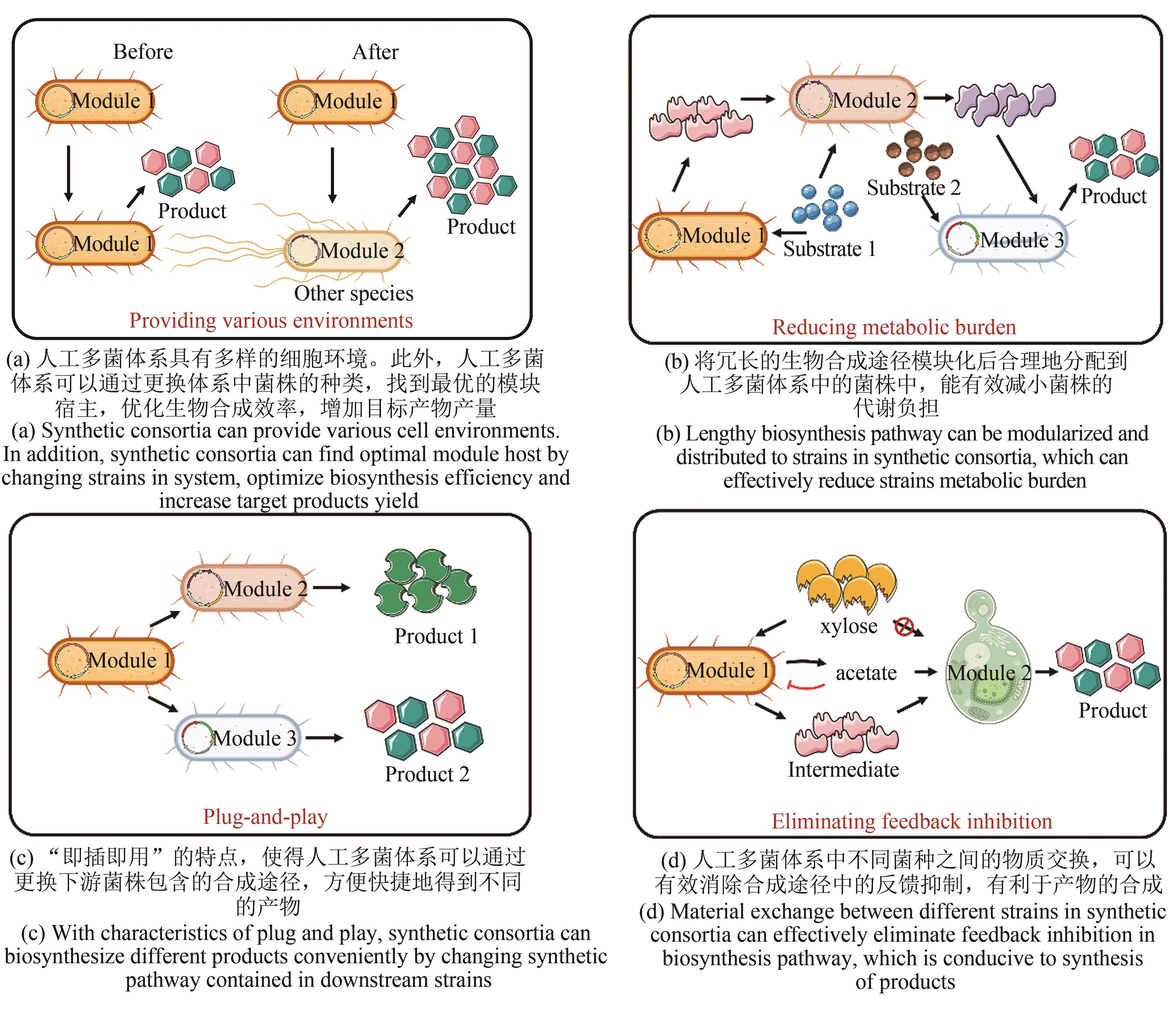
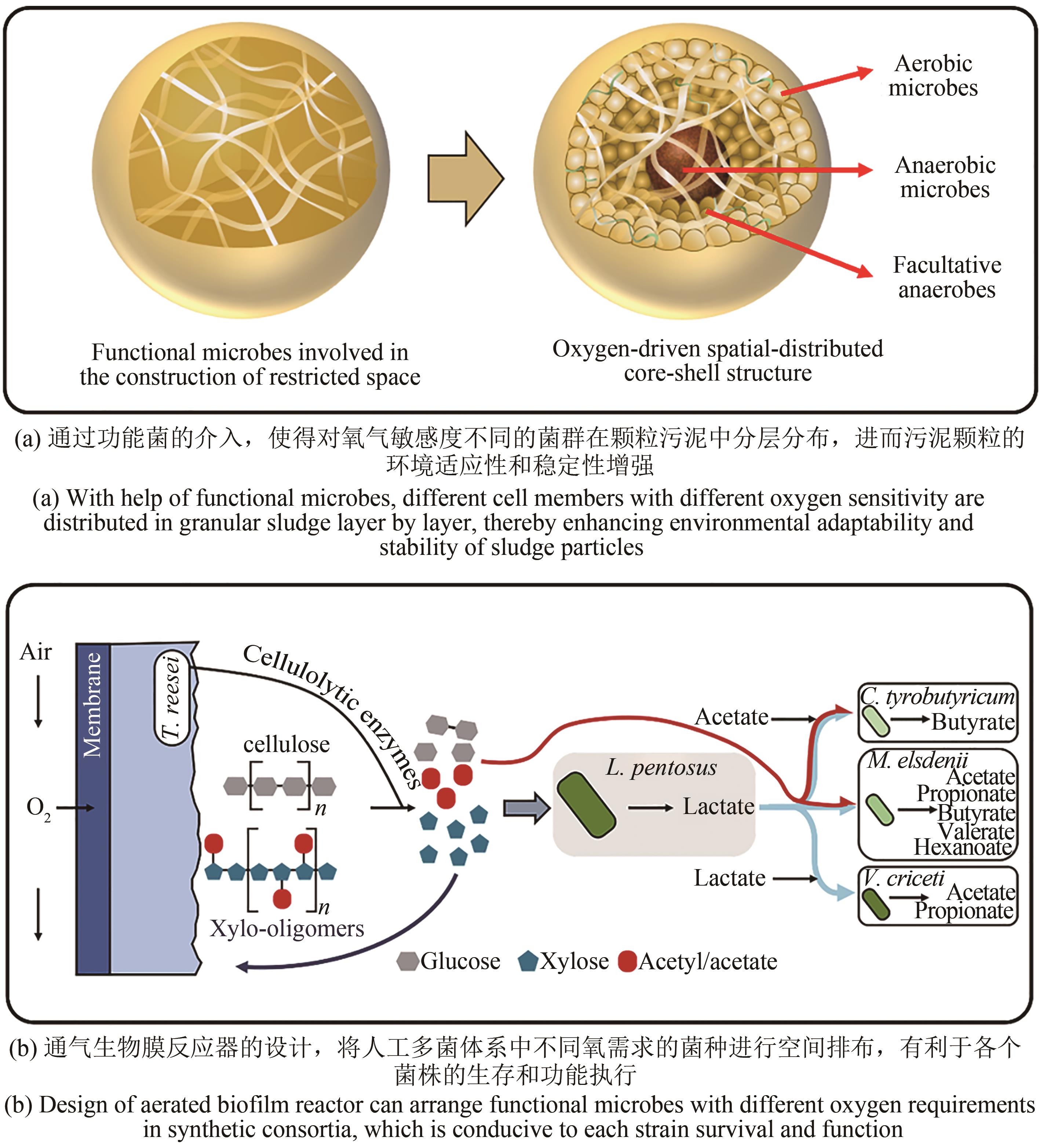
Synthetic Biology Journal ›› 2021, Vol. 2 ›› Issue (4): 635-650.DOI: 10.12211/2096-8280.2021-031
• Invited Review • Previous Articles Next Articles
Design and progress of synthetic consortia: a new frontier in synthetic biology
LIU Yu1, WEI Huiling1, LIU Jixiang1, WANG Shaojie1,2, SU Haijia1
- 1.State Key Laboratory of Chemical Resource Engineering,Beijing Key Laboratory of Bioprocess,Beijing Advanced Innovation Center for Soft Matter Science and Engineering,Beijing University of Chemical Technology,Beijing 100029,China
2.Institute of Nano Biomedicine and Engineering,School of Electronic Information and Electrical Engineering,Shanghai Jiao Tong University,Shanghai 200240,China
-
Received:2021-03-10Revised:2021-05-17Online:2021-09-10Published:2021-08-31 -
Contact:WANG Shaojie, SU Haijia
人工多菌体系的设计与构建:合成生物学研究新前沿
刘裕1, 韦惠玲1, 刘骥翔1, 王少杰1,2, 苏海佳1
- 1.北京化工大学化学资源工程国家重点实验室,北京生物过程重点实验室,北京软物质科学与工程先进创新中心,北京 100029
2.上海交通大学电子信息与电子工程学院纳米生物医学与工程研究所,上海 200240
-
通讯作者:王少杰,苏海佳 -
作者简介:刘裕 (1997—),女,硕士研究生。研究方向为基于合成生物学的人工多菌体系构建。E-mail:983381329@qq.com王少杰 (1991—),男,博士后。研究方向为合成生物学与生物能源。E-mail:wangshaojie0711@sjtu.edu.cn苏海佳 (1970—),女,教授,博士生导师。研究方向为合成生物学、生物分离、工业水处理、生物环境材料等。E-mail:suhj@mail.buct.edu.cn -
基金资助:国家自然科学基金(21838001);国家重点研发计划(2018YFA0902200);中央高校基础研究经费(JD2007);中国博士后科学基金(2020M681302)
CLC Number:
Cite this article
LIU Yu, WEI Huiling, LIU Jixiang, WANG Shaojie, SU Haijia. Design and progress of synthetic consortia: a new frontier in synthetic biology[J]. Synthetic Biology Journal, 2021, 2(4): 635-650.
刘裕, 韦惠玲, 刘骥翔, 王少杰, 苏海佳. 人工多菌体系的设计与构建:合成生物学研究新前沿[J]. 合成生物学, 2021, 2(4): 635-650.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2021-031
| 1 | BITTIHN P, DIN M O, TSIMRING L S, et al. Rational engineering of synthetic microbial systems: from single cells to consortia[J]. Current Opinion in Microbiology, 2018, 45: 92-99. |
| 2 | WU Jiwen, DONG Lili, ZHOU Chunshuang, et al. Developing a coculture for enhanced butanol production by Clostridium beijerinckii and Saccharomyces cerevisiae [J]. Bioresource Technology Reports, 2019, 6: 223-228. |
| 3 | LINDEMANN S R, BERNSTEIN H C, SONG H S, et al. Engineering microbial consortia for controllable outputs[J]. The ISME Journal, 2016, 10(9): 2077-2084. |
| 4 | DENG Yu, MA Lizhou, MAO Yin. Biological production of adipic acid from renewable substrates: current and future methods [J]. Biochemical Engineering Journal, 2016, 105: 16-26. |
| 5 | KHALIL A S, COLLINS J J. Synthetic biology: applications come of age [J]. Nature Reviews Genetics, 2010, 11(5): 367-379. |
| 6 | CHAE T U, CHOI S Y, KIM J W, et al. Recent advances in systems metabolic engineering tools and strategies [J]. Current Opinion in Biotechnology, 2017, 47: 67-82. |
| 7 | ZHANG H R, WANG X N. Modular co-culture engineering, a new approach for metabolic engineering [J]. Metabolic Engineering, 2016, 37: 114-121. |
| 8 | LEE C R, KIM C, SONG Y E, et al. Co-culture-based biological carbon monoxide conversion by Citrobacter amalonaticus Y19 and Sporomusa ovata via a reducing-equivalent transfer mediator [J]. Bioresource Technology, 2018, 259: 128-135. |
| 9 | LIU Y Q, TU X H, XU Q, et al. Engineered monoculture and co-culture of methylotrophic yeast for de novo production of monacolin J and lovastatin from methanol [J]. Metabolic Engineering, 2018, 45: 189-199. |
| 10 | SGOBBA E, WENDISCH V F. Synthetic microbial consortia for small molecule production [J]. Current Opinion in Biotechnology, 2020, 62: 72-79. |
| 11 | GAO L, XU T S, HUANG G, et al. Oral microbiomes: more and more importance in oral cavity and whole body [J]. Protein & Cell, 2018, 9(5): 488-500. |
| 12 | LI Z H, WANG X N, ZHANG H R. Balancing the non-linear rosmarinic acid biosynthetic pathway by modular co-culture engineering [J]. Metabolic Engineering, 2019, 54: 1-11. |
| 13 | SAID S BEN, OR D. Synthetic microbial ecology: engineering habitats for modular consortia [J]. Frontiers in Microbiology, 2017, 8: 1125. |
| 14 | SHEN Y P, NIU F X, YAN Z B, et al. Recent advances in metabolically engineered microorganisms for the production of aromatic chemicals derived from aromatic amino acids[J]. Frontiers in Bioengineering and Biotechnology, 2020, 8: 407. |
| 15 | ZHANG C Z, HONG K. Production of terpenoids by synthetic biology approaches [J]. Frontiers in Bioengineering and Biotechnology, 2020, 8: 347. |
| 16 | PONTRELLI S, CHIU T Y, LAN E I, et al. Escherichia coli as a host for metabolic engineering [J]. Metabolic Engineering, 2018, 50: 16-46. |
| 17 | XU P, VANSIRI A, BHAN N, et al. ePathBrick: a synthetic biology platform for engineering metabolic pathways in E. coli [J]. ACS Synthetic Biology, 2012, 1(7): 256-266. |
| 18 | WU Y F, SHEN X L, YUAN Q P, et al. Metabolic engineering strategies for co-utilization of carbon sources in microbes [J]. Bioengineering, 2016, 3(1): 10. |
| 19 | DHARMADI Y, MURARKA A, GONZALEZ R. Anaerobic fermentation of glycerol by Escherichia coli: a new platform for metabolic engineering [J]. Biotechnology and Bioengineering, 2006, 94(5): 821-829. |
| 20 | ATSUMI S, CANN A F, CONNOR M R, et al. Metabolic engineering of Escherichia coli for 1-butanol production [J]. Metabolic Engineering, 2008, 10(6): 305-311. |
| 21 | CLOMBURG J M, GONZALEZ R. Biofuel production in Escherichia coli: the role of metabolic engineering and synthetic biology [J]. Applied Microbiology and Biotechnology, 2010, 86(2): 419-434. |
| 22 | GAO C, WANG S H, HU G P, et al. Engineering Escherichia coli for malate production by integrating modular pathway characterization with CRISPRi-guided multiplexed metabolic tuning [J]. Biotechnology and Bioengineering, 2018, 115(3): 661-672. |
| 23 | CHEN T T, ZHOU Y Y, LU Y H, et al. Advances in heterologous biosynthesis of plant and fungal natural products by modular co-culture engineering [J]. Biotechnology Letters, 2019, 41(1): 27-34. |
| 24 | ZHANG H R, STEPHANOPOULOS G. Co-culture engineering for microbial biosynthesis of 3-amino-benzoic acid in Escherichia coli [J]. Biotechnology Journal, 2016, 11(7): 981-987. |
| 25 | ZHOU K, QIAO K J, EDGAR S, et al. Distributing a metabolic pathway among a microbial consortium enhances production of natural products [J]. Nature Biotechnology, 2015, 33(4): 377-383. |
| 26 | ROELL G W, ZHA J, CARR R R, et al. Engineering microbial consortia by division of labor [J]. Microbial Cell Factories, 2019, 18(1): 35. |
| 27 | NAKAGAWA A, MATSUMURA E, KOYANAGI T, et al. Total biosynthesis of opiates by stepwise fermentation using engineered Escherichia coli [J]. Nature Communications, 2016, 7: 10390. |
| 28 | WU G, YAN Q, JONES J A, et al. Metabolic burden: cornerstones in synthetic biology and metabolic engineering applications [J]. Trends in Biotechnology, 2016, 34(8): 652-664. |
| 29 | JONES J A, VERNACCHIO V R, COLLINS S M, et al. Complete biosynthesis of anthocyanins using E. coli polycultures [J]. mBio, 2017, 8(3): 00621-17. |
| 30 | JONES J A, WANG X. Use of bacterial co-cultures for the efficient production of chemicals [J]. Current Opinion in Biotechnology, 2018, 53: 33-38. |
| 31 | JIANG W, QIAO J B, BENTLEY G J, et al. Modular pathway engineering for the microbial production of branched-chain fatty alcohols [J]. Biotechnology for Biofuels, 2017, 10: 244. |
| 32 | XU P, GU Q, WANG W Y, et al. Modular optimization of multi-gene pathways for fatty acids production in E. coli [J]. Nature Communications, 2013, 4: 1409. |
| 33 | WU J J, DU G C, ZHOU J W, et al. Metabolic engineering of Escherichia coli for (2S)-pinocembrin production from glucose by a modular metabolic strategy [J]. Metabolic Engineering, 2013, 16: 48-55. |
| 34 | LAYTON D S, TRINH C T. Engineering modular ester fermentative pathways in Escherichia coli [J]. Metabolic Engineering, 2014, 26: 77-88. |
| 35 | GUO Xiaoyun, WANG Xiaonan, CHEN Tingting, et al. Comparing E. coli mono-cultures and co-cultures for biosynthesis of protocatechuic acid and hydroquinone [J]. Biochemical Engineering Journal, 2020, 156: 107518. |
| 36 | SAINI M, LIN L J, CHIANG C J, et al. Synthetic consortium of Escherichia coli for n-butanol production by fermentation of the glucose-xylose mixture[J]. Journal of Agricultural and Food Chemistry, 2017, 65(46): 10040-10047. |
| 37 | FLORES A, AYLA E Z, NIELSEN D R, et al. Engineering a synthetic, catabolically orthogonal coculture system for enhanced conversion of lignocellulose-derived sugars to ethanol[J]. ACS Synthetic Biology, 2019, 8(5): 1089-1099. |
| 38 | VARDON D R, RORRER N A, SALVACHÚA D, et al. cis,cis-Muconic acid: separation and catalysis to bio-adipic acid for nylon-6,6 polymerization [J]. Green Chemistry, 2016, 18(11): 3397-3413. |
| 39 | TONDRO H, MUSIVAND S, ZILOUEI H, et al. Biological production of hydrogen and acetone- butanol-ethanol from sugarcane bagasse and rice straw using co-culture of Enterobacter aerogenes and Clostridium acetobutylicum [J]. Biomass and Bioenergy, 2020, 142: 105818. |
| 40 | WANG S J, TANG H Z, PENG F, et al. Metabolite-based mutualism enhances hydrogen production in a two-species microbial consortium [J]. Communications Biology, 2019, 2: 82. |
| 41 | QIAN X J, CHEN L, SUI Y, et al. Biotechnological potential and applications of microbial consortia [J]. Biotechnology Advances, 2020, 40: 107500. |
| 42 | JIA X Q, LIU C, SONG H, et al. Design, analysis and application of synthetic microbial consortia [J]. Synthetic and Systems Biotechnology, 2016, 1(2): 109-117. |
| 43 | ZHANG H R, PEREIRA B, LI Z J, et al. Engineering Escherichia coli coculture systems for the production of biochemical products [J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(27): 8266-8271. |
| 44 | LIU Y R, YANG S Y, JIA X Q. Construction of a "nutrition supply-detoxification" coculture consortium for medium-chain-length polyhydroxyalkanoate production with a glucose-xylose mixture [J]. Journal of Industrial Microbiology & Biotechnology, 2020, 47(3): 343-354. |
| 45 | LU H Y, VILLADA J C, LEE P K H. Modular metabolic engineering for biobased chemical production [J]. Trends in Biotechnology, 2019, 37(2): 152-166. |
| 46 | FENG Y Y, YAO M D, WANG Y, et al. Advances in engineering UDP-sugar supply for recombinant biosynthesis of glycosides in microbes [J]. Biotechnology Advances, 2020, 41: 107538. |
| 47 | CHEN Z Y, SUN X X, LI Y, et al. Metabolic engineering of Escherichia coli for microbial synthesis of monolignols [J]. Metabolic Engineering, 2017, 39: 102-109. |
| 48 | ZHOU Y Y, LI Z H, WANG X N, et al. Establishing microbial co-cultures for 3-hydroxybenzoic acid biosynthesis on glycerol [J]. Engineeing in Life Sciences, 2019, 19(5): 389-395. |
| 49 | ZHANG H R, LI Z J, PEREIRA B, et al. Engineering E. coli — E. coli cocultures for production of muconic acid from glycerol [J]. Microbial Cell Factories, 2015, 14(1): 134. |
| 50 | GOERS L, FREEMONT P, POLIZZI K M. Co-culture systems and technologies: taking synthetic biology to the next level [J]. Journal of the Royal Society Interface, 2014, 11(96): 20140065. |
| 51 | LIU Yue, DING Mingzhu, LING Wei, et al. A three-species microbial consortium for power generation [J]. Energy & Environmental Science, 2017, 10(7): 1600-1609. |
| 52 | JAWED K, YAZDANI S S, KOFFAS M A. Advances in the development and application of microbial consortia for metabolic engineering[J]. Metabolic Engineering Communications, 2019, 9: e00095. |
| 53 | XIAO Y, BOWEN C H, LIU D, et al. Exploiting nongenetic cell-to-cell variation for enhanced biosynthesis [J]. Nature Chemical Biology, 2016, 12(5): 339-344. |
| 54 | WEN Z Q, LEDESMA-AMARO R, LU M R, et al. Combined evolutionary engineering and genetic manipulation improve low pH tolerance and butanol production in a synthetic microbial Clostridium community [J]. Biotechnology and Bioengineering, 2020, 117(7): 2008-2022. |
| 55 | PARK H, PATEL A, HUNT K A, et al. Artificial consortium demonstrates emergent properties of enhanced cellulosic-sugar degradation and biofuel synthesis[J]. NPJ Biofilms and Microbiomes, 2020, 6(1): 59. |
| 56 | MINTY J J, SINGER M E, SCHOLZ S A, et al. Design and characterization of synthetic fungal-bacterial consortia for direct production of isobutanol from cellulosic biomass [J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(36): 14592-14597. |
| 57 | SGOBBA E, STUMPF A K, VORTMANN M, et al. Synthetic Escherichia coli-Corynebacterium glutamicum consortia for L-lysine production from starch and sucrose [J]. Bioresource Technology, 2018, 260: 302-310. |
| 58 | KONG W, MELDGIN D R, COLLINS J J, et al. Designing microbial consortia with defined social interactions [J]. Nature Chemical Biology, 2018, 14(8): 821-829. |
| 59 | ZHANG W, LIU H, LI X, et al. Production of naringenin from D-xylose with co-culture of E. coli and S. cerevisiae [J]. Engineering in Life Sciences, 2017, 17(9): 1021-1029. |
| 60 | LIU X, LI L L, LIU J C, et al. Metabolic engineering Escherichia coli for efficient production of icariside D2 [J]. Biotechnology for Biofuels, 2019, 12(1): 1-12. |
| 61 | DOUGLAS A E. The microbial exometabolome: ecological resource and architect of microbial communities [J]. Philosophical Transactions of the Royal Society of London Series B, Biological Sciences, 2020, 375(1798): 20190250. |
| 62 | LIU X, LI X B, JIANG J, et al. Convergent engineering of syntrophic Escherichia coli coculture for efficient production of glycosides [J]. Metabolic Engineering, 2018, 47: 243-253. |
| 63 | MEE M T, COLLINS J J, CHURCH G M, et al. Syntrophic exchange in synthetic microbial communities[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(20): E2149-E2156. |
| 64 | SONG H, DING M Z, JIA X Q, et al. Synthetic microbial consortia: from systematic analysis to construction and applications [J]. Chemical Society Reviews, 2014, 43(20): 6954-6981. |
| 65 | SANGANI A A, MCCULLY A L, LASARRE B, et al. Fermentative Escherichia coli makes a substantial contribution to H2 production in coculture with phototrophic Rhodopseudomonas palustris [J]. FEMS Microbiology Letters, 2019, 366(14): fnz162. |
| 66 | BOURDAKOS N, MARSILI E, MAHADEVAN R. A defined co-culture of Geobacter sulfurreducens and Escherichia coli in a membrane-less microbial fuel cell [J]. Biotechnology and Bioengineering, 2014, 111(4): 709-18. |
| 67 | HARCOMBE W R, CHACÓN J M, ADAMOWICZ E M, et al. Evolution of bidirectional costly mutualism from byproduct consumption[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(47): 12000-12004. |
| 68 | WANG Xiaonan, CABALES A, LI Zhenghong, et al. Biosensor-assisted high performing cell selection using an E. coli toxin/antitoxin system [J]. Biochemical Engineering Journal, 2019, 144: 110-118. |
| 69 | WANG X N, POLICARPIO L, PRAJAPATI D, et al. Developing E. coli-E. coli co-cultures to overcome barriers of heterologous tryptamine biosynthesis [J]. Metabolic Engineering Communications, 2020, 10: e00110. |
| 70 | ZENG J, BANERJEE A, KIM J, et al. A novel bioelectronic reporter system in living cells tested with a synthetic biological comparator [J]. Scientific Reports, 2019, 9(1): 7275. |
| 71 | GUO X Y, LI Z H, WANG X N, et al. De novo phenol bioproduction from glucose using biosensor-assisted microbial coculture engineering [J]. Biotechnology and Bioengineering, 2019, 116(12): 3349-3359. |
| 72 | LAGANENKA L, SOURJIK V. Autoinducer 2-dependent Escherichia coli biofilm formation is enhanced in a dual-species coculture [J]. Applied and Environmental Microbiology, 2018, 84(5). |
| 73 | SCOTT S R, HASTY J. Quorum sensing communication modules for microbial consortia[J]. ACS Synthetic Biology, 2016, 5(9): 969-977. |
| 74 | KYLILIS N, TUZA Z A, STAN G B, et al. Tools for engineering coordinated system behaviour in synthetic microbial consortia [J]. Nature Communications, 2018, 9(1): 2677. |
| 75 | SCOTT S R, DIN M O, BITTIHN P, et al. A stabilized microbial ecosystem of self-limiting bacteria using synthetic quorum-regulated lysis [J]. Nature Microbiology, 2017, 2: 17083. |
| 76 | STEPHENS K, BENTLEY W E. Synthetic biology for manipulating quorum sensing in microbial consortia[J]. Trends in Microbiology, 2020, 28(8): 633-643. |
| 77 | STEPHENS K, POZO M, TSAO C Y, et al. Bacterial co-culture with cell signaling translator and growth controller modules for autonomously regulated culture composition [J]. Nature Communications, 2019, 10(1): 4129. |
| 78 | DINH C V, CHEN X Y, PRATHER K L J. Development of a quorum-sensing based circuit for control of coculture population composition in a naringenin production system[J]. ACS Synthetic Biology, 2020, 9(3): 590-597. |
| 79 | JOHNS N I, BLAZEJEWSKI T, GOMES A L, et al. Principles for designing synthetic microbial communities [J]. Current Opinion in Microbiology, 2016, 31: 146-153. |
| 80 | WANG Shaojie, MA Zhihong, SU Haijia. Two-step continuous hydrogen production by immobilized mixed culture on corn stalk [J]. Renewable Energy, 2018, 121: 230-235. |
| 81 | JOHNSTON T G, YUAN S F, WAGNER J M, et al. Compartmentalized microbes and co-cultures in hydrogels for on-demand bioproduction and preservation [J]. Nature Communications, 2020, 11(1): 563. |
| 82 | CHEN Y Y, GE J Y, WANG S J, et al. Insight into formation and biological characteristics of Aspergillus tubingensis-based aerobic granular sludge (AT-AGS) in wastewater treatment [J]. The Science of the Total Environment, 2020, 739: 140128. |
| 83 | XIONG W, WANG S J, ZHOU N, et al. Granulation enhancement and microbial community shift of tylosin-tolerant aerobic granular sludge on the treatment of tylosin wastewater [J]. Bioresource Technology, 2020, 318: 124041. |
| 84 | SHAHAB R L, BRETHAUER S, DAVEY M P, et al. A heterogeneous microbial consortium producing short-chain fatty acids from lignocellulose [J]. Science, 2020, 369(6507): eabb1214. |
| 85 | MOUTINHO T J JR, PANAGIDES J C, BIGGS M B, et al. Novel co-culture plate enables growth dynamic-based assessment of contact-independent microbial interactions[J]. PLoS One, 2017, 12(8): e0182163. |
| 86 | ZOMORRODI A R, SEGRÈ D. Synthetic ecology of microbes: Mathematical models and applications [J]. Journal of Molecular Biology, 2016, 428(5pt b): 837-861. |
| 87 | CHANDRASEKARAN S. A Protocol for the construction and curation of genome-scale integrated metabolic and regulatory network models [M]// SANTOS C N S, AJIKUMAR P K. Microbial metabolic engineering: methods and protocols. New York: Springer New York, 2019: 203-214. |
| 88 | GOMEZ J A, HÖFFNER K, BARTON P I. DFBAlab: a fast and reliable MATLAB code for dynamic flux balance analysis [J]. BMC Bioinformatics, 2014, 15: 409. |
| 89 | CAI J Y, TAN T W, CHAN S H J. Bridging traditional evolutionary game theory and metabolic models for predicting Nash equilibrium of microbial metabolic interactions[EB/OL]. bioRxiv, 2019, . DOI:10.1101/623173 . |
| 90 | JADHAV A, SHANMUGHAM B, RAJENDIRAN A, et al. Unraveling novel broad-spectrum antibacterial targets in food and waterborne pathogens using comparative genomics and protein interaction network analysis[J]. Infection, Genetics and Evolution, 2014, 27: 300-308. |
| 91 | JONES J A, VERNACCHIO V R, SINKOE A L, et al. Experimental and computational optimization of an Escherichia coli co-culture for the efficient production of flavonoids [J]. Metabolic Engineering, 2016, 35: 55-63. |
| 92 | HARCOMBE W R, RIEHL W J, DUKOVSKI I, et al. Metabolic resource allocation in individual microbes determines ecosystem interactions and spatial dynamics [J]. Cell Reports, 2014, 7(4): 1104-1115. |
| 93 | SANTALA S, KARP M, SANTALA V. Rationally engineered synthetic coculture for improved biomass and product formation [J]. PLoS One, 2014, 9(12): e113786. |
| 94 | MCCARTY N S, LEDESMA-AMARO R. Synthetic biology tools to engineer microbial communities for biotechnology[J]. Trends in Biotechnology, 2019, 37(2): 181-197. |
| 95 | LÜ X M, GU J L, WANG F, et al. Combinatorial pathway optimization in Escherichia coli by directed co-evolution of rate-limiting enzymes and modular pathway engineering [J]. Biotechnology and Bioengineering, 2016, 113(12): 2661-2669. |
| 96 | HANLY T J, URELLO M, HENSON M A. Dynamic flux balance modeling of S. cerevisiae and E. coli co-cultures for efficient consumption of glucose/xylose mixtures [J]. Applied Microbiology and Biotechnology, 2012, 93(6): 2529-2541. |
| 97 | RODRIGUES J L, GOMES D, RODRIGUES L R. A combinatorial approach to optimize the production of curcuminoids from tyrosine in Escherichia coli [J]. Frontiers in Bioengineering and Biotechnology, 2020, 8: 59. |
| 98 | CHEN Ye, KIM J K, HIRNING A J, et al. Emergent genetic oscillations in a synthetic microbial consortium [J]. Science, 2015, 349(6251): 986-989. |
| 99 | FIORE D, SALZANO D, CRISTÒBAL-CÓPPULO E, et al. Multicellular feedback control of a genetic toggle-switch in microbial consortia[J]. IEEE Control Systems Letters, 2021, 5(1): 151-156. |
| 100 | XIU Y, JANG S, JONES J A, et al. Naringenin-responsive riboswitch-based fluorescent biosensor module for Escherichia coli co-cultures [J]. Biotechnology and Bioengineering, 2017, 114(10): 2235-2244. |
| 101 | XIA Tian, ALTMAN E, EITEMAN M A. Succinate production from xylose-glucose mixtures using a consortium of engineered Escherichia coli [J]. Engineering in Life Sciences, 2015, 15(1): 65-72. |
| 102 | LI T Z, ZHOU W, BI H P, et al. Production of caffeoylmalic acid from glucose in engineered Escherichia coli [J]. Biotechnology Letters, 2018, 40(7): 1057-1065. |
| 103 | GANESAN V, LI Z H, WANG X N, et al. Heterologous biosynthesis of natural product naringenin by co-culture engineering [J]. Synthetic and Systems Biotechnology, 2017, 2(3): 236-242. |
| 104 | SUN J, RAZA M, SUN X X, et al. Biosynthesis of adipic acid via microaerobic hydrogenation of cis,cis-muconic acid by oxygen-sensitive enoate reductase [J]. Biotechnology Journal, 2018, 280: 49-54. |
| [1] | GUO Shuyuan, ZHANG Qiannan, Gulikezi· MAIMAITIREXIATI, YANG Yiqun, YU Tao. Advances in microbial production of liquid biofuels [J]. Synthetic Biology Journal, 2025, 6(1): 18-44. |
| [2] | ZHONG Quanzhou, SHAN Yiyi, PEI Qingyun, JIN Yanyun, WANG Yihan, MENG Luyuan, WANG Xinyun, ZHANG Yuxin, LIU Kunyuan, WANG Huizhong, FENG Shangguo. Research progress in the production of α-arbutin through biosynthesis [J]. Synthetic Biology Journal, 2025, 6(1): 118-135. |
| [3] | SHAO Mingwei, SUN Simian, YANG Shimao, CHEN Guoqiang. Bioproduction based on extremophiles [J]. Synthetic Biology Journal, 2024, 5(6): 1419-1436. |
| [4] | ZHAO Liang, LI Zhenshuai, FU Liping, LYU Ming, WANG Shi’an, ZHANG Quan, LIU Licheng, LI Fuli, LIU Ziyong. Progress in biomanufacturing of lipids and single cell protein from one-carbon compounds [J]. Synthetic Biology Journal, 2024, 5(6): 1300-1318. |
| [5] | ZHU Fanghuan, CEN Xuecong, CHEN Zhen. Research progress of diols production by microbes [J]. Synthetic Biology Journal, 2024, 5(6): 1367-1385. |
| [6] | LIU Yining, PU Wei, YANG Jinxing, WANG Yu. Recent advances in the biosynthesis of ω-amino acids and lactams [J]. Synthetic Biology Journal, 2024, 5(6): 1350-1366. |
| [7] | ZHENG Haotian, LI Chaofeng, LIU Liangxu, WANG Jiawei, LI Hengrun, NI Jun. Design, optimization and application of synthetic carbon-negative phototrophic community [J]. Synthetic Biology Journal, 2024, 5(5): 1189-1210. |
| [8] | CHENG Xiaolei, LIU Tiangang, TAO Hui. Recent research progress in non-canonical biosynthesis of terpenoids [J]. Synthetic Biology Journal, 2024, 5(5): 1050-1071. |
| [9] | CHENG Zhongyu, LI Fuzhuo. Recent advances in chemoenzymatic synthesis of natural products via site- selective P450 oxidation [J]. Synthetic Biology Journal, 2024, 5(5): 960-980. |
| [10] | LIU Zijian, MU Baiyang, DUAN Zhiqiang, WANG Xuan, LU Xiaojie. Advances in the development of DNA-compatible chemistries [J]. Synthetic Biology Journal, 2024, 5(5): 1102-1124. |
| [11] | YU Wei, GAO Jiaoqi, ZHOU Yongjin. Bioconversion of one carbon feedstocks for producing organic acids [J]. Synthetic Biology Journal, 2024, 5(5): 1169-1188. |
| [12] | ZHANG Shouqi, WANG Tao, KONG Yao, ZOU Jiasheng, LIU Yuanning, XU Zhengren. Chemoenzymatic synthesis of natural products: evolution of synthetic methodology and strategy [J]. Synthetic Biology Journal, 2024, 5(5): 913-940. |
| [13] | XIE Xiangqian, GUO Wen, WANG Huan, LI Jin. Biosynthesis and chemical synthesis of ribosomally synthesized and post-translationally modified peptides containing aminovinyl cysteine [J]. Synthetic Biology Journal, 2024, 5(5): 981-996. |
| [14] | TANG Zhijun, HU Youcai, LIU Wen. Enzymatic (4+2)- and (2+2)-cycloaddition reactions: fundamentals and applications of regio- and stereoselectivity [J]. Synthetic Biology Journal, 2024, 5(3): 401-407. |
| [15] | ZHANG Jun, JIN Shixue, YUN Qian, QU Xudong. Biosynthesis of the unnatural extender units with polyketides and their structural modifications for applications in medicines [J]. Synthetic Biology Journal, 2024, 5(3): 561-570. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||