Synthetic Biology Journal ›› 2025, Vol. 6 ›› Issue (5): 998-1024.DOI: 10.12211/2096-8280.2025-065
• Invited Review • Previous Articles Next Articles
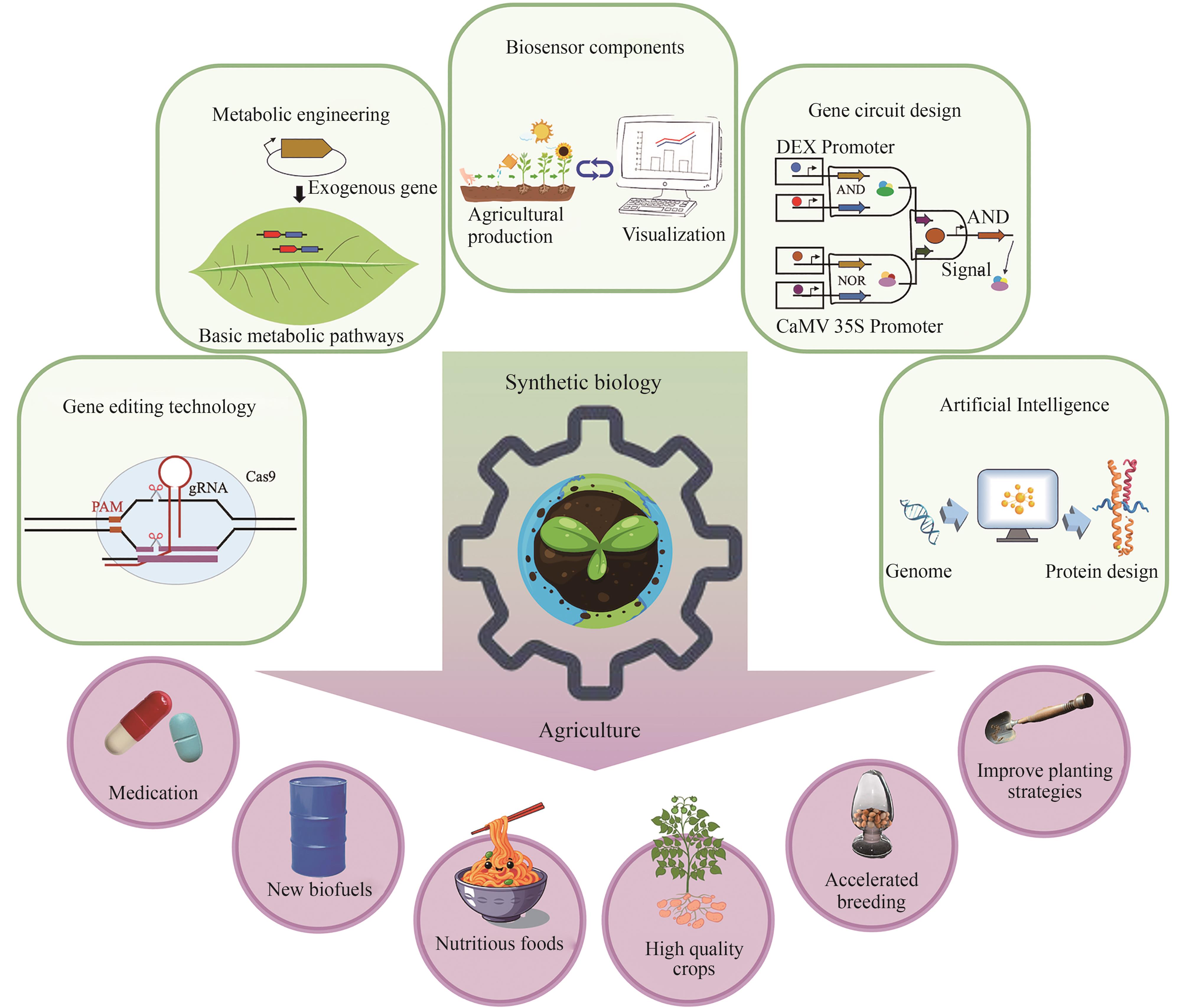
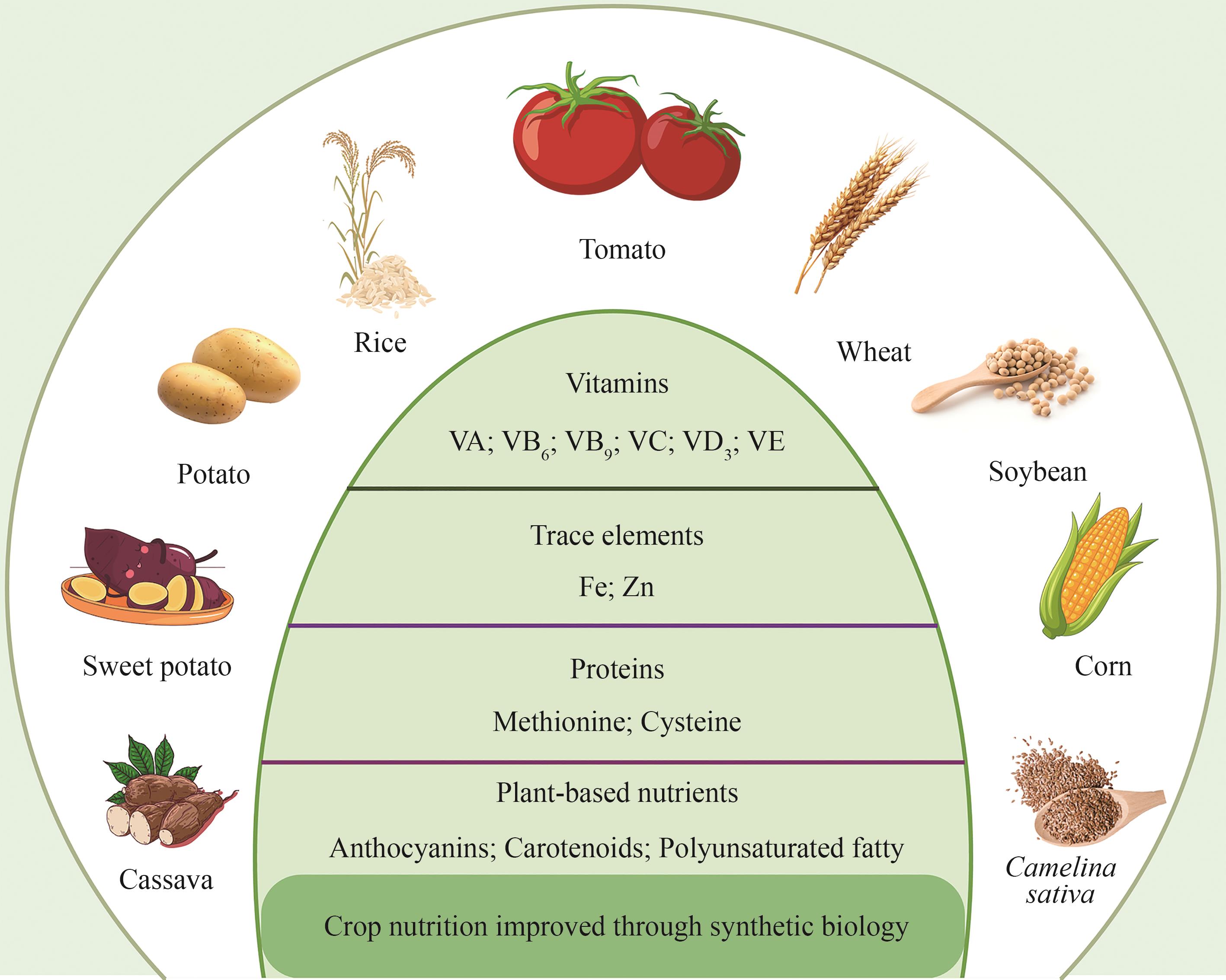
Progress and challenges of synthetic biology in agriculture
LIU Jie1, GAO Yu1, MA Yongshuo1,2, SHANG Yi1
- 1.Key Laboratory for Potato Biology of Yunnan Province,The CAAS-YNNU-YINMORE Joint Academy of Potato Science,Yunnan Normal University,Kunming 650500,Yunnan,China
2.Department of Chemical Engineering,Massachusetts Institute of Technology,Cambridge 02142,Massachusetts,USA
-
Received:2025-06-23Revised:2025-09-07Online:2025-11-05Published:2025-10-31 -
Contact:MA Yongshuo, SHANG Yi
合成生物学在农业中的进展及挑战
刘婕1, 郜钰1, 马永硕1,2, 尚轶1
- 1.云南省马铃薯生物学重点实验室,马铃薯科学研究院,云南师范大学,云南 昆明 650500
2.麻省理工学院化学工程系,美国 马萨诸塞州 剑桥市 02142
-
通讯作者:马永硕,尚轶 -
作者简介:刘婕 (1996—),女,博士研究生。研究方向为植物天然产物在植物以及微生物的高效合成。E-mail:liujie19960901@163.com马永硕 (1986—),男,研究员,博士生导师。研究方向为合成生物学与代谢工程。E-mail:mayongshuo2000@163.com尚轶 (1982—),男,研究员,博士生导师。研究方向为作物营养与风味品质,合成生物学。E-mail:shangyi@ynnu.edu.cn -
基金资助:国家自然科学基金委联合基金重点项目(U2202206);云南省“兴滇英才支持计划”云岭学者专项(XDYC-YLXZ-2022-0019)
CLC Number:
Cite this article
LIU Jie, GAO Yu, MA Yongshuo, SHANG Yi. Progress and challenges of synthetic biology in agriculture[J]. Synthetic Biology Journal, 2025, 6(5): 998-1024.
刘婕, 郜钰, 马永硕, 尚轶. 合成生物学在农业中的进展及挑战[J]. 合成生物学, 2025, 6(5): 998-1024.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2025-065
| Year | Findings | References |
|---|---|---|
| 2005 | 二代黄金大米的总胡萝卜素得到23倍的增加 | [ |
| 2011 | 植物传感器开始发展 | [ |
| 2012 | 番茄中抗坏血酸含量的增加 | [ |
| 2014 | 谷类作物中固氮途径的引入,增加作物固氮量 | [ |
| 2014 | 油料作物种子中高效合成ω-3不饱和脂肪酸 | [ |
| 2016 | 烟草叶片中青蒿素的合成 | [ |
| 2017 | 番茄果实中GABA的大量积累 | [ |
| 2017 | 大麦中引入固氮系统,提高氮利用效率 | [ |
| 2018 | 水稻胚乳中虾青素的生物合成 | [ |
| 2019 | 烟草中引入光呼吸途径,增加C3作物的产量 | [ |
| 2019 | 水稻中引入光呼吸旁路,增加光合效率 | [ |
| 2020 | 初级编辑器在作物中的应用 | [ |
| 2021 | C3作物中C4高光效特征的模拟 | [ |
| 2022 | 烟草中马钱子碱的生物合成途径重构 | [ |
| 2024 | 烟草中疫苗佐剂QS-21的生物合成途径重构 | [ |
Table1 Key achievements of synthetic biology in agriculture
| Year | Findings | References |
|---|---|---|
| 2005 | 二代黄金大米的总胡萝卜素得到23倍的增加 | [ |
| 2011 | 植物传感器开始发展 | [ |
| 2012 | 番茄中抗坏血酸含量的增加 | [ |
| 2014 | 谷类作物中固氮途径的引入,增加作物固氮量 | [ |
| 2014 | 油料作物种子中高效合成ω-3不饱和脂肪酸 | [ |
| 2016 | 烟草叶片中青蒿素的合成 | [ |
| 2017 | 番茄果实中GABA的大量积累 | [ |
| 2017 | 大麦中引入固氮系统,提高氮利用效率 | [ |
| 2018 | 水稻胚乳中虾青素的生物合成 | [ |
| 2019 | 烟草中引入光呼吸途径,增加C3作物的产量 | [ |
| 2019 | 水稻中引入光呼吸旁路,增加光合效率 | [ |
| 2020 | 初级编辑器在作物中的应用 | [ |
| 2021 | C3作物中C4高光效特征的模拟 | [ |
| 2022 | 烟草中马钱子碱的生物合成途径重构 | [ |
| 2024 | 烟草中疫苗佐剂QS-21的生物合成途径重构 | [ |
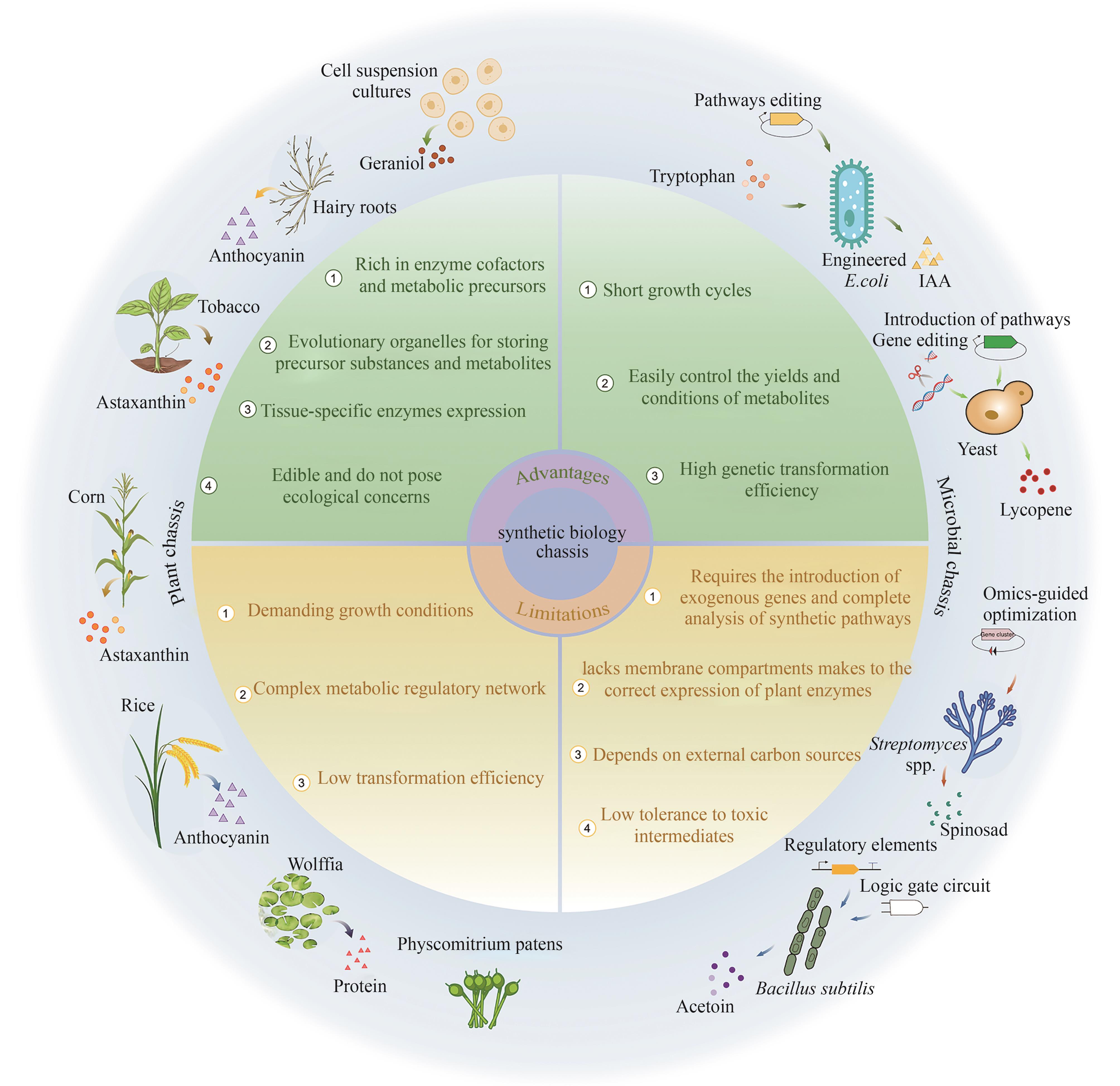
| 优/缺点 | 植物底盘 | 微生物底盘 |
|---|---|---|
| 优点 | 富含辅酶因子和前体物质 高度进化的细胞器可以储存前体物质和代谢物 组织特异性的酶表达模式 可以直接食用 | 生长周期短 培养条件和产量可控性强 基因转化效率高 |
| 缺点 | 生长环境要求高 代谢调控网络复杂 转化效率低 | 需要外源基因引入且依赖完整的途径解析 缺乏植物来源的酶表达所需的隔膜系统 依赖外部碳源 对有毒中间产物耐受性低 |
Table2 Advantages and disadvantages of plant chassis and microbial chassis
| 优/缺点 | 植物底盘 | 微生物底盘 |
|---|---|---|
| 优点 | 富含辅酶因子和前体物质 高度进化的细胞器可以储存前体物质和代谢物 组织特异性的酶表达模式 可以直接食用 | 生长周期短 培养条件和产量可控性强 基因转化效率高 |
| 缺点 | 生长环境要求高 代谢调控网络复杂 转化效率低 | 需要外源基因引入且依赖完整的途径解析 缺乏植物来源的酶表达所需的隔膜系统 依赖外部碳源 对有毒中间产物耐受性低 |
| [106] | WU R, DUAN L N, PRUNEDA-PAZ J L, et al. The 6xABRE synthetic promoter enables the spatiotemporal analysis of ABA-mediated transcriptional regulation[J]. Plant Physiology, 2018, 177(4): 1650-1665. |
| [107] | MÜLLER B, SHEEN J. Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis[J]. Nature, 2008, 453(7198): 1094-1097. |
| [108] | RIZZA A, WALIA A, TANG B J, et al. Visualizing cellular gibberellin levels using the nlsGPS1 Förster resonance energy transfer (FRET) biosensor[J]. Journal of Visualized Experiments, 2019(143): e58739. |
| [109] | LARRIEU A, CHAMPION A, LEGRAND J, et al. A fluorescent hormone biosensor reveals the dynamics of jasmonate signalling in plants[J]. Nature Communications, 2015, 6: 6043. |
| [110] | POUVREAU B, VANHERCKE T, SINGH S. From plant metabolic engineering to plant synthetic biology: the evolution of the design/build/test/learn cycle[J]. Plant Science, 2018, 273: 3-12. |
| [111] | WONG M H, GIRALDO J P, KWAK S Y, et al. Nitroaromatic detection and infrared communication from wild-type plants using plant nanobionics[J]. Nature Materials, 2017, 16(2): 264-272. |
| [112] | PENG Y H, ALLEN S, MILLWOOD R J, et al. ‘Fukusensor’: a genetically engineered plant for reporting DNA damage in response to gamma radiation[J]. Plant Biotechnology Journal, 2014, 12(9): 1329-1332. |
| [113] | JEZ J M, LEE S G, SHERP A M. The next green movement: Plant biology for the environment and sustainability[J]. Science, 2016, 353(6305): 1241-1244. |
| [114] | FICHMAN Y, MILLER G, MITTLER R. Whole-plant live imaging of reactive oxygen species[J]. Molecular Plant, 2019, 12(9): 1203-1210. |
| [115] | MCADAM E L, REID J B, FOO E. Gibberellins promote nodule organogenesis but inhibit the infection stages of nodulation[J]. Journal of Experimental Botany, 2018, 69(8): 2117-2130. |
| [116] | CRUZ A P Z, FERREIRA V, PIANZZOLA M J, et al. A novel, sensitive method to evaluate potato germplasm for bacterial wilt resistance using a luminescent Ralstonia solanacearum reporter strain[J]. Molecular Plant-Microbe Interactions, 2014, 27(3): 277-285. |
| [117] | MOHAMMAD-RAZDARI A, ROUSSEAU D, BAKHSHIPOUR A, et al. Recent advances in E-monitoring of plant diseases[J]. Biosensors and Bioelectronics, 2022, 201: 113953. |
| [118] | JUGDER B E, ERTAN H, BOHL S, et al. Organohalide respiring bacteria and reductive dehalogenases: key tools in organohalide bioremediation[J]. Frontiers in Microbiology, 2016, 7: 249. |
| [119] | SAHA G, SHAHRIN F, KHAN F H, et al. Smart IoT-driven precision agriculture: land mapping, crop prediction, and irrigation system[J]. PLoS One, 2025, 20(3): e0319268. |
| [120] | CUZICK A, MAGUIRE K, HAMMOND-KOSACK K E. Lack of the plant signalling component SGT1b enhances disease resistance to Fusarium culmorum in Arabidopsis buds and flowers[J]. New Phytologist, 2009, 181(4): 901-912. |
| [121] | JEONG H J, JUNG K H. Rice tissue-specific promoters and condition-dependent promoters for effective translational application[J]. Journal of Integrative Plant Biology, 2015, 57(11): 913-924. |
| [122] | JUSIAK B, CLETO S, PEREZ-PIÑERA P, et al. Engineering synthetic gene circuits in living cells with CRISPR technology[J]. Trends in Biotechnology, 2016, 34(7): 535-547. |
| [123] | LLOYD J P B, LY F, GONG P, et al. Synthetic memory circuits for stable cell reprogramming in plants[J]. Nature Biotechnology, 2022, 40(12): 1862-1872. |
| [124] | KHAN M A, HERRING G, ZHU J Y, et al. CRISPRi-based circuits to control gene expression in plants[J]. Nature Biotechnology, 2025, 43(3): 416-430. |
| [125] | WEINBERG B H, HANG PHAM N T, CARABALLO L D, et al. Large-scale design of robust genetic circuits with multiple inputs and outputs for mammalian cells[J]. Nature Biotechnology, 2017, 35(5): 453-462. |
| [126] | GUIZIOU S, MARANAS C J, CHU J C, et al. An integrase toolbox to record gene-expression during plant development[J]. Nature Communications, 2023, 14: 1844. |
| [127] | LI S S, LI Z L, TAN G Y, et al. In vitro allosteric transcription factor-based biosensing[J]. Trends in Biotechnology, 2023, 41(8): 1080-1095. |
| [128] | FERREIRA S S, ANTUNES M S. Genetically encoded Boolean logic operators to sense and integrate phenylpropanoid metabolite levels in plants[J]. New Phytologist, 2024, 243(2): 674-687. |
| [129] | LIANG Y, RICHARDSON S, YAN J W, et al. Endoribonuclease-based two-component repressor systems for tight gene expression control in plants[J]. ACS Synthetic Biology, 2017, 6(5): 806-816. |
| [1] | LUCIDO A, BASALLO O, MARIN-SANGUINO A, et al. Multiscale mathematical modeling in systems biology: a framework to boost plant synthetic biology[J]. Plants, 2025, 14(3): 470. |
| [2] | KE J, WANG B, YOSHIKUNI Y. Microbiome engineering: synthetic biology of plant-associated microbiomes in sustainable agriculture[J]. Trends in Biotechnology, 2021, 39(3): 244-261. |
| [3] | 张博, 马永硕, 尚轶, 等. 植物合成生物学研究进展[J]. 合成生物学, 2020, 1(2): 121-140. |
| ZHANG B, MA Y S, SHANG Y, et al. Recent advances in plant synthetic biology[J]. Synthetic Biology Journal, 2020, 1(2): 121-140. | |
| [4] | BROPHY J A N, MAGALLON K J, DUAN L N, et al. Synthetic genetic circuits as a means of reprogramming plant roots[J]. Science, 2022, 377(6607): 747-751. |
| [5] | ZHANG X N, LIU C L, DAI J B, et al. Enabling technology and core theory of synthetic biology[J]. Science China Life Sciences, 2023, 66(8): 1742-1785. |
| [6] | DOUDNA J A, CHARPENTIER E. The new frontier of genome engineering with CRISPR-Cas9[J]. Science, 2014, 346(6213): 1258096. |
| [7] | TIAN Y S, WANG B, PENG R H, et al. Enhancing carotenoid biosynthesis in rice endosperm by metabolic engineering[J]. Plant Biotechnology Journal, 2019, 17(5): 849-851. |
| [8] | XIONG D L. Perspectives of improving rice photosynthesis for higher grain yield[J]. Crop and Environment, 2024, 3(3): 123-137. |
| [9] | 燕永亮, 田长富, 杨建国, 等. 人工高效生物固氮体系创建及其农业应用[J]. 生命科学, 2021, 33(12): 1532-1543. |
| YAN Y L, TIAN C F, YANG J G, et al. Establishment of artificial efficiency biological nitrogen fixation system and its agricultural application[J]. Chinese Bulletin of Life Sciences, 2021, 33(12): 1532-1543. | |
| [10] | 潘明慧, 杨雪, 杜国忠, 等. 合成生物学助力链霉菌天然产物农药创制与产业化[J]. 植物保护, 2023, 49(5): 371-389. |
| [130] | KASENIIT K E, KATZ N, KOLBER N S, et al. Modular, programmable RNA sensing using ADAR editing in living cells[J]. Nature Biotechnology, 2023, 41(4): 482-487. |
| [131] | KONERMANN S, LOTFY P, BRIDEAU N J, et al. Transcriptome engineering with RNA-targeting type Ⅵ-D CRISPR effectors[J]. Cell, 2018, 173(3): 665-676.e14. |
| [132] | MARAND A P, CHEN Z L, GALLAVOTTI A, et al. A cis-regulatory atlas in maize at single-cell resolution[J]. Cell, 2021, 184(11): 3041-3055.e21. |
| [133] | HE Z H, LUO Y T, ZHOU X K, et al. scPlantDB: a comprehensive database for exploring cell types and markers of plant cell atlases[J]. Nucleic Acids Research, 2024, 52(D1): D1629-D1638. |
| [134] | RAI K, WANG Y D, O’CONNELL R W, et al. Using machine learning to enhance and accelerate synthetic biology[J]. Current Opinion in Biomedical Engineering, 2024, 31: 100553. |
| [135] | LAM H Y I, ONG X E, MUTWIL M. Large language models in plant biology[J]. Trends in Plant Science, 2024, 29(10): 1145-1155. |
| [136] | ZHU W C, HAN R, SHANG X Y, et al. The CropGPT project: Call for a global, coordinated effort in precision design breeding driven by AI using biological big data[J]. Molecular Plant, 2024, 17(2): 215-218. |
| [137] | JI Y R, ZHOU Z H, LIU H, et al. DNABERT: pre-trained Bidirectional Encoder Representations from Transformers model for DNA-language in genome[J]. Bioinformatics, 2021, 37(15): 2112-2120. |
| [138] | ABRAMSON J, ADLER J, DUNGER J, et al. Addendum: accurate structure prediction of biomolecular interactions with AlphaFold 3[J]. Nature, 2024, 636(8042): E4. |
| [139] | CUI H T, WANG C, MAAN H, et al. scGPT: toward building a foundation model for single-cell multi-omics using generative AI[J]. Nature Methods, 2024, 21(8): 1470-1480. |
| [140] | WANG J, ZHANG L, WANG S D, et al. AlphaFold-guided bespoke gene editing enhances field-grown soybean oil contents[J]. Advanced Science, 2025, 12(23): 2500290. |
| [141] | 宋成治, 林一瀚. AI+定向进化赋能蛋白改造及优化[J]. 合成生物学, 2025, 6(3): 617-635. |
| [10] | PAN M H, YANG X, DU G Z, et al. The exploitation and bio-manufacture of natural product pesticides from Streptomyces by synthetic biology[J]. Plant Protection, 2023, 49(5): 371-389. |
| [11] | GARDNER T S, CANTOR C R, COLLINS J J. Construction of a genetic toggle switch in Escherichia coli [J]. Nature, 2000, 403(6767): 339-342. |
| [12] | YE X, AL-BABILI S, KLÖTI A, et al. Engineering the provitamin A (beta-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm[J]. Science, 2000, 287(5451): 303-305. |
| [13] | BHULLAR S, CHAKRAVARTHY S, ADVANI S, et al. Strategies for development of functionally equivalent promoters with minimum sequence homology for transgene expression in plants: cis-elements in a novel DNA context versus domain swapping[J]. Plant Physiology, 2003, 132(2): 988-998. |
| [14] | ENGLER C, KANDZIA R, MARILLONNET S. A one pot, one step, precision cloning method with high throughput capability[J]. PLoS One, 2008, 3(11): e3647. |
| [15] | ENGLER C, GRUETZNER R, KANDZIA R, et al. Golden Gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes[J]. PLoS One, 2009, 4(5): e5553. |
| [16] | AMOS M. Population-based microbial computing: a third wave of synthetic biology?[J]. International Journal of General Systems, 2014, 43(7): 770-782. |
| [17] | FISHER A K, FREEDMAN B G, BEVAN D R, et al. A review of metabolic and enzymatic engineering strategies for designing and optimizing performance of microbial cell factories[J]. Computational and Structural Biotechnology Journal, 2014, 11(18): 91-99. |
| [18] | LU X F, VORA H, KHOSLA C. Overproduction of free fatty acids in E. coli: implications for biodiesel production[J]. Metabolic Engineering, 2008, 10(6): 333-339. |
| [19] | CONG L, RAN F A, COX D, et al. Multiplex genome engineering using CRISPR/Cas systems[J]. Science, 2013, 339(6121): 819-823. |
| [20] | SHAN Q W, WANG Y P, LI J, et al. Targeted genome modification of crop plants using a CRISPR-Cas system[J]. Nature Biotechnology, 2013, 31(8): 686-688. |
| [21] | CHAN A N, WANG L L, ZHU Y J, et al. Identification through fine mapping and verification using CRISPR/Cas9-targeted mutagenesis for a minor QTL controlling grain weight in rice[J]. Theoretical and Applied Genetics, 2021, 134(1): 327-337. |
| [141] | SONG C Z, LIN Y H. AI-enabled directed evolution for protein engineering and optimization[J]. Synthetic Biology Journal, 2025, 6(3): 617-635. |
| [142] | SHARMA A, JAIN A, GUPTA P, et al. Machine learning applications for precision agriculture: a comprehensive review[J]. IEEE Access, 2021, 9: 4843-4873. |
| [143] | LIAKOS K G, BUSATO P, MOSHOU D, et al. Machine learning in agriculture: a review[J]. Sensors, 2018, 18(8): 2674. |
| [144] | ESLAMI M, ADLER A, CACERES R S, et al. Artificial intelligence for synthetic biology[J]. Communications of the ACM, 2022, 65(5): 88-97. |
| [145] | YANG J S, REYNA-LLORENS I. Plant synthetic biology: exploring the frontiers of sustainable agriculture and fundamental plant biology[J]. Journal of Experimental Botany, 2023, 74(13): 3787-3790. |
| [146] | ORR D J, ALCÂNTARA A, KAPRALOV M V, et al. Surveying Rubisco diversity and temperature response to improve crop photosynthetic efficiency[J]. Plant Physiology, 2016, 172(2): 707-717. |
| [147] | LIN M T, OCCHIALINI A, ANDRALOJC P J, et al. A faster Rubisco with potential to increase photosynthesis in crops[J]. Nature, 2014, 513(7519): 547-550. |
| [148] | MANNING T, BIRCH R, STEVENSON T, et al. Bacterial Form Ⅱ Rubisco can support wild-type growth and productivity in Solanum tuberosum cv. Desiree (potato) under elevated CO2 [J]. PNAS Nexus, 2023, 2(2): pgac305. |
| [149] | GUNN L H, MARTIN AVILA E, BIRCH R, et al. The dependency of red Rubisco on its cognate activase for enhancing plant photosynthesis and growth[J]. Proceedings of the National Academy of Sciences of the United States of America, 2020, 117(41): 25890-25896. |
| [150] | MATSUMURA H, SHIOMI K, YAMAMOTO A, et al. Hybrid rubisco with complete replacement of rice Rubisco small subunits by sorghum counterparts confers C4 plant-like high catalytic activity[J]. Molecular Plant, 2020, 13(11): 1570-1581. |
| [151] | MAO Y W, CATHERALL E, DÍAZ-RAMOS A, et al. The small subunit of Rubisco and its potential as an engineering target[J]. Journal of Experimental Botany, 2023, 74(2): 543-561. |
| [152] | ADLER L, DÍAZ-RAMOS A, MAO Y W, et al. New horizons for building pyrenoid-based CO2-concentrating mechanisms in plants to improve yields[J]. Plant Physiology, 2022, 190(3): 1609-1627. |
| [22] | JARVIS D E, HO Y S, LIGHTFOOT D J, et al. The genome of Chenopodium quinoa [J]. Nature, 2017, 542(7641): 307-312. |
| [23] | ZIMIN A V, PUIU D, HALL R, et al. The first near-complete assembly of the hexaploid bread wheat genome, Triticum aestivum [J]. GigaScience, 2017, 6(11): 1-7. |
| [24] | YU W C, YAU Y Y, BIRCHLER J A. Plant artificial chromosome technology and its potential application in genetic engineering[J]. Plant Biotechnology Journal, 2016, 14(5): 1175-1182. |
| [25] | XU C H, BIRCHLER J A. Editorial: Plant artificial chromosomes: progress and perspectives[J]. Frontiers in Plant Science, 2023, 14: 1290386. |
| [26] | RAVIKUMAR S, BAYLON M G, PARK S J, et al. Engineered microbial biosensors based on bacterial two-component systems as synthetic biotechnology platforms in bioremediation and biorefinery[J]. Microbial Cell Factories, 2017, 16(1): 62. |
| [27] | ARACIC S, MANNA S, PETROVSKI S, et al. Innovative biological approaches for monitoring and improving water quality[J]. Frontiers in Microbiology, 2015, 6: 826. |
| [28] | NEETHIRAJAN S, RAGAVAN V, WENG X, et al. Biosensors for sustainable food engineering: challenges and perspectives[J]. Biosensors, 2018, 8(1): 23. |
| [29] | 刘华梅, 许国建. 微生物农药苏云金杆菌G033A[J]. 农药科学与管理, 2018, 39(4): 59-60. |
| LIU H M, XU G J. Microbial pesticide Bacillus thuringiensis G033A[J]. Pesticide Science and Administration, 2018, 39(4): 59-60. | |
| [30] | 王晓梅, 李辉尚, 杨小薇. 全球农业合成生物学发展现状及对中国的启示[J]. 农业展望, 2023, 19(4): 71-76. |
| WANG X M, LI H S, YANG X W. Development status of global agricultural synthetic biology and its enlightenment to China[J]. Agricultural Outlook, 2023, 19(4): 71-76. | |
| [31] | WALTZ E. GABA-enriched tomato is first CRISPR-edited food to enter market[J]. Nature Biotechnology, 2022, 40(1): 9-11. |
| [153] | GONG H Y, LI Y, FANG G, et al. Transgenic rice expressing ictb and FBP/sbpase derived from cyanobacteria exhibits enhanced photosynthesis and mesophyll conductance to CO2 [J]. PLoS One, 2015, 10(10): e0140928. |
| [154] | LONG B M, HEE W Y, SHARWOOD R E, et al. Carboxysome encapsulation of the CO2-fixing enzyme Rubisco in tobacco chloroplasts[J]. Nature Communications, 2018, 9: 3570. |
| [155] | CHEN T Y, HOJKA M, DAVEY P, et al. Engineering α-carboxysomes into plant chloroplasts to support autotrophic photosynthesis[J]. Nature Communications, 2023, 14: 2118. |
| [156] | FURBANK R T. Evolution of the C4 photosynthetic mechanism: are there really three C4 acid decarboxylation types?[J]. Journal of Experimental Botany, 2011, 62(9): 3103-3108. |
| [157] | FURBANK R, KELLY S, VON CAEMMERER S. Photosynthesis and food security: the evolving story of C4 rice[J]. Photosynthesis Research, 2023, 158(2): 121-130. |
| [158] | ERMAKOVA M, DANILA F R, FURBANK R T, et al. On the road to C4 rice: advances and perspectives[J]. The Plant Journal, 2020, 101(4): 940-950. |
| [159] | ERMAKOVA M, ARRIVAULT S, GIULIANI R, et al. Installation of C4 photosynthetic pathway enzymes in rice using a single construct[J]. Plant Biotechnology Journal, 2021, 19(3): 575-588. |
| [160] | SMITH E N, VAN AALST M, TOSENS T, et al. Improving photosynthetic efficiency toward food security: strategies, advances, and perspectives[J]. Molecular Plant, 2023, 16(10): 1547-1563. |
| [161] | LIN X L, LONG Y M, YAO Z, et al. Synthetic photorespiratory bypass more stably increases potato yield per plant by improving photosynthesis[J]. Plant Biotechnology Journal, 2025, 23(7): 2526-2536. |
| [162] | MO B Q, CHEN X F, YANG J J, et al. Engineering of photorespiration-dependent glycine betaine biosynthesis improves photosynthetic carbon fixation and panicle architecture in rice[J]. Journal of Integrative Plant Biology, 2025, 67(4): 979-992. |
| [163] | WALTZ E. A new crop of microbe startups raises big bucks, takes on the establishment [J]. Nature Biotechnology, 2017, 35(12): 1120-1122. |
| [164] | ALLEN R S, TILBROOK K, WARDEN A C, et al. Expression of 16 nitrogenase proteins within the plant mitochondrial matrix[J]. Frontiers in Plant Science, 2017, 8: 287. |
| [32] | PAINE J A, SHIPTON C A, CHAGGAR S, et al. Improving the nutritional value of Golden Rice through increased pro-vitamin A content[J]. Nature Biotechnology, 2005, 23(4): 482-487. |
| [33] | ANTUNES M S, MOREY K J, SMITH J J, et al. Programmable ligand detection system in plants through a synthetic signal transduction pathway[J]. PLoS One, 2011, 6(1): e16292. |
| [34] | BULLEY S, WRIGHT M, ROMMENS C, et al. Enhancing ascorbate in fruits and tubers through over-expression of the L-galactose pathway gene GDP-L-galactose phosphorylase[J]. Plant Biotechnology Journal, 2012, 10(4): 390-397. |
| [35] | ROGERS C, OLDROYD G E D. Synthetic biology approaches to engineering the nitrogen symbiosis in cereals[J]. Journal of Experimental Botany, 2014, 65(8): 1939-1946. |
| [36] | RUIZ-LOPEZ N, HASLAM R P, NAPIER J A, et al. Successful high-level accumulation of fish oil omega-3 long-chain polyunsaturated fatty acids in a transgenic oilseed crop[J]. The Plant Journal, 2014, 77(2): 198-208. |
| [37] | WANG B, KASHKOOLI A B, SALLETS A, et al. Transient production of artemisinin in Nicotiana benthamiana is boosted by a specific lipid transfer protein from A. annua [J]. Metabolic Engineering, 2016, 38: 159-169. |
| [38] | NONAKA S, ARAI C, TAKAYAMA M, et al. Efficient increase of γ-aminobutyric acid (GABA) content in tomato fruits by targeted mutagenesis[J]. Scientific Reports, 2017, 7: 7057. |
| [39] | PERCHLIK M, TEGEDER M. Improving plant nitrogen use efficiency through alteration of amino acid transport processes[J]. Plant Physiology, 2017, 175(1): 235-247. |
| [40] | ZHU Q L, ZENG D C, YU S Z, et al. From golden rice to aSTARice: bioengineering astaxanthin biosynthesis in rice endosperm[J]. Molecular Plant, 2018, 11(12): 1440-1448. |
| [41] | SOUTH P F, CAVANAGH A P, LIU H W, et al. Synthetic glycolate metabolism pathways stimulate crop growth and productivity in the field[J]. Science, 2019, 363(6422): eaat9077. |
| [42] | SHEN B R, WANG L M, LIN X L, et al. Engineering a new chloroplastic photorespiratory bypass to increase photosynthetic efficiency and productivity in rice[J]. Molecular Plant, 2019, 12(2): 199-214. |
| [43] | LIN Q P, ZONG Y, XUE C X, et al. Prime genome editing in rice and wheat[J]. Nature Biotechnology, 2020, 38(5): 582-585. |
| [165] | BIRCHLER J A, SWYERS N C. Engineered minichromosomes in plants [J]. Experimental Cell Research, 2020, 388(2): 111852. |
| [166] | OLDROYD G E, DIXON R. Biotechnological solutions to the nitrogen problem[J]. Current Opinion in Biotechnology, 2014, 26: 19-24. |
| [167] | BAGESHWAR U K, SRIVASTAVA M, PARDHA-SARADHI P, et al. An environmentally friendly engineered Azotobacter strain that replaces a substantial amount of urea fertilizer while sustaining the same wheat yield[J]. Applied and Environmental Microbiology, 2017, 83(15): e00590-17. |
| [168] | GEDDES B A, PARAMASIVAN P, JOFFRIN A, et al. Engineering transkingdom signalling in plants to control gene expression in rhizosphere bacteria[J]. Nature Communications, 2019, 10: 3430. |
| [169] | BLOCH S E, CLARK R, GOTTLIEB S S, et al. Biological nitrogen fixation in maize: optimizing nitrogenase expression in a root-associated diazotroph[J]. Journal of Experimental Botany, 2020, 71(15): 4591-4603. |
| [170] | CHRISTIAENS O, TARDAJOS M G, MARTINEZ REYNA Z L, et al. Increased RNAi efficacy in Spodoptera exigua via the formulation of dsRNA with guanylated polymers[J]. Frontiers in Physiology, 2018, 9: 316. |
| [171] | HUAN Y C, KONG Q, MOU H J, et al. Antimicrobial peptides: classification, design, application and research progress in multiple fields[J]. Frontiers in Microbiology, 2020, 11: 582779. |
| [172] | MARCOS J F, MUÑOZ A, PÉREZ-PAYÁ E, et al. Identification and rational design of novel antimicrobial peptides for plant protection[J]. Annual Review of Phytopathology, 2008, 46: 273-301. |
| [173] | MONTESINOS E. Functional peptides for plant disease control[J]. Annual Review of Phytopathology, 2023, 61: 301-324. |
| [174] | ZEITLER B, BERNHARD A, MEYER H, et al. Production of a de-novo designed antimicrobial peptide in Nicotiana benthamiana [J]. Plant Molecular Biology, 2013, 81(3): 259-272. |
| [175] | HOLÁSKOVÁ E, GALUSZKA P, MIČÚCHOVÁ A, et al. Molecular farming in barley: development of a novel production platform to produce human antimicrobial peptide LL-37[J]. Biotechnology Journal, 2018, 13(6): e1700628. |
| [176] | MIRZAEE M, HOLÁSKOVÁ E, MIČÚCHOVÁ A, et al. Long-lasting stable expression of human LL-37 antimicrobial peptide in transgenic barley plants[J]. Antibiotics, 2021, 10(8): 898. |
| [44] | ROELL M S, SCHADA VON BORZYKOWSKI L, WESTHOFF P, et al. A synthetic C4 shuttle via the β-hydroxyaspartate cycle in C3 plants[J]. Proceedings of the National Academy of Sciences of the United States of America, 2021, 118(21): e2022307118. |
| [45] | HONG B K, GRZECH D, CAPUTI L, et al. Biosynthesis of strychnine[J]. Nature, 2022, 607(7919): 617-622. |
| [46] | MARTIN L B B, KIKUCHI S, REJZEK M, et al. Complete biosynthesis of the potent vaccine adjuvant QS-21[J]. Nature Chemical Biology, 2024, 20(4): 493-502. |
| [47] | WU H X, YANG J H, SHEN P J, et al. High-level production of indole-3-acetic acid in the metabolically engineered Escherichia coli [J]. Journal of Agricultural and Food Chemistry, 2021, 69(6): 1916-1924. |
| [48] | ZHANG Y, CHIU T Y, ZHANG J T, et al. Systematical engineering of synthetic yeast for enhanced production of lycopene[J]. Bioengineering, 2021, 8(1): 14. |
| [49] | ZHANG Y, YUAN M D, WU X X, et al. The construction and optimization of engineered yeast chassis for efficient biosynthesis of 8-hydroxygeraniol[J]. mLife, 2023, 2(4): 438-449. |
| [50] | ZHU Y, LI J X, PENG L Y, et al. High-yield production of protopanaxadiol from sugarcane molasses by metabolically engineered Saccharomyces cerevisiae [J]. Microbial Cell Factories, 2022, 21(1): 230. |
| [51] | TAN G Y, DENG K H, LIU X H, et al. Heterologous biosynthesis of spinosad: an omics-guided large polyketide synthase gene cluster reconstitution in Streptomyces [J]. ACS Synthetic Biology, 2017, 6(6): 995-1005. |
| [52] | WANG Q, BAO T, HU M K, et al. Efficient acetoin production in Bacillus subtilis by multivariate modular metabolic engineering with spatiotemporal modulation[J]. ACS Sustainable Chemistry & Engineering, 2025, 13(5): 1927-1936. |
| [53] | SABATE R, DE GROOT N S, VENTURA S. Protein folding and aggregation in bacteria[J]. Cellular and Molecular Life Sciences, 2010, 67(16): 2695-2715. |
| [54] | TIAN Y, KONG L Z, LI Q, et al. Structural diversity, evolutionary origin, and metabolic engineering of plant specialized benzylisoquinoline alkaloids[J]. Natural Product Reports, 2024, 41(11): 1787-1810. |
| [55] | SUN Q Y, DING L W, LOMONOSSOFF G P, et al. Improved expression and purification of recombinant human serum albumin from transgenic tobacco suspension culture[J]. Journal of Biotechnology, 2011, 155(2): 164-172. |
| [177] | BUNDÓ M, SHI X Q, VERNET M, et al. Rice seeds as biofactories of rationally designed and cell-penetrating antifungal PAF peptides[J]. Frontiers in Plant Science, 2019, 10: 731. |
| [178] | KOCH A, KUMAR N, WEBER L, et al. Host-induced gene silencing of cytochrome P450 lanosterol C14α-demethylase-encoding genes confers strong resistance to Fusarium species[J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(48): 19324-19329. |
| [179] | SÚNICO V, HIGUERA J J, MOLINA-HIDALGO F J, et al. The intragenesis and synthetic biology approach towards accelerating genetic gains on strawberry: development of new tools to improve fruit quality and resistance to pathogens[J]. Plants, 2022, 11(1): 57. |
| [180] | MOORE B D, ANDREW R L, KÜLHEIM C, et al. Explaining intraspecific diversity in plant secondary metabolites in an ecological context[J]. New Phytologist, 2014, 201(3): 733-750. |
| [181] | AHUJA I, KISSEN R, BONES A M. Phytoalexins in defense against pathogens[J]. Trends in Plant Science, 2012, 17(2): 73-90. |
| [182] | HU H, LI J J, DELATTE T, et al. Modification of chrysanthemum odour and taste with chrysanthemol synthase induces strong dual resistance against cotton aphids[J]. Plant Biotechnology Journal, 2018, 16(8): 1434-1445. |
| [183] | MITTLER R, BLUMWALD E. Genetic engineering for modern agriculture: challenges and perspectives[J]. Annual Review of Plant Biology, 2010, 61: 443-462. |
| [184] | SHI H T, YE T T, CHAN Z L. Comparative proteomic responses of two bermudagrass (Cynodon dactylon (L). Pers.) varieties contrasting in drought stress resistance[J]. Plant Physiology and Biochemistry, 2014, 82: 218-228. |
| [185] | HU H H, XIONG L Z. Genetic engineering and breeding of drought-resistant crops[J]. Annual Review of Plant Biology, 2014, 65: 715-741. |
| [186] | CHEN S J, XU K, KONG D Y, et al. Ubiquitin ligase OsRINGzf1 regulates drought resistance by controlling the turnover of OsPIP2;1[J]. Plant Biotechnology Journal, 2022, 20(9): 1743-1755. |
| [187] | DING L, MILHIET T, PARENT B, et al. The plasma membrane aquaporin ZmPIP2;5 enhances the sensitivity of stomatal closure to water deficit[J]. Plant, Cell & Environment, 2022, 45(4): 1146-1156. |
| [188] | XU Y, HU W, LIU J H, et al. An aquaporin gene MaPIP2-7 is involved in tolerance to drought, cold and salt stresses in transgenic banana (Musa acuminata L.)[J]. Plant Physiology and Biochemistry, 2020, 147: 66-76. |
| [56] | VASILEV N, SCHMITZ C, DONG L M, et al. Comparison of plant-based expression platforms for the heterologous production of geraniol[J]. Plant Cell, Tissue and Organ Culture (PCTOC), 2014, 117(3): 373-380. |
| [57] | YOUSEFIAN S, LOHRASEBI T, FARHADPOUR M, et al. Effect of methyl jasmonate on phenolic acids accumulation and the expression profile of their biosynthesis-related genes in Mentha spicata hairy root cultures[J]. Plant Cell, Tissue and Organ Culture (PCTOC), 2020, 142(2): 285-297. |
| [58] | WU S J, XIE X G, FENG K M, et al. Transcriptome sequencing and signal transduction for the enhanced tanshinone production in Salvia miltiorrhiza hairy roots induced by Trichoderma atroviride D16 polysaccharide fraction[J]. Bioscience, Biotechnology, and Biochemistry, 2022, 86(8): 1049-1059. |
| [59] | KUŹMA Ł, KISIEL W, KRÓLICKA A, et al. Genetic transformation of Salvia austriaca by Agrobacterium rhizogenes and diterpenoid isolation[J]. Die Pharmazie, 2011, 66(11): 904-907. |
| [60] | LIU J, ZHAO Y X, ZHANG J M, et al. Production of species-specific anthocyanins through an inducible system in plant hairy roots[J]. Metabolic Engineering, 2024, 81: 182-196. |
| [61] | NOGUEIRA M, ENFISSI E M A, WELSCH R, et al. Construction of a fusion enzyme for astaxanthin formation and its characterisation in microbial and plant hosts: a new tool for engineering ketocarotenoids[J]. Metabolic Engineering, 2019, 52: 243-252. |
| [62] | BEYRAGHDAR KASHKOOLI A, VAN DER KROL A R, RABE P, et al. Substrate promiscuity of enzymes from the sesquiterpene biosynthetic pathways from Artemisia annua and Tanacetum parthenium allows for novel combinatorial sesquiterpene production[J]. Metabolic Engineering, 2019, 54: 12-23. |
| [63] | FORESTIER E C F, CZECHOWSKI T, CORDING A C, et al. Developing a Nicotiana benthamiana transgenic platform for high-value diterpene production and candidate gene evaluation[J]. Plant Biotechnology Journal, 2021, 19(8): 1614-1623. |
| [64] | ZHU Q L, YU S Z, ZENG D C, et al. Development of “purple endosperm rice” by engineering anthocyanin biosynthesis in the endosperm with a high-efficiency transgene stacking system[J]. Molecular Plant, 2017, 10(7): 918-929. |
| [65] | LIU X Q, MA X H, WANG H, et al. Metabolic engineering of astaxanthin-rich maize and its use in the production of biofortified eggs[J]. Plant Biotechnology Journal, 2021, 19(9): 1812-1823. |
| [66] | LAM E, MICHAEL T P. Wolffia, a minimalist plant and synthetic biology chassis[J]. Trends in Plant Science, 2022, 27(5): 430-439. |
| [67] | BI G Q, ZHAO S J, YAO J W, et al. Near telomere-to-telomere genome of the model plant Physcomitrium patens [J]. Nature Plants, 2024, 10(2): 327-343. |
| [189] | SAJA-GARBARZ D, LIBIK-KONIECZNY M, FELLNER M, et al. Silicon-induced alterations in the expression of aquaporins and antioxidant system activity in well-watered and drought-stressed oilseed rape[J]. Plant Physiology and Biochemistry, 2022, 174: 73-86. |
| [190] | HOSSEINIFARD M, STEFANIAK S, GHORBANI JAVID M, et al. Contribution of exogenous proline to abiotic stresses tolerance in plants: a review[J]. International Journal of Molecular Sciences, 2022, 23(9): 5186. |
| [191] | ARAÚJO W L, NUNES-NESI A, OSORIO S, et al. Antisense inhibition of the iron-sulphur subunit of succinate dehydrogenase enhances photosynthesis and growth in tomato via an organic acid-mediated effect on stomatal aperture[J]. The Plant Cell, 2011, 23(2): 600-627. |
| [192] | MOVAHEDI A, DZINYELA R, AGHAEI-DARGIRI S, et al. Advanced study of drought-responsive protein pathways in plants[J]. Agronomy, 2023, 13(3): 849. |
| [193] | EDWARDS R A, NG X Y, TUCKER M R, et al. Plant synthetic biology as a tool to help eliminate hidden hunger[J]. Current Opinion in Biotechnology, 2024, 88: 103168. |
| [194] | BEYENE G, SOLOMON F R, CHAUHAN R D, et al. Provitamin A biofortification of cassava enhances shelf life but reduces dry matter content of storage roots due to altered carbon partitioning into starch[J]. Plant Biotechnology Journal, 2018, 16(6): 1186-1200. |
| [195] | LOW J W, MWANGA R O M, ANDRADE M, et al. Tackling vitamin A deficiency with biofortified sweetpotato in sub-Saharan Africa[J]. Global Food Security, 2017, 14: 23-30. |
| [196] | LI K T, MOULIN M, MANGEL N, et al. Increased bioavailable vitamin B6 in field-grown transgenic cassava for dietary sufficiency[J]. Nature Biotechnology, 2015, 33(10): 1029-1032. |
| [197] | DE LEPELEIRE J, STROBBE S, VERSTRAETE J, et al. Folate biofortification of potato by Tuber-specific expression of four folate biosynthesis genes[J]. Molecular Plant, 2018, 11(1): 175-188. |
| [198] | BULLEY S, LAING W. The regulation of ascorbate biosynthesis[J]. Current Opinion in Plant Biology, 2016, 33: 15-22. |
| [199] | WANG L Y, MENG X, YANG D Y, et al. Overexpression of tomato GDP-L-galactose phosphorylase gene in tobacco improves tolerance to chilling stress[J]. Plant Cell Reports, 2014, 33(9): 1441-1451. |
| [200] | ALI B, PANTHA S, ACHARYA R, et al. Enhanced ascorbate level improves multi-stress tolerance in a widely grown indica rice variety without compromising its agronomic characteristics[J]. Journal of Plant Physiology, 2019, 240: 152998. |
| [201] | MA L C, WANG Y R, LIU W X, et al. Overexpression of an alfalfa GDP-mannose 3,5-epimerase gene enhances acid, drought and salt tolerance in transgenic Arabidopsis by increasing ascorbate accumulation[J]. Biotechnology Letters, 2014, 36(11): 2331-2341. |
| [202] | LI X J, YE J, MUNIR S, et al. Biosynthetic gene pyramiding leads to ascorbate accumulation with enhanced oxidative stress tolerance in tomato[J]. International Journal of Molecular Sciences, 2019, 20(7): 1558. |
| [203] | ZHANG G Y, LIU R R, ZHANG C Q, et al. Manipulation of the rice L-galactose pathway: evaluation of the effects of transgene overexpression on ascorbate accumulation and abiotic stress tolerance[J]. PLoS One, 2015, 10(5): e0125870. |
| [204] | LI J, SCARANO A, GONZALEZ N M, et al. Biofortified tomatoes provide a new route to vitamin D sufficiency[J]. Nature Plants, 2022, 8(6): 611-616. |
| [205] | UPADHYAYA D C, BAGRI D S, UPADHYAYA C P, et al. Genetic engineering of potato (Solanum tuberosum L.) for enhanced α-tocopherols and abiotic stress tolerance[J]. Physiologia Plantarum, 2021, 173(1): 116-128. |
| [206] | BOONYAVES K, WU T Y, GRUISSEM W, et al. Enhanced grain iron levels in rice expressing an IRON-REGULATED METAL TRANSPORTER, NICOTIANAMINE SYNTHASE, and FERRITIN gene cassette[J]. Frontiers in Plant Science, 2017, 8: 130. |
| [207] | NARAYANAN N, BEYENE G, CHAUHAN R D, et al. Biofortification of field-grown cassava by engineering expression of an iron transporter and ferritin[J]. Nature Biotechnology, 2019, 37(2): 144-151. |
| [208] | HARRINGTON S A, CONNORTON J M, NYANGOMA N I M, et al. A two-gene strategy increases iron and zinc concentrations in wheat flour, improving mineral bioaccessibility[J]. Plant Physiology, 2023, 191(1): 528-541. |
| [209] | NAGESH C R, PRASHAT G R, GOSWAMI S, et al. Sulfate transport and metabolism: strategies to improve the seed protein quality[J]. Molecular Biology Reports, 2024, 51(1): 242. |
| [210] | KIM W S, SUN-HYUNG J, OEHRLE N W, et al. Overexpression of ATP sulfurylase improves the sulfur amino acid content, enhances the accumulation of Bowman-Birk protease inhibitor and suppresses the accumulation of the β-subunit of β-conglycinin in soybean seeds[J]. Scientific Reports, 2020, 10: 14989. |
| [211] | HUANG Y C, WANG H H, ZHU Y D, et al. THP9 enhances seed protein content and nitrogen-use efficiency in maize[J]. Nature, 2022, 612(7939): 292-300. |
| [212] | LEE S, PARK J, LEE J, et al. OsASN1 overexpression in rice increases grain protein content and yield under nitrogen-limiting conditions[J]. Plant & Cell Physiology, 2020, 61(7): 1309-1320. |
| [213] | LIU X Q, LI S Z, YANG W Z, et al. Synthesis of seed-specific bidirectional promoters for metabolic engineering of anthocyanin-rich maize[J]. Plant & Cell Physiology, 2018, 59(10): 1942-1955. |
| [214] | GONZALI S, PERATA P. Anthocyanins from purple tomatoes as novel antioxidants to promote human health[J]. Antioxidants, 2020, 9(10): 1017. |
| [215] | BEYER P, AL-BABILI S, YE X D, et al. Golden Rice: introducing the beta-carotene biosynthesis pathway into rice endosperm by genetic engineering to defeat vitamin A deficiency[J]. The Journal of Nutrition, 2002, 132(3): 506S-510S. |
| [216] | HAN L H, SILVESTRE S, SAYANOVA O, et al. Using field evaluation and systematic iteration to rationalize the accumulation of omega-3 long-chain polyunsaturated fatty acids in transgenic Camelina sativa [J]. Plant Biotechnology Journal, 2022, 20(9): 1833-1852. |
| [217] | RYU M H, ZHANG J, TOTH T, et al. Control of nitrogen fixation in bacteria that associate with cereals[J]. Nature Microbiology, 2020, 5(2): 314-330. |
| [218] | SHULSE C N, CHOVATIA M, AGOSTO C, et al. Engineered root bacteria release plant-available phosphate from phytate[J]. Applied and Environmental Microbiology, 2019, 85(18): e01210-19. |
| [219] | SHAO J H, LI S Q, ZHANG N, et al. Analysis and cloning of the synthetic pathway of the phytohormone indole-3-acetic acid in the plant-beneficial Bacillus amyloliquefaciens SQR9[J]. Microbial Cell Factories, 2015, 14: 130. |
| [220] | ZÚÑIGA A, DE LA FUENTE F, FEDERICI F, et al. An engineered device for indoleacetic acid production under quorum sensing signals enables Cupriavidus pinatubonensis JMP134 to stimulate plant growth[J]. ACS Synthetic Biology, 2018, 7(6): 1519-1527. |
| [221] | TRDÁ L, BAREŠOVÁ M, ŠAŠEK V, et al. Cytokinin metabolism of pathogenic fungus Leptosphaeria maculans involves isopentenyltransferase, adenosine kinase and cytokinin oxidase/dehydrogenase[J]. Frontiers in Microbiology, 2017, 8: 1374. |
| [222] | SALOMON M V, BOTTINI R, DE SOUZA FILHO G A, et al. Bacteria isolated from roots and rhizosphere of Vitis vinifera retard water losses, induce abscisic acid accumulation and synthesis of defense-related terpenes in in vitro cultured grapevine[J]. Physiologia Plantarum, 2014, 151(4): 359-374. |
| [223] | MULLINS A J, MURRAY J A H, BULL M J, et al. Genome mining identifies cepacin as a plant-protective metabolite of the biopesticidal bacterium Burkholderia ambifaria [J]. Nature Microbiology, 2019, 4(6): 996-1005. |
| [224] | LI Z L, HUANG P J, WANG M Y, et al. Stepwise increase of thaxtomins production in Streptomyces albidoflavus J1074 through combinatorial metabolic engineering[J]. Metabolic Engineering, 2021, 68: 187-198. |
| [225] | LIU Y P, ZHU A P, TAN H M, et al. Engineering banana endosphere microbiome to improve Fusarium wilt resistance in banana[J]. Microbiome, 2019, 7(1): 74. |
| [226] | SANATI NEZHAD A. Microfluidic platforms for plant cells studies[J]. Lab on a Chip, 2014, 14(17): 3262-3274. |
| [227] | MASSALHA H, KORENBLUM E, MALITSKY S, et al. Live imaging of root-bacteria interactions in a microfluidics setup[J]. Proceedings of the National Academy of Sciences of the United States of America, 2017, 114(17): 4549-4554. |
| [228] | KEHE J, KULESA A, ORTIZ A, et al. Massively parallel screening of synthetic microbial communities[J]. Proceedings of the National Academy of Sciences of the United States of America, 2019, 116(26): 12804-12809. |
| [229] | ZENGLER K, HOFMOCKEL K, BALIGA N S, et al. EcoFABs: advancing microbiome science through standardized fabricated ecosystems[J]. Nature Methods, 2019, 16(7): 567-571. |
| [230] | LIU P, PANDA K, EDWARDS S A, et al. Transposase-assisted target-site integration for efficient plant genome engineering[J]. Nature, 2024, 631(8021): 593-600. |
| [231] | 邵洁, 刘海利, 王勇. 植物合成生物学的现在与未来[J]. 合成生物学, 2020, 1(4): 395-412. |
| SHAO J, LIU H L, WANG Y. Present and future of plant synthetic biology[J]. Synthetic Biology Journal, 2020, 1(4): 395-412. | |
| [232] | SANDHYA D, JOGAM P, ALLINI V R, et al. The present and potential future methods for delivering CRISPR/Cas9 components in plants[J]. Journal of Genetic Engineering and Biotechnology, 2020, 18(1): 25. |
| [233] | CHEN L, LIU G Q, ZHANG T. Integrating machine learning and genome editing for crop improvement[J]. aBIOTECH, 2024, 5(2): 262-277. |
| [234] | BAUER-PANSKUS A, MIYAZAKI J, KAWALL K, et al. Risk assessment of genetically engineered plants that can persist and propagate in the environment[J]. Environmental Sciences Europe, 2020, 32(1): 32. |
| [235] | LI J, ZHAO H M, ZHENG L X, et al. Advances in synthetic biology and biosafety governance[J]. Frontiers in Bioengineering and Biotechnology, 2021, 9: 598087. |
| [236] | 王盼娣, 熊小娟, 付萍, 等. 《生物安全法》实施背景下对合成生物学的监管[J]. 华中农业大学学报, 2021, 40(6): 231-245. |
| WANG P D, XIONG X J, FU P, et al. Regulation of synthetic biology under background of implementing Biosafety Law[J]. Journal of Huazhong Agricultural University, 2021, 40(6): 231-245. | |
| [237] | DE SOUZA A P, BURGESS S J, DORAN L, et al. Soybean photosynthesis and crop yield are improved by accelerating recovery from photoprotection[J]. Science, 2022, 377(6608): 851-854. |
| [238] | SPRONCKEN C C M, LIU P, MONNEY J, et al. Large-area, self-healing block copolymer membranes for energy conversion[J]. Nature, 2024, 630(8018): 866-871. |
| [239] | HE Y, NING T T, XIE T T, et al. Large-scale production of functional human serum albumin from transgenic rice seeds[J]. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(47): 19078-19083. |
| [68] | LI Y Z, LIANG J, DENG B F, et al. Applications and prospects of CRISPR/Cas9-mediated base editing in plant breeding[J]. Current Issues in Molecular Biology, 2023, 45(2): 918-935. |
| [69] | GAUDELLI N M, KOMOR A C, REES H A, et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage[J]. Nature, 2017, 551(7681): 464-471. |
| [70] | LI C, ZHANG R, MENG X B, et al. Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors[J]. Nature Biotechnology, 2020, 38(7): 875-882. |
| [71] | MIKAMI M, TOKI S, ENDO M. In planta processing of the SpCas9-gRNA complex[J]. Plant & Cell Physiology, 2017, 58(11): 1857-1867. |
| [72] | CAMPA C C, WEISBACH N R, SANTINHA A J, et al. Multiplexed genome engineering by Cas12a and CRISPR arrays encoded on single transcripts[J]. Nature Methods, 2019, 16(9): 887-893. |
| [73] | LI C, ZONG Y, JIN S, et al. SWISS: multiplexed orthogonal genome editing in plants with a Cas9 nickase and engineered CRISPR RNA scaffolds[J]. Genome Biology, 2020, 21(1): 141. |
| [74] | LIU H J, JIAN L M, XU J T, et al. High-throughput CRISPR/Cas9 mutagenesis streamlines trait gene identification in maize[J]. The Plant Cell, 2020, 32(5): 1397-1413. |
| [75] | BAI M Y, YUAN J H, KUANG H Q, et al. Generation of a multiplex mutagenesis population via pooled CRISPR-Cas9 in soya bean[J]. Plant Biotechnology Journal, 2020, 18(3): 721-731. |
| [76] | JACOBS T B, ZHANG N, PATEL D, et al. Generation of a collection of mutant tomato lines using pooled CRISPR libraries[J]. Plant Physiology, 2017, 174(4): 2023-2037. |
| [77] | KUANG Y J, LI S F, REN B, et al. Base-editing-mediated artificial evolution of OsALS1 in planta to develop novel herbicide-tolerant rice germplasms[J]. Molecular Plant, 2020, 13(4): 565-572. |
| [78] | HU N, TIAN H L, LI Y H, et al. pHNRhCas9NG, single expression cassette-based dual-component dual-transcription unit CRISPR/Cas9 system for plant genome editing[J]. Trends in Biotechnology, 2025, 43(7): 1788-1808. |
| [79] | TATSIS E C, O’CONNOR S E. New developments in engineering plant metabolic pathways[J]. Current Opinion in Biotechnology, 2016, 42: 126-132. |
| [80] | WU Q Y, HUANG Z Y, WANG J Y, et al. Construction of an Escherichia coli cell factory to synthesize taxadien-5α-ol, the key precursor of anti-cancer drug paclitaxel[J]. Bioresources and Bioprocessing, 2022, 9(1): 82. |
| [81] | IGNEA C, ATHANASAKOGLOU A, IOANNOU E, et al. Carnosic acid biosynthesis elucidated by a synthetic biology platform[J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(13): 3681-3686. |
| [82] | TAN H X, CHEN X H, LIANG N, et al. Transcriptome analysis reveals novel enzymes for apo-carotenoid biosynthesis in saffron and allows construction of a pathway for crocetin synthesis in yeast[J]. Journal of Experimental Botany, 2019, 70(18): 4819-4834. |
| [83] | ZHANG J, HANSEN L G, GUDICH O, et al. A microbial supply chain for production of the anti-cancer drug vinblastine[J]. Nature, 2022, 609(7926): 341-347. |
| [84] | LIU J Q, WANG X, DAI G Z, et al. Microbial chassis engineering drives heterologous production of complex secondary metabolites[J]. Biotechnology Advances, 2022, 59: 107966. |
| [85] | KWAN B D, SELIGMANN B, NGUYEN T D, et al. Leveraging synthetic biology and metabolic engineering to overcome obstacles in plant pathway elucidation[J]. Current Opinion in Plant Biology, 2023, 71: 102330. |
| [86] | BUTELLI E, TITTA L, GIORGIO M, et al. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors[J]. Nature Biotechnology, 2008, 26(11): 1301-1308. |
| [87] | MALHOTRA K, SUBRAMANIYAN M, RAWAT K, et al. Compartmentalized metabolic engineering for artemisinin biosynthesis and effective malaria treatment by oral delivery of plant cells[J]. Molecular Plant, 2016, 9(11): 1464-1477. |
| [88] | POLTURAK G, BREITEL D, GROSSMAN N, et al. Elucidation of the first committed step in betalain biosynthesis enables the heterologous engineering of betalain pigments in plants[J]. New Phytologist, 2016, 210(1): 269-283. |
| [89] | MAJER E, LLORENTE B, RODRÍGUEZ-CONCEPCIÓN M, et al. Rewiring carotenoid biosynthesis in plants using a viral vector[J]. Scientific Reports, 2017, 7: 41645. |
| [90] | FUCHS L K, HOLLAND A H, LUDLOW R A, et al. Genetic manipulation of biosynthetic pathways in mint[J]. Frontiers in Plant Science, 2022, 13: 928178. |
| [91] | D’ANDREA L, SIMON-MOYA M, LLORENTE B, et al. Interference with Clp protease impairs carotenoid accumulation during tomato fruit ripening[J]. Journal of Experimental Botany, 2018, 69(7): 1557-1568. |
| [92] | PETRIE J R, SHRESTHA P, BELIDE S, et al. Metabolic engineering Camelina sativa with fish oil-like levels of DHA[J]. PLoS One, 2014, 9(1): e85061. |
| [93] | KIM J Y, KIM J H, JANG Y H, et al. Transcriptome and metabolite profiling of tomato SGR-knockout null lines using the CRISPR/Cas9 system[J]. International Journal of Molecular Sciences, 2023, 24(1): 109. |
| [94] | TU M X, FANG J H, ZHAO R K, et al. CRISPR/Cas9-mediated mutagenesis of VvbZIP36 promotes anthocyanin accumulation in grapevine (Vitis vinifera)[J]. Horticulture Research, 2022, 9: uhac022. |
| [95] | WEN D, WU L, WANG M Y, et al. CRISPR/Cas9-mediated targeted mutagenesis of FtMYB45 promotes flavonoid biosynthesis in Tartary buckwheat (Fagopyrum tataricum)[J]. Frontiers in Plant Science, 2022, 13: 879390. |
| [96] | DEVIREDDY A R, ZANDALINAS S I, FICHMAN Y, et al. Integration of reactive oxygen species and hormone signaling during abiotic stress[J]. The Plant Journal, 2021, 105(2): 459-476. |
| [97] | MIYAWAKI A, LLOPIS J, HEIM R, et al. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin[J]. Nature, 1997, 388(6645): 882-887. |
| [98] | BRUNOUD G, WELLS D M, OLIVA M, et al. A novel sensor to map auxin response and distribution at high spatio-temporal resolution[J]. Nature, 2012, 482(7383): 103-106. |
| [99] | LIAO C Y, SMET W, BRUNOUD G, et al. Reporters for sensitive and quantitative measurement of auxin response[J]. Nature Methods, 2015, 12(3): 207-210. |
| [100] | JONES A M. A new look at stress: abscisic acid patterns and dynamics at high-resolution[J]. New Phytologist, 2016, 210(1): 38-44. |
| [101] | FERNANDEZ-MORENO J P, STEPANOVA A N. Monitoring ethylene in plants: genetically encoded reporters and biosensors[J]. Small Methods, 2020, 4(8): 1900260. |
| [102] | RIZZA A, JONES A M. The makings of a gradient: spatiotemporal distribution of gibberellins in plant development[J]. Current Opinion in Plant Biology, 2019, 47: 9-15. |
| [103] | ORTEGA-VILLASANTE C, BURÉN S, BARÓN-SOLA Á, et al. In vivo ROS and redox potential fluorescent detection in plants: present approaches and future perspectives[J]. Methods, 2016, 109: 92-104. |
| [104] | ZÜRCHER E, TAVOR-DESLEX D, LITUIEV D, et al. A robust and sensitive synthetic sensor to monitor the transcriptional output of the cytokinin signaling network in planta[J]. Plant Physiology, 2013, 161(3): 1066-1075. |
| [105] | STEINER E, LIVNE S, KOBINSON-KATZ T, et al. The putative O-linked N-acetylglucosamine transferase SPINDLY inhibits class Ⅰ TCP proteolysis to promote sensitivity to cytokinin[J]. Plant Physiology, 2016, 171(2): 1485-1494. |
| [1] | SONG Kainan, ZHANG Liwen, WANG Chao, TIAN Pingfang, LI Guangyue, PAN Guohui, XU Yuquan. Advances in small-molecule biopesticides and their biosynthesis [J]. Synthetic Biology Journal, 2025, 6(5): 1203-1223. |
| [2] | ZHAO Xinyu, SHENG Qi, LIU Kaifang, LIU Jia, LIU Liming. Construction of microbial cell factories for aspartate-family feed amino acids [J]. Synthetic Biology Journal, 2025, 6(5): 1184-1202. |
| [3] | HE Yangyu, YANG Kai, WANG Weilin, HUANG Qian, QIU Ziying, SONG Tao, HE Liushang, YAO Jinxin, GAN Lu, HE Yuchi. Design and practice of plant synthetic biology theme in the International Genetically Engineered Machine Competition [J]. Synthetic Biology Journal, 2025, 6(5): 1243-1254. |
| [4] | ZHENG Lei, ZHENG Qiteng, ZHANG Tianjiao, DUAN Kun, ZHANG Ruifu. Engineering rhizosphere synthetic microbial communities to enhance crop nutrient use efficiency [J]. Synthetic Biology Journal, 2025, 6(5): 1058-1071. |
| [5] | FANG Xinyi, SUN Lichao, HUO Yixin, WANG Ying, YUE Haitao. Trends and challenges in microbial synthesis of higher alcohols [J]. Synthetic Biology Journal, 2025, 6(4): 873-898. |
| [6] | LI Yongzhu, CHEN Yu. Advances and prospects in genome-scale models of yeast [J]. Synthetic Biology Journal, 2025, 6(3): 585-602. |
| [7] | XIA Chenliang, ZHANG Zecheng, GUAN Xingyue, TANG Qianyuan. Protein structural bioinformatics empowered by statistical physics and artificial intelligence [J]. Synthetic Biology Journal, 2025, 6(3): 547-565. |
| [8] | GAO Qi, XIAO Wenhai. Advances in the biosynthesis of monoterpenes by yeast [J]. Synthetic Biology Journal, 2025, 6(2): 357-372. |
| [9] | XIAO Sen, HU Litao, SHI Zhicheng, WANG Fayin, YU Siting, DU Guocheng, CHEN Jian, KANG Zhen. Research advances in biosynthesis of hyaluronic acid with controlled molecular weights [J]. Synthetic Biology Journal, 2025, 6(2): 445-460. |
| [10] | WANG Qian, GUO Shiting, XIN Bo, ZHONG Cheng, WANG Yu. Advances in biosynthesis of L-arginine using engineered microorganisms [J]. Synthetic Biology Journal, 2025, 6(2): 290-305. |
| [11] | ZUO Yimeng, ZHANG Jiaojiao, LIAN Jiazhang. Enabling technology for the biosynthesis of cosmetic raw materials with Saccharomyces cerevisiae [J]. Synthetic Biology Journal, 2025, 6(2): 233-253. |
| [12] | HUANG Shuhan, MA He, LUO Yunzi. Research progress in the biosynthesis of salidroside [J]. Synthetic Biology Journal, 2025, 6(2): 391-407. |
| [13] | GUO Shuyuan, ZHANG Qiannan, Gulikezi· MAIMAITIREXIATI, YANG Yiqun, YU Tao. Advances in microbial production of liquid biofuels [J]. Synthetic Biology Journal, 2025, 6(1): 18-44. |
| [14] | WEN Yanhua, LIU Hedong, CAO Chunlai, WU Ruibo. Applications of protein engineering in pharmaceutical industry [J]. Synthetic Biology Journal, 2025, 6(1): 65-86. |
| [15] | SHAO Mingwei, SUN Simian, YANG Shimao, CHEN Guoqiang. Bioproduction based on extremophiles [J]. Synthetic Biology Journal, 2024, 5(6): 1419-1436. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||