotechnology, 2015, 99(11): 4917-4930. |
| 27 | JIANG L, WANG J F, LIANG S Z, et al. Enhanced butyric acid tolerance and bioproduction by Clostridium tyrobutyricum immobilized in a fibrous bed bioreactor[J]. Biotechnology and Bioengineering, 2011, 108(1): 31-40. |
| 28 | LIU X G, ZHU Y, YANG S T. Construction and characterization of ack deleted mutant of Clostridium tyrobutyricum for enhanced butyric acid and hydrogen production[J]. Biotechnology Progress, 2006, 22(5): 1265-1275. |
| 29 | ZHANG J Z, YU L, XU M M, et al. Metabolic engineering of Clostridium tyrobutyricum for n-butanol production from sugarcane juice[J]. Applied Microbiology and Biotechnology, 2017, 101(10): 4327-4337. |
| 30 | YU L, XU M M, TANG I C, et al. Metabolic engineering of Clostridium tyrobutyricum for n-butanol production from maltose and soluble starch by overexpressing alpha-glucosidase[J]. Applied Microbiology and Biotechnology, 2015, 99(14): 6155-6165. |
| 31 | GUO X L, FU H X, FENG J, et al. Direct conversion of untreated cane molasses into butyric acid by engineered Clostridium tyrobutyricum [J]. Bioresource Technology, 2020, 301: 122764. |
| 32 | WEI D, LIU X G, YANG S T. Butyric acid production from sugarcane bagasse hydrolysate by Clostridium tyrobutyricum immobilized in a fibrous-bed bioreactor[J]. Bioresource Technology, 2013, 129: 553-560. |
| 33 | PYNE M E, MOO-YOUNG M, CHUNG D A, et al. Antisense-RNA-mediated gene downregulation in Clostridium pasteurianum [J]. Fermentation, 2015, 1(1): 113-126. |
| 34 | XIAO Z P, CHENG C, BAO T, et al. Production of butyric acid from acid hydrolysate of corn husk in fermentation by Clostridium tyrobutyricum: kinetics and process economic analysis[J]. Biotechnology for Biofuels, 2018, 11: 164. |
| 35 | BAROI G N, BAUMANN I, WESTERMANN P, et al. Butyric acid fermentation from pretreated and hydrolysed wheat straw by an adapted Clostridium tyrobutyricum strain[J]. Microbial Biotechnology, 2015, 8(5): 874-882. |
| 36 | SJÖBLOM M, MATSAKAS L, CHRISTAKOPOULOS P, et al. Production of butyric acid by Clostridium tyrobutyricum (ATCC25755) using sweet sorghum stalks and beet molasses[J]. Industrial Crops and Products, 2015, 74: 535-544. |
| 37 | ZHOU X, LU X H, LI X H, et al. Radiation induces acid tolerance of Clostridium tyrobutyricum and enhances bioproduction of butyric acid through a metabolic switch[J]. Biotechnology for Biofuels, 2014, 7(1): 22. |
| 38 | LIU T T, JIANG C, ZHU L Y, et al. Fe3O4@chitosan microspheres coating as cytoprotective exoskeletons for the enhanced production of butyric acid with Clostridium tyrobutyricum under acid stress[J]. Frontiers in Bioengineering and Biotechnology, 2020, 8: 449. |
| 39 | 常瀚文, 郑鑫铃, 骆健美, 等. 抗逆元件及其在高效微生物细胞工厂构建中的应用进展[J]. 生物技术通报, 2020, 36(6): 13-34. |
| CHANG H W, ZHENG X L, LUO J M, et al. Tolerance elements and their application progress on the construction of highly-efficient microbial cell factory[J]. Biotechnology Bulletin, 2020, 36(6): 13-34. | |
| 40 | ZHU Y, LIU X G, YANG S T. Construction and characterization of pta gene-deleted mutant of Clostridium tyrobutyricum for enhanced butyric acid fermentation[J]. Biotechnology and Bioengineering, 2005, 90(2): 154-166. |
| 41 | WU Q, ZHU L Y, XU Q, et al. Tailoring the oxidative stress tolerance of Clostridium tyrobutyricum CCTCC W428 by introducing trehalose biosynthetic capability[J]. Journal of Agricultural and Food Chemistry, 2017, 65(40): 8892-8901. |
| 42 | LIU Z H, WANG K, CHEN Y, et al. Third-generation biorefineries as the means to produce fuels and chemicals from CO2 [J]. Nature Catalysis, 2020, 3(3): 274-288. |
| 43 | PIERCE E, XIE G, BARABOTE R D, et al. The complete genome sequence of Moorella thermoacetica (f. Clostridium thermoaceticum)[J]. Environmental Microbiology, 2008, 10(10): 2550-2573. |
| 44 | FAST A G, PAPOUTSAKIS E T. Functional expression of the Clostridium ljungdahlii acetyl-coenzyme A synthase in Clostridium acetobutylicum as demonstrated by a novel in vivo CO exchange activity en route to heterologous installation of a functional Wood-Ljungdahl pathway[J]. Applied and Environmental Microbiology, 2018, 84(7): e02307-e02317. |
| 45 | CARLSON E D, PAPOUTSAKIS E T. Heterologous expression of the Clostridium carboxidivorans CO dehydrogenase alone or together with the acetyl coenzyme A synthase enables both reduction of CO2 and oxidation of CO by Clostridium acetobutylicum [J]. Applied and Environmental Microbiology, 2017, 83(16): e00829-e00817. |
| 46 | 卞化, 孙新晓, 袁其朋. 代谢工程改造异养微生物固定CO2研究进展[J]. 生物工程学报, 2019, 35(2): 195-203. |
| BIAN H, SUN X X, YUAN Q P. Advances in metabolic engineering of heterotrophic microorganisms for CO2 fixation: a review[J]. Chinese Journal of Biotechnology, 2019, 35(2): 195-203. | |
| 47 | DOBBEK H, SVETLITCHNYI V, GREMER L, et al. Crystal structure of a carbon monoxide dehydrogenase reveals a [Ni-4Fe-5S] cluster[J]. Science, 2001, 293(5533): 1281-1285. |
| 48 | ALTAŞ N, ASLAN A S, KARATAŞ E, et al. Heterologous production of extreme alkaline thermostable NAD+-dependent formate dehydrogenase with wide-range pH activity from Myceliophthora thermophila [J]. Process Biochemistry, 2017, 61: 110-118. |
| 49 | BANG J, LEE S Y. Assimilation of formic acid and CO2 by engineered Escherichia coli equipped with reconstructed one-carbon assimilation pathways[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(40): E9271-E9279. |
| 50 | FUCHS G. Alternative pathways of carbon dioxide fixation: insights into the early evolution of life?[J]. Annual Review of Microbiology, 2011, 65: 631-658. |
| 51 | HÜGLER M, MENENDEZ C, SCHÄGGER H, et al. Malonyl-coenzyme A reductase from Chloroflexus aurantiacus, a key enzyme of the 3-hydroxypropionate cycle for autotrophic CO2 fixation[J]. Journal of Bacteriology, 2002, 184(9): 2404-2410. |
| 52 | STRAUSS G, FUCHS G. Enzymes of a novel autotrophic CO2 fixation pathway in the phototrophic bacterium Chloroflexus aurantiacus, the 3-hydroxypropionate cycle[J]. European Journal of Biochemistry, 1993, 215(3): 633-643. |
| 53 | 王媛媛. 艰难梭菌电子歧化酶NfnAB性质和功能的初步研究[D]. 济南:济南大学, 2018. |
| WANG Y Y. The preliminary study on the properties and functions of the electron-bifucating NfnAB from Clostridium difficile [D]. Jinan: University of Jinan, 2018. | |
| 54 | LI F L, HINDERBERGER J, SEEDORF H, et al. Coupled ferredoxin and crotonyl coenzyme A (CoA) reduction with NADH catalyzed by the butyryl-CoA dehydrogenase/Etf complex from Clostridium kluyveri [J]. Journal of Bacteriology, 2008, 190(3): 843-850. |
| 55 | WANG S N, HUANG H Y, MOLL J, et al. NADP+ reduction with reduced ferredoxin and NADP+ reduction with NADH are coupled via an electron-bifurcating enzyme complex in Clostridium kluyveri [J]. Journal of Bacteriology, 2010, 192(19): 5115-5123. |
| 56 | MAYER A, WEUSTER-BOTZ D. Reaction engineering analysis of the autotrophic energy metabolism of Clostridium aceticum [J]. FEMS Microbiology Letters, 2017, 364(22): fnx219. |
| 57 | LIU T T, ZHU L Y, ZHU Z M, et al. Genome sequence analysis of Clostridium tyrobutyricum, a promising microbial host for human health and industrial applications[J]. Current Microbiology, 2020, 77(11): 3685-3694. |
| 58 | 禹伟, 高教琪, 周雍进. 蛋白质组学和代谢组学在微生物代谢工程中的应用[J]. 色谱, 2019, 37(8): 798-805. |
| YU W, GAO J Q, ZHOU Y J. Application of proteomics and metabolomics in microbial metabolic engineering[J]. Chinese Journal of Chromatography, 2019, 37(8): 798-805. | |
| 59 | LI W M, CHENG C, CAO G L, et al. Comparative transcriptome analysis of Clostridium tyrobutyricum expressing a heterologous uptake hydrogenase[J]. Science of the Total Environment, 2020, 749: 142022. |
| 60 | JANSSEN H, DÖRING C, EHRENREICH A, et al. A proteomic and transcriptional view of acidogenic and solventogenic steady-state cells of Clostridium acetobutylicum in a chemostat culture[J]. Applied Microbiology and Biotechnology, 2010, 87(6): 2209-2226. |
| 61 | MAO S M, LUO Y M, ZHANG T R, et al. Proteome reference map and comparative proteomic analysis between a wild type Clostridium acetobutylicum DSM 1731 and its mutant with enhanced butanol tolerance and butanol yield[J]. Journal of Proteome Research, 2010, 9(6): 3046-3061. |
| 62 | JIA K Z, ZHANG Y P, LI Y. Identification and characterization of two functionally unknown genes involved in butanol tolerance of Clostridium acetobutylicum [J]. PLoS One, 2012, 7(6): e38815. |
| 63 | SIVAGNANAM K, RAGHAVAN V G S, SHAH M, et al. Shotgun proteomic monitoring of Clostridium acetobutylicum during stationary phase of butanol fermentation using xylose and comparison with the exponential phase[J]. Journal of Industrial Microbiology and Biotechnology, 2012, 39(6): 949-955. |
| 64 | MA C, KOJIMA K, XU N, et al. Comparative proteomics analysis of high n-butanol producing metabolically engineered Clostridium tyrobutyricum [J]. Journal of Biotechnology, 2015, 193: 108-119. |
| 65 | JONES S W, PAREDES C J, TRACY B, et al. The transcriptional program underlying the physiology of clostridial sporulation[J]. Genome Biology, 2008, 9(7): R114. |
| 66 | AMADOR-NOGUEZ D, BRASG I A, FENG X J, et al. Metabolome remodeling during the acidogenic-solventogenic transition in Clostridium acetobutylicum [J]. Applied and Environmental Microbiology, 2011, 77(22): 7984-7997. |
| 67 | WANG Y F, TIAN J, JI Z H, et al. Intracellular metabolic changes of Clostridium acetobutylicum and promotion to butanol tolerance during biobutanol fermentation[J]. The International Journal of Biochemistry & Cell Biology, 2016, 78: 297-306. |
| 68 | ZU T N K, LIU S C, GERLACH E S, et al. Real-time metabolite monitoring of glucose-fed Clostridium acetobutylicum fermentations using Raman assisted metabolomics[J]. Journal of Raman Spectroscopy, 2017, 48(12): 1852-1862. |
| 69 | IDLE J R, GONZALEZ F J. Metabolomics[J]. Cell Metabolism, 2007, 6(5): 348-351. |
| 70 | KIM T Y, SOHN S B, KIM Y B, et al. Recent advances in reconstruction and applications of genome-scale metabolic models[J]. Current Opinion in Biotechnology, 2012, 23(4): 617-623. |
| 71 | PAPOUTSAKIS E T. Equations and calculations for fermentations of butyric acid bacteria[J]. Biotechnology and Bioengineering, 1984, 26(2): 174-187. |
| 72 | DESAI R P, HARRIS L M, WELKER N E, et al. Metabolic flux analysis elucidates the importance of the acid-formation pathways in regulating solvent production by Clostridium acetobutylicum [J]. Metabolic Engineering, 1999, 1(3): 206-213. |
| 73 | DASH S, MUELLER T J, VENKATARAMANAN K P, et al. Capturing the response of Clostridium acetobutylicum to chemical stressors using a regulated genome-scale metabolic model[J]. Biotechnology for Biofuels, 2014, 7(1): 144. |
| 74 | THOMPSON R A, DAHAL S, GARCIA S, et al. Exploring complex cellular phenotypes and model-guided strain design with a novel genome-scale metabolic model of Clostridium thermocellum DSM 1313 implementing an adjustable cellulosome[J]. Biotechnology for Biofuels, 2016, 9(1): 194. |
| 75 | GARCIA S, THOMPSON R A, GIANNONE R J, et al. Development of a genome-scale metabolic model of Clostridium thermocellum and its applications for integration of multi-omics datasets and computational strain design[J]. Frontiers in Bioengineering and Biotechnology, 2020, 8: 772. |
| 76 | THIELE I, PALSSON B Ø. A protocol for generating a high-quality genome-scale metabolic reconstruction[J]. Nature Protocols, 2010, 5(1): 93-121. |
| 77 | 董风晴. 凝结芽孢杆菌36D1全基因组代谢网络模型的构建和验证[D]. 上海: 华东理工大学, 2017. |
| DONG F Q. Reconstruction and verification of the genome-scale metabolic model of Bacillus coagulans 36D1[D]. Shanghai: East China University of Science and Technology, 2017. | |
| 78 | 叶超. 新一代工业微生物生物网络模型的构建及应用[D]. 无锡: 江南大学, 2019. |
| YE C. Construction and application of the new generation industrial microorganisms biological network models[D]. Wuxi: Jiangnan University, 2019. | |
| 79 | 林璐, 吕雪芹, 刘延峰, 等. 枯草芽孢杆菌底盘细胞的设计、构建与应用[J]. 合成生物学, 2020, 1(2): 247-265. |
| LIN L, LV X Q, LIU Y F, et al. Advances in design, construction and applications of Bacillus subtilis chassis cells[J]. Synthetic Biology Journal, 2020, 1(2): 247-265. | |
| 80 | TAO X Y, XU T, KEMPHER M L, et al. Precise promoter integration improves cellulose bioconversion and thermotolerance in Clostridium cellulolyticum [J]. Metabolic Engineering, 2020, 60: 110-118. |
| 81 | FU H X, YU L, LIN M, et al. Metabolic engineering of Clostridium tyrobutyricum for enhanced butyric acid production from glucose and xylose[J]. Metabolic Engineering, 2017, 40: 50-58. |
| 82 | LEE J, JANG Y S, PAPOUTSAKIS E T, et al. Stable and enhanced gene expression in Clostridium acetobutylicum using synthetic untranslated regions with a stem-loop[J]. Journal of Biotechnology, 2016, 230: 40-43. |
| 83 | GORWA M F, CROUX C, SOUCAILLE P. Molecular characterization and transcriptional analysis of the putative hydrogenase gene of Clostridium acetobutylicum ATCC 824[J]. Journal of Bacteriology, 1996, 178(9): 2668-2675. |
| 84 | ZHANG L, LEYN S A, GU Y, et al. Ribulokinase and transcriptional regulation of arabinose metabolism in Clostridium acetobutylicum [J]. Journal of Bacteriology, 2012, 194(5): 1055-1064. |
| 85 | SIZEMORE C, BUCHNER E, RYGUS T, et al. Organization, promoter analysis and transcriptional regulation of the Staphylococcus xylosus xylose utilization operon[J]. Molecular & General Genetics, 1991, 227(3): 377-384. |
| 86 | HARTMAN A H, LIU H L, MELVILLE S B. Construction and characterization of a lactose-inducible promoter system for controlled gene expression in Clostridium perfringens [J]. Applied and Environmental Microbiology, 2011, 77(2): 471-478. |
| 87 | MEARLS E B, OLSON D G, HERRING C D, et al. Development of a regulatable plasmid-based gene expression system for Clostridium thermocellum [J]. Applied Microbiology and Biotechnology, 2015, 99(18): 7589-7599. |
| 88 | DONG H J, TAO W W, ZHANG Y P, et al. Development of an anhydrotetracycline-inducible gene expression system for solvent-producing Clostridium acetobutylicum: A useful tool for strain engineering[J]. Metabolic Engineering, 2012, 14(1): 59-67. |
| 89 | YU M R, DU Y M, JIANG W Y, et al. Effects of different replicons in conjugative plasmids on transformation efficiency, plasmid stability, gene expression and n-butanol biosynthesis in Clostridium tyrobutyricum [J]. Applied Microbiology and Biotechnology, 2012, 93(2): 881-889. |
| 90 | ZHANG X D, YANG S T. High-throughput 3-D cell-based proliferation and cytotoxicity assays for drug screening and bioprocess development[J]. Journal of Biotechnology, 2011, 151(2): 186-193. |
| 91 | DREPPER T, EGGERT T, CIRCOLONE F, et al. Reporter proteins for in vivo fluorescence without oxygen[J]. Nature Biotechnology, 2007, 25(4): 443-445. |
| 92 | CHENG C, LIN M, JIANG W Y, et al. Development of an in vivo fluorescence based gene expression reporter system for Clostridium tyrobutyricum [J]. Journal of Biotechnology, 2019, 305: 18-22. |
| 93 | BAO T, ZHAO J B, ZHANG Q X, et al. Development of a shuttle plasmid without host restriction sites for efficient transformation and heterologous gene expression in Clostridium cellulovorans [J]. Applied Microbiology and Biotechnology, 2019, 103(13): 5391-5400. |
| 94 | HEAP J T, PENNINGTON O J, CARTMAN S T, et al. A modular system for Clostridium shuttle plasmids[J]. Journal of Microbiological Methods, 2009, 78(1): 79-85. |
| 95 | OH Y H, EOM G T, KANG K H, et al. Optimized transformation of newly constructed Escherichia coli-Clostridia shuttle vectors into Clostridium beijerinckii [J]. Applied Biochemistry and Biotechnology, 2015, 177(1): 226-236. |
| 96 | 张晋龙. 3株放线菌的分离鉴定及其遗传转化系统的构建与优化[D]. 咸阳: 西北农林科技大学, 2017. |
| ZHANG J L. Isolation and identification of three Actinomyces strains and construction and optimization of their genetic transformation system[D]. Xianyang: Northwest A&F University, 2017. | |
| 97 | 周隽. 电穿孔转化沙眼衣原体建立衣原体载体实验研究[D]. 武汉: 华中科技大学, 2006. |
| ZHOU J. Research on electroporation transfection Chlamydia trachomatis to build vector of Chlamydia [D]. Wuhan: Huazhong University of Science and Technology, 2006. | |
| 98 | WOODS C, HUMPHREYS C M, RODRIGUES R M, et al. A novel conjugal donor strain for improved DNA transfer into Clostridium spp.[J]. Anaerobe, 2019, 59: 184-191. |
| 99 | SZOSTKOVÁ M, HORÁKOVÁ D, NĚMEC M. The influence of the growth phase of enteric bacteria on electrotransformation with plasmid DNA[J]. Folia Microbiologica, 1999, 44(2): 177-180. |
| 100 | ZHANG J, HONG W, GUO L, et al. Enhancing plasmid transformation efficiency and enabling CRISPR-Cas9/Cpf1-based genome editing in Clostridium tyrobutyricum [J]. Biotechnology and Bioengineering, 2020, 117(9): 2911-2917. |
| 101 | MURRAY N E. Type I restriction systems: Sophisticated molecular machines (a legacy of Bertani and Weigle)[J]. Microbiology and Molecular Biology Reviews, 2000, 64(2): 412-434. |
| 102 | ROBERTS R J, BELFORT M, BESTOR T, et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes[J]. Nucleic Acids Research, 2003, 31(7): 1805-1812. |
| 103 | LOENEN W A M, DRYDEN D T F, RALEIGH E A, et al. Type I restriction enzymes and their relatives[J]. Nucleic Acids Research, 2013, 42(1): 20-44. |
| 104 | CUI G Z, HONG W, ZHANG J, et al. Targeted gene engineering in Clostridium cellulolyticum H10 without methylation[J]. Journal of Microbiological Methods, 2012, 89(3): 201-208. |
| 105 | BARRANGOU R, FREMAUX C, DEVEAU H, et al. CRISPR provides acquired resistance against viruses in prokaryotes[J]. Science, 2007, 315(5819): 1709-1712. |
| 106 | LIAN J Z, HAMEDIRAD M, HU S M, et al. Combinatorial metabolic engineering using an orthogonal tri-functional CRISPR system[J]. Nature Communications, 2017, 8: 1688. |
| 107 | 李洋, 申晓林, 孙新晓, 等. CRISPR基因编辑技术在微生物合成生物学领域的研究进展[J]. 合成生物学, 2021, 2(1): 106-120. |
| LI Y, SHEN X L, SUN X X, et al. Advances of CRISPR gene editing in microbial synthetic biology[J]. Synthetic Biology Journal, 2021, 2(1): 106-120. | |
| 108 | SHMAKOV S, ABUDAYYEH O O, MAKAROVA K S, et al. Discovery and functional characterization of diverse Class 2 CRISPR-Cas systems[J]. Molecular Cell, 2015, 60(3): 385-397. |
| 109 | 柳柯,林桂虹, 刘坤,等. CRISPR/Cas系统的挖掘、改造与功能拓展[J/OL]. 合成生物学, 2021-04-30[2022-05-24]. . |
| LIU K, LIN G H, LIU S, et al. Mining, engineering and functional expansion of CRISPR/Cas systems[J/OL]. Synthetic Biology Journal, 2021-04-30[2022-05-24]. . | |
| 110 | PYNE M E, BRUDER M R, MOO-YOUNG M, et al. Harnessing heterologous and endogenous CRISPR-Cas machineries for efficient markerless genome editing in Clostridium [J]. Scientific Reports, 2016, 6: 25666. |
| 111 | ZHANG J, ZONG W M, HONG W, et al. Exploiting endogenous CRISPR-Cas system for multiplex genome editing in Clostridium tyrobutyricum and engineer the strain for high-level butanol production[J]. Metabolic Engineering, 2018, 47: 49-59. |
| 112 | MAIKOVA A, KREIS V, BOUTSERIN A, et al. Using an endogenous CRISPR-Cas system for genome editing in the human pathogen Clostridium difficile [J]. Applied and Environmental Microbiology, 2019, 85(20): e01416-e01419. |
| 113 | ZHOU X Q, WANG X L, LUO H Y, et al. Exploiting heterologous and endogenous CRISPR-Cas systems for genome editing in the probiotic Clostridium butyricum [J]. Biotechnology and Bioengineering, 2021, 118(7): 2448-2459. |
| 114 | 刘洋, 牟庆璇, 石雅南, 等. 微生物细胞工厂的代谢调控[J]. 生物工程学报, 2021, 37(5): 1541-1563. |
| LIU Y, MOU Q X, SHI Y N, et al. Metabolic regulation in constructing microbial cell factories[J]. Chinese Journal of Biotechnology, 2021, 37(5): 1541-1563. | |
| 115 | XU N, WEI L, LIU J. Recent advances in the applications of promoter engineering for the optimization of metabolite biosynthesis[J]. World Journal of Microbiology & Biotechnology, 2019, 35(2): 33. |
| 116 | YU M R, ZHANG Y L, TANG I C, et al. Metabolic engineering of Clostridium tyrobutyricum for n-butanol production[J]. Metabolic Engineering, 2011, 13(4): 373-382. |
| 117 | FENG J, ZHANG J, MA Y C, et al. Renewable fatty acid ester production in Clostridium [J]. Nature Communications, 2021, 12: 4368. |
| 118 | YANG G H, JIA D C, JIN L, et al. Rapid generation of universal synthetic promoters for controlled gene expression in both gas-fermenting and saccharolytic Clostridium species[J]. ACS Synthetic Biology, 2017, 6(9): 1672-1678. |
| 119 | HOLTZ W J, KEASLING J D. Engineering static and dynamic control of synthetic pathways[J]. Cell, 2010, 140(1): 19-23. |
| 120 | LOWERY C A, DICKERSON T J, JANDA K D. Interspecies and interkingdom communication mediated by bacterial quorum sensing[J]. Chemical Society Reviews, 2008, 37(7): 1337-1346. |
| 121 | QSIM M, ASHFAQ U A, YOUSAF M Z, et al. Genetically modified aedes aegypti to control dengue: a review[J]. Critical Reviews in Eukaryotic Gene Expression, 2017, 27(4): 331-340. |
| 122 | NOVICK R P, GEISINGER E. Quorum sensing in staphylococci[J]. Annual Review of Genetics, 2008, 42: 541-564. |
| 123 | OLSON M E, TODD D A, SCHAEFFER C R, et al. Staphylococcus epidermidis agr quorum-sensing system: signal identification, cross talk, and importance in colonization[J]. Journal of Bacteriology, 2014, 196(19): 3482-3493. |
| 124 | XUE C, ZHAO J B, CHEN L J, et al. Recent advances and state-of-the-art strategies in strain and process engineering for biobutanol production by Clostridium acetobutylicum [J]. Biotechnology Advances, 2017, 35(2): 310-322. |
| 125 | STEINER E, SCOTT J, MINTON N P, et al. An agr quorum sensing system that regulates granulose formation and sporulation in Clostridium acetobutylicum [J]. Applied and Environmental Microbiology, 2012, 78(4): 1113-1122. |
| 126 | NEIDITCH M B, CAPODAGLI G C, PREHNA G, et al. Genetic and structural analyses of RRNPP intercellular peptide signaling of gram-positive bacteria[J]. Annual Review of Genetics, 2017, 51: 311-333. |
| 127 | DO H, KUMARASWAMI M. Structural mechanisms of peptide recognition and allosteric modulation of gene regulation by the RRNPP family of quorum-sensing regulators[J]. Journal of Molecular Biology, 2016, 428(14): 2793-2804. |
| 128 | PEREZ-PASCUAL D, MONNET V, GARDAN R. Bacterial cell-cell communication in the host via RRNPP peptide-binding regulators[J]. Frontiers in Microbiology, 2016, 7: 706. |
| 129 | FENG J, ZONG W M, WANG P X, et al. RRNPP-type quorum-sensing systems regulate solvent formation, sporulation and cell motility in Clostridium saccharoperbutylacetonicum [J]. Biotechnology for Biofuels, 2020, 13: 84. |
| 130 | JO J H, JEON C O, LEE S Y, et al. Molecular characterization and homologous overexpression of [FeFe]-hydrogenase in Clostridium tyrobutyricum JM1[J]. International Journal of Hydrogen Energy, 2010, 35(3): 1065-1073. |
| 131 | ZHANG Z T, TAYLOR S, WANG Y. In situ esterification and extractive fermentation for butyl butyrate production with Clostridium tyrobutyricum [J]. Biotechnology and Bioengineering, 2017, 114(7): 1428-1437. |
| 132 | WANG J F, LIN M, XU M M, et al. Anaerobic fermentation for production of carboxylic acids as bulk chemicals from renewable biomass[M]// Hatti-Kaul R., Mamo G., Mattiasson B. ed. Anaerobes in Biotechnology. Advances in Biochemical Engineering/Biotechnology, Cham, Swizerland: Springer, 2016, 156: 323-361. |
| 133 | DU J J, MCGRAW A, HESTEKIN J. Modeling of Clostridium tyrobutyricum for butyric acid selectivity in continuous fermentation[J]. Energies, 2014, 7(4): 2421-2435. |
| 134 | 张亚南. 调控胞内NADH/NAD+对酪丁酸梭菌丁酸生物合成的影响[D]. 广州: 华南理工大学, 2018. |
| ZHANG Y N. Effects of intracellular NADH/NAD+ ratio regulation on butyric acid biosynthesis in Clostridium tyrobutyricum ATCC 25755[D]. Guangzhou: South China University of Technology, 2018. | |
| 135 | AMIRI H, KARIMI K. Pretreatment and hydrolysis of lignocellulosic wastes for butanol production: challenges and perspectives[J]. Bioresource Technology, 2018, 270: 702-721. |
| 136 | XUE C, ZHAO X Q, LIU C G, et al. Prospective and development of butanol as an advanced biofuel[J]. Biotechnology Advances, 2013, 31(8): 1575-1584. |
| 137 | SINHA P, PANDEY A. An evaluative report and challenges for fermentative biohydrogen production[J]. International Journal of Hydrogen Energy, 2011, 36(13): 7460-7478. |
| 138 | JIANG L, WU Q, XU Q, et al. Fermentative hydrogen production from Jerusalem artichoke by Clostridium tyrobutyricum expressing exo-inulinase gene[J]. Scientific Reports, 2017, 7: 7940. |
| 139 | PALSSON B O, FATHI-AFSHAR S, RUDD D F, et al. Biomass as a source of chemical feedstocks: an economic evaluation[J]. Science, 1981, 213(4507): 513-517. |
| 140 | RODRIGUEZ G M, TASHIRO Y, ATSUMI S. Expanding ester biosynthesis in Escherichia coli [J]. Nature Chemical Biology, 2014, 10(4): 259-265. |
| 141 | CUI Y H, HE J Z, YANG K L, et al. Production of isopropyl and butyl esters by Clostridium mono-culture and co-culture[J]. Journal of Industrial Microbiology & Biotechnology, 2020, 47(6/7): 543-550. |
| 142 | HEIRENDT L, ARRECKX S, PFAU T, et al. Creation and analysis of biochemical constraint-based models using the COBRA Toolbox v.3.0[J]. Nature Protocols, 2019, 14(3): 639-702. |
| 143 | HU G P, LI Y, YE C, et al. Engineering microorganisms for enhanced CO2 sequestration[J]. Trends in Biotechnology, 2019, 37(5): 532-547. |
| 144 | LI X L, ZHOU Z, LI W N, et al. Design of stable and self-regulated microbial consortia for chemical synthesis[J]. Nature Communications, 2022, 13: 1554. |
| 145 | ERTL P, STICKER D, CHARWAT V, et al. Lab-on-a-chip technologies for stem cell analysis[J]. Trends in Biotechnology, 2014, 32(5): 245-253. |
| 146 | JOHNSTON C D, COTTON S L, RITTLING S R, et al. Systematic evasion of the restriction-modification barrier in bacteria[J]. Proceedings of the National Academy of Sciences of the United States of America, 2019, 116(23): 11454-11459. |
| [1] | GUO Shuyuan, ZHANG Qiannan, Gulikezi· MAIMAITIREXIATI, YANG Yiqun, YU Tao. Advances in microbial production of liquid biofuels [J]. Synthetic Biology Journal, 2025, 6(1): 18-44. |
| [2] | SHAO Mingwei, SUN Simian, YANG Shimao, CHEN Guoqiang. Bioproduction based on extremophiles [J]. Synthetic Biology Journal, 2024, 5(6): 1419-1436. |
| [3] | ZHAO Liang, LI Zhenshuai, FU Liping, LYU Ming, WANG Shi’an, ZHANG Quan, LIU Licheng, LI Fuli, LIU Ziyong. Progress in biomanufacturing of lipids and single cell protein from one-carbon compounds [J]. Synthetic Biology Journal, 2024, 5(6): 1300-1318. |
| [4] | ZHU Fanghuan, CEN Xuecong, CHEN Zhen. Research progress of diols production by microbes [J]. Synthetic Biology Journal, 2024, 5(6): 1367-1385. |
| [5] | ZHENG Haotian, LI Chaofeng, LIU Liangxu, WANG Jiawei, LI Hengrun, NI Jun. Design, optimization and application of synthetic carbon-negative phototrophic community [J]. Synthetic Biology Journal, 2024, 5(5): 1189-1210. |
| [6] | YU Wei, GAO Jiaoqi, ZHOU Yongjin. Bioconversion of one carbon feedstocks for producing organic acids [J]. Synthetic Biology Journal, 2024, 5(5): 1169-1188. |
| [7] | CHEN Xiwei, ZHANG Huaran, ZOU Yi. Biosynthesis and metabolic engineering of fungal non-ribosomal peptides [J]. Synthetic Biology Journal, 2024, 5(3): 571-592. |
| [8] | XIE Huang, ZHENG Yilei, SU Yiting, RUAN Jingyi, LI Yongquan. An overview on reconstructing the biosynthetic system of actinomycetes for polyketides production [J]. Synthetic Biology Journal, 2024, 5(3): 612-630. |
| [9] | ZHAO Jingyu, ZHANG Jian, QI Qingsheng, WANG Qian. Research progress in biosensors based on bacterial two-component systems [J]. Synthetic Biology Journal, 2024, 5(1): 38-52. |
| [10] | SUN Huili, CUI Jinyu, LUAN Guodong, LYU Xuefeng. Progress of cyanobacterial synthetic biotechnology for efficient light-driven carbon fixation and ethanol production [J]. Synthetic Biology Journal, 2023, 4(6): 1161-1177. |
| [11] | YAN Xiongying, WANG Zhen, LOU Jiyun, ZHANG Haoyu, HUANG Xingyu, WANG Xia, YANG Shihui. Progress in the construction of microbial cell factories for efficient biofuel production [J]. Synthetic Biology Journal, 2023, 4(6): 1082-1121. |
| [12] | CHEN Yongcan, SI Tong, ZHANG Jianzhi. Applications of automated synthetic biotechnology in DNA assembly and microbial chassis manipulation [J]. Synthetic Biology Journal, 2023, 4(5): 857-876. |
| [13] | CHENG Zhenzhen, ZHANG Jian, GAO Cong, LIU Liming, CHEN Xiulai. Progress in metabolic engineering of microorganisms for the utilization of formate [J]. Synthetic Biology Journal, 2023, 4(4): 756-778. |
| [14] | SUN Zhi, YANG Ning, LOU Chunbo, TANG Chao, YANG Xiaojing. Rational design for functional topology and its applications in synthetic biology [J]. Synthetic Biology Journal, 2023, 4(3): 444-463. |
| [15] | Shuyuan GUO, Lianghuan WU, Xiangjian LIU, Bo WANG, Tao YU. Developing C1-based metabolic network in methylotrophy for biotransformation [J]. Synthetic Biology Journal, 2022, 3(1): 116-137. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||