Synthetic Biology Journal ›› 2020, Vol. 1 ›› Issue (4): 454-469.DOI: 10.12211/2096-8280.2020-026
• Invited Review • Previous Articles Next Articles
Progress in the regulatory tools of gene expression for model microorganisms
TIAN Rongzhen1,2, LIU Yanfeng1,2, LI Jianghua2, LIU Long1,2, DU Guocheng1,2
- 1.Key Laboratory of Carbohydrate Chemistry and Biotechnology,Jiangnan University,Wuxi 214122,Jiangsu,China
2.Key Laboratory of Industrial Biotechnology,Ministry of Education,Jiangnan University,Wuxi 214122,Jiangsu,China
-
Received:2020-03-15Revised:2020-06-21Online:2020-10-09Published:2020-08-31 -
Contact:DU Guocheng
典型模式微生物基因表达精细调控工具的研究进展
田荣臻1,2, 刘延峰1,2, 李江华2, 刘龙1,2, 堵国成1,2
- 1.江南大学糖化学与生物技术教育部重点实验室,江苏 无锡 214122
2.江南大学工业生物技术教育部重点实验室,江苏 无锡 214122
-
通讯作者:堵国成 -
作者简介:田荣臻(1995—),男,博士研究生,研究方向为发酵工程。E-mail:rz_tian@stu.jiangnan.edu.cn
作者简介:堵国成(1965—),男,博士生导师,教授,研究方向为发酵工程与酶工程。E-mail:gcdu@jiangnan.edu.cn -
基金资助:国家重点研发计划(2018YFA0900300);国家自然科学基金(31972854)
CLC Number:
Cite this article
TIAN Rongzhen, LIU Yanfeng, LI Jianghua, LIU Long, DU Guocheng. Progress in the regulatory tools of gene expression for model microorganisms[J]. Synthetic Biology Journal, 2020, 1(4): 454-469.
田荣臻, 刘延峰, 李江华, 刘龙, 堵国成. 典型模式微生物基因表达精细调控工具的研究进展[J]. 合成生物学, 2020, 1(4): 454-469.
share this article
Add to citation manager EndNote|Ris|BibTeX
URL: https://synbioj.cip.com.cn/EN/10.12211/2096-8280.2020-026
| 调控工具 | 调控水平 | 调控机理 | 典型实例 | 参考 文献 |
|---|---|---|---|---|
| 合成启动子 | 转录起始 | 实现天然启动子难以达到的特性,如更高的强度、更短的序列等 | 枯草芽孢杆菌混合启动子文库的构建 | [ |
| 广谱启动子 | 转录起始 | 实现启动子在所有模式微生物中的通用性 | 大肠杆菌、枯草芽孢杆菌、酿酒酵母广谱启动子 | [ |
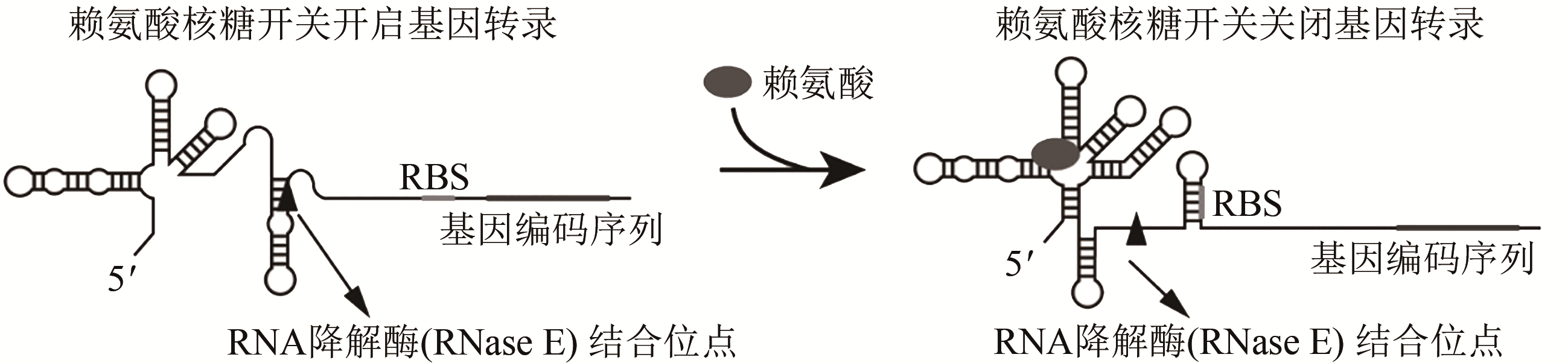
| 核糖开关 | 转录终止, 翻译起始, mRNA丰度 | 通过响应特定代谢物的结合而改变自身结构,可以调节转录终止、翻译起始和mRNA降解速率 | 大肠杆菌赖氨酸核糖开关、枯草芽孢杆菌glmS核糖开关 | [ |
| 核酸适配体 | 转录起始, 翻译起始 | 通过特异性结合响应核糖配体可以调节基因的转录起始或翻译起始 | 在枯草芽孢杆菌中使用凝血酶配体-适配体构建双功能动态代谢调控回路提高2′-岩藻糖基产量 | [ |
| sRNA | 转录延伸, 翻译起始 与延伸 | 在基因组上的反义转录可以造成目的基因转录中止,同时通过与特定mRNA进行碱基配对可以影响核糖体的结合与延伸 | 通过合成asRNA调节丙二酰辅酶A代谢,增强4-羟基香豆素、白藜芦醇和柚皮素的生物合成 | [ |
| 转录因子 | 转录起始 | 充当基因转录的阻遏物或激活物 | 构建CodY和CcpA突变文库调控枯草芽孢杆菌内碳氮代谢网络以提高β-半乳糖苷酶、木聚糖酶和肽酶产量 | [ |
| σ因子 | 转录起始 | 通过偏好性引导RNA聚合酶转录不同基因实现全局基因转录调控 | σ70的编码基因rpoD随机突变获得高性能的目标大肠杆菌 | [ |
| 基因表观 遗传修饰 | 转录起始 | 通过在宿主基因组中引入DNA修饰以实现基因转录的调控 | 通过调控DNA甲基转移酶表达提高大肠杆菌环境抗逆性 | [ |
| CRISPRa/i | 转录起始, 翻译起始 | 将dCas9蛋白融合强激活域或强抑制域靶向特定DNA或mRNA序列调控基因转录或翻译 | 在酿酒酵母中使用CRISPR/dCsd9系统同时实现对基因表达的激活、抑制与沉默 | [ |
| 群体响应 | 转录起始 | 响应细胞密度信号并将其转化为基因转录状态的变化 | 在大肠杆菌基于LuxR群体相应系统实现基因表达状态自动切换,提高了异丙醇产量 | [ |
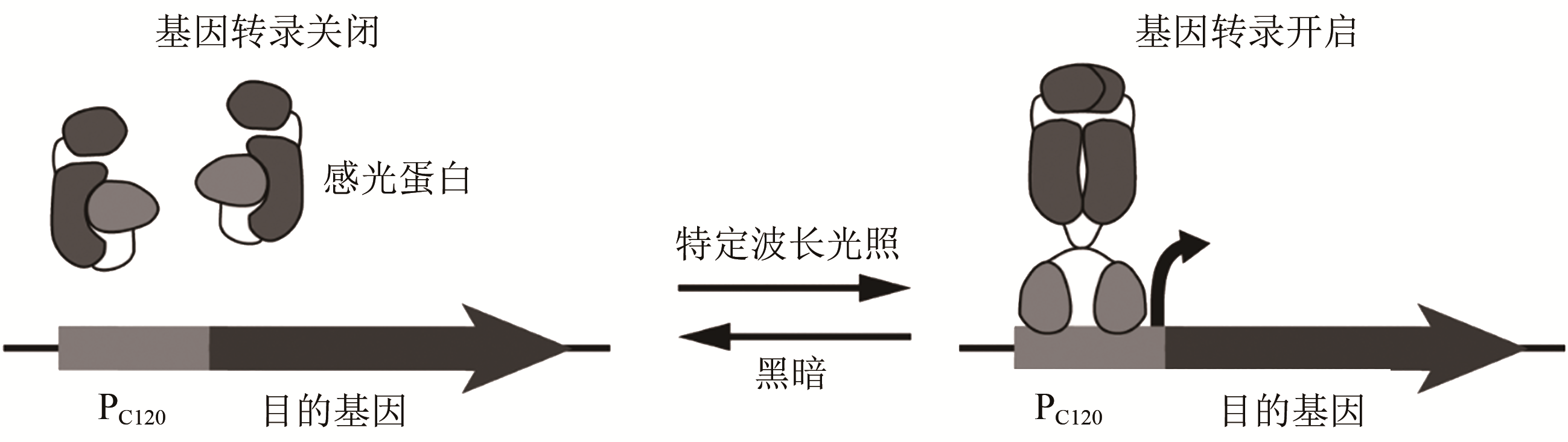
| 光遗传学调控 | 转录起始 | 响应特定波长光信号并将其转化为基因转录状态的变化 | 在酵母中实现蓝光依赖的基因表达激活与抑制以提高异丁醇和2-甲基-1-丁醇产量 | [ |
| 生物传感器 | 转录起始 | 响应特定代谢产物浓度并将其转化为基因转录状态的变化 | 在大肠杆菌中使用丙二酰-CoA生物传感器自动调控脂肪酸合成关键基因的表达,实现细胞生长和产物和成间的平衡 | [ |
| N端编码序列 | 翻译延伸 | 影响核糖体在mRNA上的延伸速率或脱落率 | 构建枯草芽孢杆菌N端编码序列文库调控NeuAc合成途径关键基因表达水平与动态模式以提高NeuAc产量 | [ |
| 蛋白降解标签 | 蛋白丰度 | 通过对目的蛋白的降解调控目的蛋白的丰度 | 将蛋白降解标签用于构建大肠杆菌正交基因回路,实现基因表达水平高强度激活与抑制 | [ |
Tab. 1 Regulatory tools of gene expression for model microorganisms
| 调控工具 | 调控水平 | 调控机理 | 典型实例 | 参考 文献 |
|---|---|---|---|---|
| 合成启动子 | 转录起始 | 实现天然启动子难以达到的特性,如更高的强度、更短的序列等 | 枯草芽孢杆菌混合启动子文库的构建 | [ |
| 广谱启动子 | 转录起始 | 实现启动子在所有模式微生物中的通用性 | 大肠杆菌、枯草芽孢杆菌、酿酒酵母广谱启动子 | [ |
| 核糖开关 | 转录终止, 翻译起始, mRNA丰度 | 通过响应特定代谢物的结合而改变自身结构,可以调节转录终止、翻译起始和mRNA降解速率 | 大肠杆菌赖氨酸核糖开关、枯草芽孢杆菌glmS核糖开关 | [ |
| 核酸适配体 | 转录起始, 翻译起始 | 通过特异性结合响应核糖配体可以调节基因的转录起始或翻译起始 | 在枯草芽孢杆菌中使用凝血酶配体-适配体构建双功能动态代谢调控回路提高2′-岩藻糖基产量 | [ |
| sRNA | 转录延伸, 翻译起始 与延伸 | 在基因组上的反义转录可以造成目的基因转录中止,同时通过与特定mRNA进行碱基配对可以影响核糖体的结合与延伸 | 通过合成asRNA调节丙二酰辅酶A代谢,增强4-羟基香豆素、白藜芦醇和柚皮素的生物合成 | [ |
| 转录因子 | 转录起始 | 充当基因转录的阻遏物或激活物 | 构建CodY和CcpA突变文库调控枯草芽孢杆菌内碳氮代谢网络以提高β-半乳糖苷酶、木聚糖酶和肽酶产量 | [ |
| σ因子 | 转录起始 | 通过偏好性引导RNA聚合酶转录不同基因实现全局基因转录调控 | σ70的编码基因rpoD随机突变获得高性能的目标大肠杆菌 | [ |
| 基因表观 遗传修饰 | 转录起始 | 通过在宿主基因组中引入DNA修饰以实现基因转录的调控 | 通过调控DNA甲基转移酶表达提高大肠杆菌环境抗逆性 | [ |
| CRISPRa/i | 转录起始, 翻译起始 | 将dCas9蛋白融合强激活域或强抑制域靶向特定DNA或mRNA序列调控基因转录或翻译 | 在酿酒酵母中使用CRISPR/dCsd9系统同时实现对基因表达的激活、抑制与沉默 | [ |
| 群体响应 | 转录起始 | 响应细胞密度信号并将其转化为基因转录状态的变化 | 在大肠杆菌基于LuxR群体相应系统实现基因表达状态自动切换,提高了异丙醇产量 | [ |
| 光遗传学调控 | 转录起始 | 响应特定波长光信号并将其转化为基因转录状态的变化 | 在酵母中实现蓝光依赖的基因表达激活与抑制以提高异丁醇和2-甲基-1-丁醇产量 | [ |
| 生物传感器 | 转录起始 | 响应特定代谢产物浓度并将其转化为基因转录状态的变化 | 在大肠杆菌中使用丙二酰-CoA生物传感器自动调控脂肪酸合成关键基因的表达,实现细胞生长和产物和成间的平衡 | [ |
| N端编码序列 | 翻译延伸 | 影响核糖体在mRNA上的延伸速率或脱落率 | 构建枯草芽孢杆菌N端编码序列文库调控NeuAc合成途径关键基因表达水平与动态模式以提高NeuAc产量 | [ |
| 蛋白降解标签 | 蛋白丰度 | 通过对目的蛋白的降解调控目的蛋白的丰度 | 将蛋白降解标签用于构建大肠杆菌正交基因回路,实现基因表达水平高强度激活与抑制 | [ |
| 1 | KENT R, DIXON N. Contemporary tools for regulating gene expression in bacteria [J]. Trends in Biotechnology, 2020, 38(3): 316-333. DOI: 10.1016/j.tibtech.2019.09.007 . |
| 2 | TYO K E, ALPER H S, STEPHANOPOULOS G N. Expanding the metabolic engineering toolbox: more options to engineer cells [J]. Trends in Biotechnology, 2007, 25(3): 132-137. DOI: 10.1016/j.tibtech.2007.01.003 . |
| 3 | YANG Sen, DU Guocheng, CHEN Jian, et al. Characterization and application of endogenous phase-dependent promoters in Bacillus subtilis [J]. Applied Microbiology and Biotechnology, 2017, 101(10): 4151-4161. DOI: 10.1007/s00253-017-8142-7 . |
| 4 | ALPER H, FISCHER C, NEVOIGT E, et al. Tuning genetic control through promoter engineering [J]. Proceedings of the National Academy of Sciences of the United States of America, 2005, 102(36): 12678. DOI: 10.1073/pnas.0504604102 . |
| 5 | REZNIKOFF W S. The lactose operon-controlling elements: a complex paradigm [J]. Molecular Microbiology, 1992, 6(17): 2419-2422. DOI: 10.1111/j.1365-2958.1992.tb01416.x . |
| 6 | GUIZIOU S, SAUVEPLANE V, CHANG Hung-Ju, et al. A part toolbox to tune genetic expression in Bacillus subtilis [J]. Nucleic Acids Research, 2016, 44(15): 7495-7508. DOI: 10.1093/nar/gkw624 . |
| 7 | SALIS H M, MIRSKY E A, VOIGT C A. Automated design of synthetic ribosome binding sites to control protein expression [J]. Nature Biotechnology, 2009, 27(10): 946-950. DOI: 10.1038/nbt.1568 . |
| 8 | SINUMVAYO J P, 杨森, 陈坚, 等. 枯草芽孢杆菌168新型转录终止子的构建与表征[J]. 生物工程学报, 2017, 33(7): 1091-1100. DOI: 10.13345/j.cjb.160484 . |
| SINUMVAYO J P, YANG S, CHEN J, et al. Engineering and characterization of new intrinsic transcriptional terminators in Bacillus subtilis 168 [J]. Chinese Journal of Biotechnology, 2017, 33(7): 1091-1100. DOI: 10.13345/j.cjb.160484 . | |
| 9 | POPE S D, MEDZHITOV R. Emerging principles of gene expression programs and their regulation [J]. Molecular Cell, 2018, 71(3): 389-397. DOI: 10.1016/j.molcel.2018.07.017 . |
| 10 | LU Zhenghui, YANG Shihui, YUAN Xin, et al. CRISPR-assisted multi-dimensional regulation for fine-tuning gene expression in Bacillus subtilis [J]. Nucleic acids research, 2019, 47(7): e40. DOI: 10.1093/nar/gkz072 . |
| 11 | YANG Sen, LIU Qingtao, ZHANG Yunfeng, et al. Construction and characterization of broad-spectrum promoters for synthetic biology [J]. ACS Synthetic Biology, 2018, 7(1): 287-291. DOI: 10.1021/acssynbio.7b00258 . |
| 12 | PAPENFORT K, VANDERPOOL C K. Target activation by regulatory RNAs in bacteria [J]. FEMS Microbiology Reviews, 2015, 39(3): 362-378. DOI: 10.1093/femsre/fuv016 . |
| 13 | BREAKER R R. Riboswitches and translation control [J]. Cold Spring Harbor Perspectives in Biology, 2018, 10(11). DOI: 10.1101/cshperspect.a032797 . |
| 14 | MANDAL M, BREAKER R R. Gene regulation by riboswitches [J]. Nature Reviews Molecular Cell Biology, 2004, 5(6): 451-463. DOI: 10.1038/nrm1403 . |
| 15 | CARON M P, BASTET L, LUSSIER A, et al. Dual-acting riboswitch control of translation initiation and mRNA decay [J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(50): E3444-E3453. DOI: 10.1073/pnas.1214024109 . |
| 16 | JANG Sungho, JUNG Gyoo Yeol. Systematic optimization of L-tryptophan riboswitches for efficient monitoring of the metabolite in Escherichia coli [J]. Biotechnology and Bioengineering, 2018, 115(1): 266-271. DOI: 10.1002/bit.26448 . |
| 17 | JANG Sungho, JANG Sungyeon, XIU Yu, et al. Development of artificial riboswitches for monitoring of naringenin in vivo [J]. ACS Synthetic Biology, 2017, 6(11): 2077-2085. DOI: 10.1021/acssynbio.7b00128 . |
| 18 | XIU Yu, JANG Sungho, JONES J A, et al. Naringenin-responsive riboswitch-based fluorescent biosensor module for Escherichia coli co-cultures [J]. Biotechnology and Bioengineering, 2017, 114(10): 2235-2244. DOI: 10.1002/bit.26340 . |
| 19 | CAI Yao, HU Huasi, PAN Ziheng, et al. Metaheuristic optimization in shielding design for neutrons and gamma rays reducing dose equivalent as much as possible [J]. Annals of Nuclear Energy, 2018, 120: 27-34. DOI: 10.1016/j.anucene.2018.05.038 . |
| 20 | BURKE-AGUERO D H. Methods in enzymology : riboswitches as targets and tools [J]. Methods in Enzymology, 2015. DOI: 10.1016/S0076-6879(15)00012-9 . |
| 21 | SEDLYAROVA N, SHAMOVSKY I, BHARATI B K, et al. sRNA-mediated control of transcription termination in E. coli [J]. Cell, 2016, 167(1): 111-121. e13. DOI: 10.1016/j.cell.2016.09.004. |
| 22 | Young Je LEE, HOYNES-O'CONNOR A, LEONG M C, et al. Programmable control of bacterial gene expression with the combined CRISPR and antisense RNA system [J]. Nucleic Acids Research, 2016, 44(5): 2462-2473. DOI: 10.1093/nar/gkw056 . |
| 23 | YANG Yaping, LIN Yuheng, LI Lingyun, et al. Regulating malonyl-CoA metabolism via synthetic antisense RNAs for enhanced biosynthesis of natural products [J]. Metabolic Engineering, 2015, 29: 217-226. DOI: 10.1016/j.ymben.2015.03.018 . |
| 24 | NAKASHIMA N, TAMURA T, GOOD L. Paired termini stabilize antisense RNAs and enhance conditional gene silencing in Escherichia coli [J]. Nucleic Acids Research, 2006, 34(20): e138-e138. DOI: 10.1093/nar/gkl697 . |
| 25 | SHERWOOD A V, HENKIN T M. Riboswitch-mediated gene regulation: novel RNA architectures dictate gene expression responses [J]. Annual Review of Microbiology, 2016, 70(1): 361-374. DOI: 10.1146/annurev-micro-091014-104306 . |
| 26 | RÖTHLISBERGER P, HOLLENSTEIN M. Aptamer chemistry [J]. Advanced Drug Delivery Reviews, 2018, 134: 3-21. DOI: 10.1016/j.addr.2018.04.007 . |
| 27 | ABOUL-ELA F, HUANG Wei, ELRAHMAN M A, et al. Linking aptamer-ligand binding and expression platform folding in riboswitches: prospects for mechanistic modeling and design [J]. Wiley Interdisciplinary Reviews. RNA, 2015, 6(6): 631-650. DOI: 10.1002/wrna.1300 . |
| 28 | WANG j, YANG Le, CUI Xun, et al. A DNA bubble-mediated gene regulation system based on thrombin-bound DNA aptamers [J]. ACS Synthetic Biology, 2017, 6(5): 758-765. DOI: 10.1021/acssynbio.6b00391 . |
| 29 | DENG Jieying, CHEN Chunmei, GU Yang, et al. Creating an in vivo bifunctional gene expression circuit through an aptamer-based regulatory mechanism for dynamic metabolic engineering in Bacillus subtilis [J]. Metabolic Engineering, 2019, 55: 179-190. DOI: 10.1016/j.ymben.2019.07.008 . |
| 30 | GOODMAN D B, CHURCH G M, KOSURI S. Causes and effects of N-terminal codon bias in bacterial genes [J]. Science, 2013, 342(6157): 475. DOI: 10.1126/science.1241934 . |
| 31 | KUDLA G, MURRAY A W, TOLLERVEY D, et al. Coding-sequence determinants of gene expression in Escherichia coli [J]. Science, 2009, 324(5924): 255. DOI: 10.1126/science.1170160 . |
| 32 | SAUER C, THEMAAT E V L VAN, BOENDER L G M, et al. Exploring the nonconserved sequence space of synthetic expression modules in Bacillus subtilis [J]. ACS Synthetic Biology, 2018, 7(7): 1773-1784. DOI: 10.1021/acssynbio.8b00110 . |
| 33 | ESPAH BORUJENI A, SALIS H M. Translation initiation is controlled by RNA folding kinetics via a ribosome drafting mechanism [J]. Journal of the American Chemical Society, 2016, 138(22): 7016-7023. DOI: 10.1021/jacs.6b01453 . |
| 34 | BORUJENI A E, CETNAR D, FARASAT I, et al. Precise quantification of translation inhibition by mRNA structures that overlap with the ribosomal footprint in N-terminal coding sequences [J]. Nucleic Acids Research, 2017, 45(9): 5437-5448. DOI: 10.1093/nar/gkx061 . |
| 35 | DOUGAN D A, TRUSCOTT K N, ZETH K. The bacterial N-end rule pathway: expect the unexpected [J]. Molecular Microbiology, 2010, 76(3): 545-558. DOI: 10.1111/j.1365-2958.2010 .07120.x. |
| 36 | LU Jianli, DEUTSCH C. Electrostatics in the ribosomal tunnel modulate chain elongation rates [J]. Journal of Molecular Biology, 2008, 384(1): 73-86. DOI: 10.1016/j.jmb.2008.08.089 . |
| 37 | SHARMA A K, BUKAU B, O'BRIEN E P. Physical origins of codon positions that strongly influence cotranslational folding: a framework for controlling nascent-protein folding [J]. Journal of the American Chemical Society, 2016, 138(4): 1180-1195. DOI: 10.1021/jacs.5b08145 . |
| 38 | TOBIAS J W, SHRADER T E, ROCAP G, et al. The N-end rule in bacteria [J]. Science, 1991, 254(5036): 1374-1377. |
| 39 | ZADEH J N, STEENBERG C D, BOIS J S, et al. NUPACK: analysis and design of nucleic acid systems [J]. Journal of Computational Chemistry, 2011, 32(1): 170-173. DOI: 10.1002/jcc.21596 . |
| 40 | CAMBRAY G, GUIMARAES J C, ARKIN A P. Evaluation of 244,000 synthetic sequences reveals design principles to optimize translation in Escherichia coli [J]. Nature Biotechnology, 2018, 36(10): 1005-1015. DOI: 10.1038/nbt.4238 . |
| 41 | TIAN Rongzhen, LIU Yanfeng, CHEN Junrong, et al. Synthetic N-terminal coding sequences for fine-tuning gene expression and metabolic engineering in Bacillus subtilis [J]. Metabolic Engineering, 2019, 55: 131-141. DOI: 10.1016/j.ymben.2019 .07.001. |
| 42 | XU Peng. Production of chemicals using dynamic control of metabolic fluxes [J]. Current Opinion in Biotechnology, 2018, 53: 12-19. DOI: 10.1016/j.copbio.2017.10.009 . |
| 43 | HOLTZ W J, KEASLING J D. Engineering static and dynamic control of synthetic pathways [J]. Cell, 2010, 140(1): 19-23. DOI: 10.1016/j.cell.2009.12.029 . |
| 44 | ALPER H, STEPHANOPOULOS G. Global transcription machinery engineering: a new approach for improving cellular phenotype [J]. Metabolic Engineering, 2007, 9(3): 258-267. DOI: 10.1016/j.ymben.2006.12.002 . |
| 45 | MY L, ACHKAR N G, VIALA J P, et al. Reassessment of the genetic regulation of fatty acid synthesis in Escherichia coli: global positive control by the dual functional regulator FadR [J]. Journal of Bacteriology, 2015, 197(11): 1862-1872. DOI: 10.1128/JB.00064-15 . |
| 46 | KURODA K, UEDA M. Engineering of global regulators and cell surface properties toward enhancing stress tolerance in Saccharomyces cerevisiae [J]. Journal of Bioscience and Bioengineering, 2017, 124(6): 599-605. DOI: 10.1016/j.jbiosc.2017 .06.010. |
| 47 | NGUYEN-VO T P, LIANG Yunxiao, SANKARANARAYANAN M, et al. Development of 3-hydroxypropionic-acid-tolerant strain of Escherichia coli W and role of minor global regulator yieP [J]. Metabolic Engineering, 2019, 53: 48-58. DOI: 10.1016/j.ymben.2019.02.001 . |
| 48 | BRINSMADE S R, ALEXANDER E L, LIVNY J, et al. Hierarchical expression of genes controlled by the Bacillus subtilis global regulatory protein CodY [J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(22): 8227. DOI: 10.1073/pnas.1321308111 . |
| 49 | CAO Haojie, KUIPERS O P. Influence of global gene regulatory networks on single cell heterogeneity of green fluorescent protein production in Bacillus subtilis [J]. Microbial Cell Factories, 2018, 17(1): 134. DOI: 10.1186/s12934-018-0985-9 . |
| 50 | ZHU Liying, GAO Shan, ZHANG Hongman, et al. Improvement of lead tolerance of Saccharomyces cerevisiae by random mutagenesis of transcription regulator SPT3 [J]. Applied Biochemistry and Biotechnology, 2018, 184(1): 155-167. DOI: 10.1007/s12010-017-2531-3 . |
| 51 | PARK Kyung-Soon, Dong-ki LEE, Horim LEE, et al. Phenotypic alteration of eukaryotic cells using randomized libraries of artificial transcription factors [J]. Nature Biotechnology, 2003, 21(10): 1208-1214. DOI: 10.1038/nbt868 . |
| 52 | ALPER H, MOXLEY J, NEVOIGT E, et al. Engineering yeast transcription machinery for improved ethanol tolerance and production [J]. Science, 2006, 314(5805): 1565. DOI: 10.1126/science.1131969 . |
| 53 | BURGESS R R, ANTHONY L. How sigma docks to RNA polymerase and what sigma does [J]. Current Opinion in Microbiology, 2001, 4(2): 126-131. DOI: 10.1016/S1369-5274(00)00177-6 . |
| 54 | GAO Xi, JIANG Ling, ZHU Liying, et al. Tailoring of global transcription sigma D factor by random mutagenesis to improve Escherichia coli tolerance towards low-pHs [J]. Journal of Biotechnology, 2016, 224: 55-63. DOI: 10.1016/j.jbiotec.2016.03.012 . |
| 55 | ADHIKARI S, CURTIS P D. DNA methyltransferases and epigenetic regulation in bacteria [J]. FEMS Microbiology Reviews, 2016, 40(5): 575-591. DOI: 10.1093/femsre/fuw023 . |
| 56 | KANG Jeong Gu, PARK Jin Suk, Jeong-Heosn KO, et al. Regulation of gene expression by altered promoter methylation using a CRISPR/Cas9-mediated epigenetic editing system [J]. Scientific Reports, 2019, 9(1): 11960. DOI: 10.1038/s41598-019-48130-3 . |
| 57 | GRUNSTEIN M, GASSER S M. Epigenetics in Saccharomyces cerevisiae [J]. Cold Spring Harbor Perspectives in Biology, 2013, 5(7): a017491. DOI: 10.1101/cshperspect.a017491 . |
| 58 | CHEN Chao, WANG Lianrong, CHEN Si, et al. Convergence of DNA methylation and phosphorothioation epigenetics in bacterial genomes [J]. Proceedings of the National Academy of Sciences of the United States of America, 2017, 114(17): 4501-4506. DOI: 10.1073/pnas.1702450114 . |
| 59 | LAL A, KRISHNA S, SESHASAYEE A S N. Regulation of global transcription in Escherichia coli by Rsd and 6S RNA [J]. G3: Genes, Genomes, Genetics, 2018, 8(6): 2079-2089. DOI: 10.1534/g3.118.200265 . |
| 60 | WASSARMAN K M, STORZ G. 6S RNA Regulates E. coli RNA polymerase activity [J]. Cell, 2000, 101(6): 613-623. DOI: 10.1016/S0092-8674(00)80873-9 . |
| 61 | WASSARMAN K M. 6S RNA, a global regulator of transcription [J]. Microbiology Spectrum, 2018, 6(3): 10.1128/microbiolspec. RWR-0019-2018. DOI: 10.1128/microbiolspec.RWR-0019-2018 . |
| 62 | CAVANAGH A T, WASSARMAN K M. 6S RNA, a global regulator of transcription in Escherichia coli, Bacillus subtilis, and beyond [J]. Annual Review of Microbiology, 2014, 68(1): 45-60. DOI: 10.1146/annurev-micro-092611-150135 . |
| 63 | QI Lei S, LARSON M H, GILBERT L A, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression [J]. Cell, 2013, 152(5): 1173-1183. DOI: 10.1016/j.cell.2013.02.022 . |
| 64 | RAN F A, HSU P D, WRIGHT J, et al. Genome engineering using the CRISPR-Cas9 system [J]. Nature Protocols, 2013, 8(11): 2281-2308. DOI: 10.1038/nprot.2013.143 . |
| 65 | HSU P D, LANDER E S, ZHANG Feng. Development and applications of CRISPR-Cas9 for genome engineering [J]. Cell, 2014, 157(6): 1262-1278. DOI: 10.1016/j.cell.2014.05.010 . |
| 66 | LIU Yang, WAN Xinyi, WANG Baojun. Engineered CRISPRa enables programmable eukaryote-like gene activation in bacteria [J]. Nature Communications, 2019, 10(1): 3693. DOI: 10.1038/s41467-019-11479-0 . |
| 67 | LIAN Jiazhang, HAMEDIRAD M, HU Sumeng, et al. Combinatorial metabolic engineering using an orthogonal tri-functional CRISPR system [J]. Nature Communications, 2017, 8(1): 1688. DOI: 10.1038/s41467-017-01695-x . |
| 68 | WU Yaokang, CHEN Taichi, LIU Yanfeng, et al. Design of a programmable biosensor-CRISPRi genetic circuits for dynamic and autonomous dual-control of metabolic flux in Bacillus subtilis [J]. Nucleic Acids Research, 2019, 48(2): 996-1009. DOI: 10.1093/nar/gkz1123 . |
| 69 | FENNO L, YIZHAR O, DEISSEROTH K. The development and application of optogenetics [J]. Annual Review of Neuroscience, 2011, 34: 389-412. DOI: 10.1146/annurev-neuro-061010-113817 . |
| 70 | BACCHUS W, FUSSENEGGER M. The use of light for engineered control and reprogramming of cellular functions [J]. Current Opinion in Biotechnology, 2012, 23(5): 695-702. DOI: 10.1016/j.copbio.2011.12.004 . |
| 71 | OLSON E J, TABOR J J. Optogenetic characterization methods overcome key challenges in synthetic and systems biology [J]. Nature Chemical Biology, 2014, 10(7): 502-511. DOI: 10.1038/nchembio.1559 . |
| 72 | PATHAK G P, VRANA J D, TUCKER C L. Optogenetic control of cell function using engineered photoreceptors [J]. Biology of the Cell, 2013, 105(2): 59-72. DOI: 10.1111/boc.201200056 . |
| 73 | ZHAO E M, ZHANG Yanfei, MEHL J, et al. Optogenetic regulation of engineered cellular metabolism for microbial chemical production [J]. Nature, 2018, 555(7698): 683-687. DOI: 10.1038/nature26141 . |
| 74 | SHIMIZU-SATO S, HUQ E, TEPPERMAN J M, et al. A light-switchable gene promoter system [J]. Nature Biotechnology, 2002, 20(10): 1041-1044. DOI: 10.1038/nbt734 . |
| 75 | MOTTA-MENA L B, READE A, MALLORY M J, et al. An optogenetic gene expression system with rapid activation and deactivation kinetics [J]. Nature Chemical Biology, 2014, 10(3): 196-202. DOI: 10.1038/nchembio.1430 . |
| 76 | SHIN Yongdae, BERRY J, PANNUCCI N, et al. Spatiotemporal control of intracellular phase transitions using light-activated optoDroplets [J]. Cell, 2017, 168(1/2): 159-171.e 14. DOI: 10.1016/j.cell.2016.11.054 . |
| 77 | TABOR J J, LEVSKAYA A, VOIGT C A. Multichromatic control of gene expression in Escherichia coli [J]. Journal of Molecular Biology, 2011, 405(2): 315-324. DOI: 10.1016/j.jmb.2010.10.038 . |
| 78 | MILIAS-ARGEITIS A, RULLAN M, AOKI S K, et al. Automated optogenetic feedback control for precise and robust regulation of gene expression and cell growth [J]. Nature Communications, 2016, 7: 12546. DOI: 10.1038/ncomms12546 . |
| 79 | CASTILLO-HAIR S M, BAERMAN E A, FUJITA M, et al. Optogenetic control of Bacillus subtilis gene expression [J]. Nature Communications, 2019, 10(1): 3099. DOI: 10.1038/s41467-019-10906-6 . |
| 80 | GAO Cong, HOU Jianshen, XU Peng, et al. Programmable biomolecular switches for rewiring flux in Escherichia coli [J]. Nature Communications, 2019, 10(1): 3751. DOI: 10.1038/s41467-019-11793-7 . |
| 81 | CAMERON D E, COLLINS J J. Tunable protein degradation in bacteria [J]. Nature Biotechnology, 2014, 32(12): 1276-1281. DOI: 10.1038/nbt.3053 . |
| 82 | CHUNG Hokyung K, JACOBS C L, HUO Yunwen, et al. Tunable and reversible drug control of protein production via a self-excising degron [J]. Nature Chemical Biology, 2015, 11(9): 713-720. DOI: 10.1038/nchembio.1869 . |
| 83 | DOONG S J, GUPTA A, PRATHER K L J. Layered dynamic regulation for improving metabolic pathway productivity in Escherichia coli [J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(12): 2964-2969. DOI: 10.1073/pnas.1716920115 . |
| 84 | MARTÍNEZ V, LAURITSEN I, HOBEL T, et al. CRISPR/Cas9-based genome editing for simultaneous interference with gene expression and protein stability [J]. Nucleic Acids Research, 2017, 45(20): e171-e171. DOI: 10.1093/nar/gkx797 . |
| 85 | FERNANDEZ-RODRIGUEZ J, VOIGT C A. Post-translational control of genetic circuits using Potyvirus proteases [J]. Nucleic Acids Research, 2016, 44(13): 6493-6502. DOI: 10.1093/nar/gkw537 . |
| 86 | TAN S Z, PRATHER K L J. Dynamic pathway regulation:recent advances and methods of construction [J]. Current Opinion in Chemical Biology, 2017, 41: 28-35. DOI: 10.1016/j.cbpa.2017.10.004 . |
| 87 | WHITELEY M, DIGGLE S P, GREENBERG E P. Progress in and promise of bacterial quorum sensing research [J]. Nature, 2017, 551(7680): 313-320. DOI: 10.1038/nature24624 . |
| 88 | SOMA Y, HANAI T. Self-induced metabolic state switching by a tunable cell density sensor for microbial isopropanol production [J]. Metabolic Engineering, 2015, 30: 7-15. DOI: 10.1016/j.ymben.2015.04.005 . |
| 89 | GUPTA A, REIZMAN I M B, REISCH C R, et al. Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit [J]. Nature biotechnology, 2017, 35(3): 273-279. DOI: 10.1038/nbt.3796 . |
| 90 | CUI Shixiu, Xueqin LÜ, WU Yaokang, et al. Engineering a bifunctional Phr60-Rap60-Spo0A quorum-sensing molecular switch for dynamic fine-tuning of menaquinone-7 synthesis in Bacillus subtilis [J]. ACS Synthetic Biology, 2019, 8(8): 1826-1837. DOI: 10.1021/acssynbio.9b00140 . |
| 91 | WILLIAMS T C, AVERESCH N, WINTER G, et al. Quorum-sensing linked RNA interference for dynamic metabolic pathway control in Saccharomyces cerevisiae [J]. Metabolic Engineering, 2015, 29: 124-134. DOI: 10.1016/j.ymben.2015.03.008 . |
| 92 | XU Peng, LI Lingyun, ZHANG Fuming, et al. Improving fatty acids production by engineering dynamic pathway regulation and metabolic control [J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(31): 11299-11304. DOI: 10.1073/pnas.1406401111 . |
| 93 | RUGBJERG P, SARUP-LYTZEN K, NAGY M, et al. Synthetic addiction extends the productive life time of engineered Escherichia coli populations [J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(10): 2347. DOI: 10.1073/pnas.1718622115 . |
| 94 | SANDBERG T E, SALAZAR M J, WENG L L, et al. The emergence of adaptive laboratory evolution as an efficient tool for biological discovery and industrial biotechnology [J]. Metabolic Engineering, 2019, 56: 1-16. DOI: 10.1016/j.ymben.2019.08.004 . |
| 95 | BUERGER J, GRONENBERG L S, GENEE H J, et al. Wiring cell growth to product formation [J]. Current Opinion in Biotechnology, 2019, 59: 85-92. DOI: 10.1016/j.copbio.2019.02.014 . |
| 96 | CHOU H H, KEASLING J D. Programming adaptive control to evolve increased metabolite production [J]. Nature Communications, 2013, 4(1): 2595. DOI: 10.1038/ncomms3595 . |
| 97 | LEAVITT J M, WAGNER J M, TU C C, et al. Biosensor-enabled directed evolution to improve muconic acid production in Saccharomyces cerevisiae [J]. Biotechnology Journal, 2017, 12(10): 1600687. DOI: 10.1002/biot.201600687 . |
| [1] | GUO Shuyuan, ZHANG Qiannan, Gulikezi· MAIMAITIREXIATI, YANG Yiqun, YU Tao. Advances in microbial production of liquid biofuels [J]. Synthetic Biology Journal, 2025, 6(1): 18-44. |
| [2] | GAO Ge, BIAN Qi, WANG Baojun. Synthetic genetic circuit engineering: principles, advances and prospects [J]. Synthetic Biology Journal, 2025, 6(1): 45-64. |
| [3] | LI Jiyuan, WU Guosheng. Two hypothesises for the origins of organisms from the synthetic biology perspective [J]. Synthetic Biology Journal, 2025, 6(1): 190-202. |
| [4] | JIAO Hongtao, QI Meng, SHAO Bin, JIANG Jinsong. Legal issues for the storage of DNA data [J]. Synthetic Biology Journal, 2025, 6(1): 177-189. |
| [5] | REN Jiawei, ZHANG Jinpeng, XU Guoqiang, ZHANG Xiaomei, XU Zhenghong, ZHANG Xiaojuan. Effect of terminators on the downstream transcript unit with gene expression in Escherichiacoli [J]. Synthetic Biology Journal, 2025, 6(1): 213-227. |
| [6] | TANG Xinghua, LU Qianneng, HU Yilin. Philosophical reflections on synthetic biology in the Anthropocene [J]. Synthetic Biology Journal, 2025, 6(1): 203-212. |
| [7] | XU Huaisheng, SHI Xiaolong, LIU Xiaoguang, XU Miaomiao. Key technologies for DNA storage: encoding, error correction, random access, and security [J]. Synthetic Biology Journal, 2025, 6(1): 157-176. |
| [8] | SHI Ting, SONG Zhan, SONG Shiyi, ZHANG Yi-Heng P. Job. In vitro BioTransformation (ivBT): a new frontier of industrial biomanufacturing [J]. Synthetic Biology Journal, 2024, 5(6): 1437-1460. |
| [9] | CHAI Meng, WANG Fengqing, WEI Dongzhi. Synthesis of organic acids from lignocellulose by biotransformation [J]. Synthetic Biology Journal, 2024, 5(6): 1242-1263. |
| [10] | SHAO Mingwei, SUN Simian, YANG Shimao, CHEN Guoqiang. Bioproduction based on extremophiles [J]. Synthetic Biology Journal, 2024, 5(6): 1419-1436. |
| [11] | ZHAO Liang, LI Zhenshuai, FU Liping, LYU Ming, WANG Shi’an, ZHANG Quan, LIU Licheng, LI Fuli, LIU Ziyong. Progress in biomanufacturing of lipids and single cell protein from one-carbon compounds [J]. Synthetic Biology Journal, 2024, 5(6): 1300-1318. |
| [12] | ZHU Fanghuan, CEN Xuecong, CHEN Zhen. Research progress of diols production by microbes [J]. Synthetic Biology Journal, 2024, 5(6): 1367-1385. |
| [13] | CHEN Yu, ZHANG Kang, QIU Yijing, CHENG Caiyun, YIN Jingjing, SONG Tianshun, XIE Jingjing. Progress of microbial electrosynthesis for conversion of CO2 [J]. Synthetic Biology Journal, 2024, 5(5): 1142-1168. |
| [14] | ZHENG Haotian, LI Chaofeng, LIU Liangxu, WANG Jiawei, LI Hengrun, NI Jun. Design, optimization and application of synthetic carbon-negative phototrophic community [J]. Synthetic Biology Journal, 2024, 5(5): 1189-1210. |
| [15] | YU Wei, GAO Jiaoqi, ZHOU Yongjin. Bioconversion of one carbon feedstocks for producing organic acids [J]. Synthetic Biology Journal, 2024, 5(5): 1169-1188. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||